Abstract
Apolipoprotein E (APOE) is a confirmed risk factor for Alzheimer disease. APOE is also involved in several other neurodegenerative disorders, including Parkinson disease and multiple sclerosis. Previous studies of amyotrophic lateral sclerosis (Lou Gehrig disease, ALS) have investigated the effect of APOE on the risk of developing ALS, age at onset, site of onset, and duration of the disease. The results have been inconsistent, possibly due to small sample sizes and complete reliance on case-control data. No family-based association studies were performed. To address these limitations, we investigated the relationship between APOE functional polymorphisms and age at onset of ALS in a large set of 508 families. We treated age at onset as a quantitative trait and performed family-based association analysis using the TDT Q5 method. APOE-2 is protective against earlier onset ( P =0.001) with an average age at onset of APOE-2 carriers approximately 3 years later than that of non-APOE-2 carriers. Similar to our previous report, we did not find APOE associated with ALS risk. Our findings suggest that APOE may express its strongest effect through age at onset rather than on risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (Lou Gehrig disease, ALS) is an age-dependent, neurodegenerative disorder of motor neurons with both sporadic and familial forms. It is a rapidly progressive disease that results in progressive paralysis until death occurs, usually within 5 years of onset. The cause of most types of ALS is unknown and the disease is untreatable. ALS arises as a dominantly inherited trait in approximately 10% of ALS cases. Approximately 25% of autosomal dominant ALS is caused by mutations in the Cu/Zn superoxide dismutase (SOD1) gene [1, 2]. A recessive form of juvenile-onset ALS has been associated with ALS in chromosome 2q33 [3, 4]. Additional chromosomal regions have been identified in chromosomes 9q [5], 15q [6], 16q [7, 8, 9], 18q [10], 20 [8], and X [11] for familial ALS. However, the underlying genetic causes of the remaining forms of ALS are still unknown.
The apolipoprotein E ( APOE) gene is a confirmed genetic risk factor that influences both risk and age at onset in Alzheimer disease [12, 13] and Parkinson disease [14, 15]. APOE functional polymorphisms have been examined for association with different phenotypes of ALS, including the risk of developing ALS, age at onset, site of onset, and duration of the disease [16, 17, 18, 19]. Most studies found no association between APOE and the risk of developing ALS [16, 17, 19]. However, for other phenotypes, no consensus has been made [17, 19, 20, 21, 22]. In particular, the studies of APOE affecting age at onset and site of onset in ALS have been the most inconsistent. Moulard et al. [19] found that bulbar-onset ALS patients with an APOE-4 allele showed an average 6 years earlier age at onset than those without APOE-4. Conversely, two later studies [17, 20] reported no significant difference in age at onset between patients with and without an APOE-4 allele in either the overall ALS dataset or the bulbar-onset ALS subset. These previous studies all used unrelated case-control samples to test allelic association between the groups of interest. No family-based association tests have been performed. In addition, most studies examining age at onset of ALS compared the average ag at onset between different APOE genotypes. This approach does not extract the full amount of information from the data.
In this study, we used family-based association approaches for quantitative and qualitative traits to assess the relationship of APOE with age at onset of ALS and the risk of developing ALS, respectively.
Materials and methods
Family data
The dataset consists of 446 singleton families with only 1 affected in each family (238 parent-child trio and 208 discordant sibpairs) and 62 multiplex families with at least 2 affected relatives (Table 1). All families have at least 1 affected individual with age at onset information available. The family ascertainment was conducted by two clinical sites, Northwestern University Medical School (396 families) and Massachussetts General Hospital (112 families). All patients diagnosed with ALS were examined by a neurologist (T.S, R.B. and colleagues) and met the El Escorial criteria [23] for the diagnosis of ALS. Clinical information, including patient’s age at onset (defined as the time at which symptoms first occurred), was obtained at the time of examination. Because ALS is characterized by sudden onset and rapid progression, age at onset is essentially equivalent to age at examination. All participants or their legal representatives gave informed consent prior to joining the study, and data were collected according to protocols approved by each contributing group’s institutional review board.
Genotyping
Genomic DNA was extracted from whole blood according to established protocols [24]. We genotyped APOE functional polymorphisms in a total of 1,462 samples. APOE genotyping was performed as previously described [25]. Substantial quality control procedures were followed [26]. Genotype data were uploaded into the PEDIGENE® database and merged into the LAPIS management system for data analysis [27].
Statistical analysis
All genotype data were examined for mendelian consistency, using the PedCheck program [28]. APOE functional polymorphisms were tested for Hardy-Weinberg equilibrium (HWE) in the affected group (1 affected from each family) and the unaffected group (1 unaffected from each family). An exact test implemented in Genetic Data Analysis (GDA) [29] was used to test HWE, in which 3,200 replicate samples were simulated for estimating the empirical P value.
To investigate the effect of APOE on age at onset of ALS, we treated age at onset as a quantitative trait. We used the TDT Q5 method [30], which utilizes parent-child triad data, implemented in the QTDT program [31] (http://www.sph.umich.edu/csg/abecasis/QTDT/) to test the allelic association between APOE and age at onset of ALS. The method proposed by Rabinowitz [32], which uses parents and multiple siblings data, served as a second method to confirm the results. To test whether ascertainment center has an effect on the results, we incorporated center as a covariate in both tests.
The pedigree disequilibrium test (PDT) [33] was employed to test APOE association with ALS risk. Two PDT statistics were used: the PDT-sum statistic [34], which examines allelic effects, and the genotype-PDT, which examines genotypic effects [35]. We performed the PDT analysis on the full dataset as well as stratified datasets based on an age at onset cutoff of 40 years. The late-onset subset (LOALS) consists of families with all affecteds having age at onset greater than or equal to 40 years. The early onset subset (EOALS) consists of families with at least 1 affected with age at onset less than 40 years.
Results
For each APOE genotype, the number of unaffected and affected individuals of all family members is summarized in Table 2. Since these counts were taken from family data, one should take into account familial correlation in interpretation of the data. Overall, APOE-33 is the most-frequent genotype. The allele frequencies in our overall ALS family dataset were 8.7% for APOE-2, 75.9% for APOE-3, and 15.4% for APOE-4. A similar range of APOE allele frequencies was observed in the overall ALS, EOALS, and LOALS subsets (Table 1).
Age at onset ranged from 18 to 81 years, with average age at onset (±SD) being 48.1±13.1 years in the overall ALS dataset. Although the average age at onset tends to be higher in multiplex than singleton families in all three subsets, a large standard deviation of age at onset is found in the multiplex families due to their small sample sizes (Table 1). No deviation from HWE was found in either affected or unaffected samples for APOE.
In the TDT Q5 analysis, APOE-2 showed the strongest evidence of positive association with age at onset ( P =0.001) and APOE-3 showed a marginally significant P value ( P =0.058). Similar results were obtained with the Rabinowitz method ( P =0.008 for APOE-2 and P =0.054 for APOE-3). Furthermore, the association between APOE-2 and age at onset was not changed after taking into account the ascertainment effect as a covariate in the tests. The positive association between APOE-2 and age at onset indicates that age at onset increases as the number of APOE-2 alleles transmitted from parents increases. This is also seen in the mean age at onset in each APOE-2 carrier group, in which the average age at onset was 48±13.2 years for non- APOE-2 carriers (433 individuals), 51±11.3 years for one- APOE-2 carriers (81 individuals), and 64±3.5 years for two- APOE-2 carriers (2 individuals). Overall, we found approximately a 3-year difference between APOE-2 carrier and non- APOE-2 carrier groups (51 vs. 48 years) for age at onset.
To update our analyses [17] of APOE and the risk of ALS, we examined all 508 families for both allelic and genotypic effects. No significant results were observed (global P =0.276 for alleles and global P =0.491 for genotypes). For the association between APOE and the risk of ALS in the stratified subsets, we found that APOE-2 significantly reduced the risk of ALS in the EOALS subset ( P =0.012), but not in the LOALS subset.
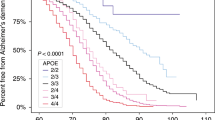
In order to investigate the relationship of APOE-2 with the risk and age at onset of ALS, we also examined the pattern of APOE-2 carriers in each dataset. We randomly selected 1 affected and 1 unaffected with APOE genotype available from each family. The percentages of APOE-2 carriers in affected and unaffected groups, respectively, for each subset are depicted in Fig. 1. The frequencies of APOE-2 carriers in unaffecteds are similar (15.9%–17.6%) among EOALS, LOALS, and the overall ALS datasets. For the affecteds, the EOALS subset clearly consists of far fewer APOE-2 carriers than the other two datasets (8.7% vs. 18.6% and 15.5%) as well as all unaffecteds. The differences in the numbers of APOE-2 carriers between affected and unaffected groups in the EOALS subset may lead to significant association between APOE-2 and the risk of ALS.
Percentages of APOE-2 carriers for affecteds and unaffecteds in each amyotrophic lateral scleosis (ALS) subset. Early onset ALS ( EOALS) consists of families with at least 1 affected with age at onset <40 years and late-onset ALS ( LOALS) consists of families with all affected with age at onset ≥40 years. We selected 1 affected and 1 unaffected individual from each family for each subset for the calculation of percentages of APOE-2 carriers
Discussion
APOE has been implicated in several neurodegenerative diseases [13, 15, 36]. To our knowledge, there are at least eight publications using unrelated samples with inconsistent conclusions on the role of APOE in ALS. Some datasets are larger than the others, for instance, no effect of APOE on age at onset of ALS was found using 360 ALS cases versus 351 controls [17] in either the overall data or bulbar-onset subset, while significant results were found for age at onset of bulbar-onset patients using 130 ALS cases versus 161 controls [19]. The current study is different from the previous studies in several respects. In particular, we used the family based association approach to dissect the effect of APOE on the age at onset of ALS. This approach protects against spurious association due to population substructure [37]. In this study, age at onset is treated as a quantitative trait, increasing the amount of information extracted from the data.
Overall, the present study showed a strong association of later onset with increasing number of APOE-2 alleles. The protective effect of APOE-2 on age at onset of ALS may have been because the early onset ALS patients tended to receive less APOE-2 from parents than the late-onset ALS patients. This leads to a plausible scenario that parents transmit APOE-2 less than expected to ALS affected individuals within the early onset group, as shown in Fig. 1. This is probably the main reason that we detected negative association between APOE-2 and ALS risk in the early onset group. We, therefore, concluded that the effect of APOE-2 on age at onset is the main cause of the APOE risk effect in early onset ALS.
Previous studies have focused on the risk effect of APOE-4 in ALS [17, 20, 21]. Clearly, the strong dose effect of APOE-4 in Alzheimer disease has impacted the direction of previous research on APOE in ALS. Overall, there are few studies of the risk of APOE-2 in ALS. Moulard et al. [19] showed a difference for APOE-23 genotype frequency between 130 cases and 540 controls (7.7% vs. 14%, respectively), but they did not stratify the data by age at onset. Our comprehensive analysis examined risk and age at onset effects of each allele in overall as well as stratified datasets, which provides the first in-depth analysis of APOE-2.
Unlike the dose effect of APOE-4 in Alzheimer disease [13], we did not find that APOE-4 affected age at onset of ALS ( P =0.75), which is consistent with our previous report of no significant differences in age at onset between patients with APOE-X/X and APOE-4/4 or APOE-4/X genotypes [17]. Furthermore, we did not detect any association between APOE and the risk of ALS, which is consistent with our previous report using sporadic ALS data [17] and several others [16]. Rather, we found that APOE affects the risk of ALS within age at onset-specific groups. The use of an age at onset cutoff of 40 years to stratify the dataset is rather arbitrary, because there is no clear definition for the early and late-onset ALS. We also investigated the subsets based on other cut-offs (e.g., 45 and 50 years), but similar results were generated. In conclusion, our findings suggest that APOE may express its strongest effect on the risk of ALS through age at onset.
References
Siddique T, Figlewicz DA, Pericak-Vance MA, Haines JL, Rouleau G, Jeffers AJ, Sapp P, Hung WY, Bebout J, McKenna-Yasek D, Deng G, Horvitz HR, Gusella JF, Brown RH Jr, Roses AD (1991) Linkage of a gene causing familial amyotrophic lateral sclerosis to chromosome 21 and evidence of genetic-locus heterogeneity. N Engl J Med 324:1381–1384
Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung WY, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella J, Horvitz HR, Brown RH Jr (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62
Hentati A, Bejaoui K, Pericak-Vance MA, Hentati F, Speer MC, Hung WY, Figlewicz DA, Haines J, Rimmler J, Ben Hamida C, Ben Hamida M, Brown RHJ, Siddique T (1994) Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33-q35. Nat Genet 7:425–428
Hosler BA, Sapp PC, Berger R, O’Neill G, Bejaoui K, Hamida MB, Hentati F, Chin W, McKenna-Yasek D, Haines JL, Patterson D, Horvitz HR, Brown RH Jr, Day CB (1998) Refined mapping and characterization of the recessive familial amyotrophic lateral sclerosis locus ( ALS2) on chromosome 2q33. Neurogenetics 2:34–42
Hosler BA, Siddique T, Sapp PC, Sailor W, Huang MC, Hossain A, Daube JR, Nance M, Fan C, Kaplan J, Hung WY, McKenna-Yasek D, Haines JL, Pericak-Vance MA, Horvitz HR, Brown RH Jr (2000) Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA 284:1664–1669
Hentati A, Ouahchi K, Pericak-Vance MA, Nijhawan D, Ahmad A, Yi Y, Rimmler J, Hung WY, Schlotter B, Ahmed A, Ben Hamida M, Hentati F, Siddique T (1998) Linkage of a common locus for recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics 2:55–60
Abalkhail H, Mitchell J, Habgood J, Orrell R, Belleroche J de (2003) A new familial amyotrophic lateral sclerosis locus on chromosome 16q12.1–16q12.2. Am J Hum Genet 73:383–389
Sapp PC, Hosler BA, McKenna-Yasek D, Chin W, Gann A, Genise H, Gorenstein J, Huang M, Sailer W, Scheffler M, Valesky M, Haines JL, Pericak-Vance M, Siddique T, Horvitz HR, Brown RH Jr (2003) Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am J Hum Genet 73:397–403
Ruddy DM, Parton MJ, Al Chalabi A, Lewis CM, Vance C, Smith BN, Leigh PN, Powell JF, Siddique T, Meyjes EP, Baas F, Shaw CE (2003) Two families with familial amyotrophic lateral sclerosis are linked to a novel locus on chromosome 16q. Am J Hum Genet 73:390–396
Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AAS, Mayeux PV, Brown RH, Meininger V, Camu W, Rouleau GA (2002) A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet 70:251–256
Siddique T, Hong S-T, Brooks BR, Hung WY, Siddique NA, Rimmler J, Kaplan JP, Haines JL, Brown RH Jr, Pericak-Vance MA (2004) X-linked dominant locus for late-onset familial amyotrophic lateral sclerosis. Am J Hum Genet:A308
Pericak-Vance MA, St George-Hyslop PH, Gaskell PC Jr, Growdon J, Crain BJ, Hulette C, Gusella JF, Yamaoka L, Tanzi RE, Roses AD, Haines JL (1993) Linkage analysis in familial Alzheimer disease: description of the Duke and Boston data sets. Genet Epidemiol 10:361–364
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261:921–923
Zareparsi S, Kaye J, Camicioli R, Grimslid H, Oken B, Litt M, Nutt J, Bird T, Schellenberg G, Payami H (1997) Modulation of the age at onset of Parkinson’s disease by apolipoprotein E genotypes. Ann Neurol 42:655–658
Li YJ, Hauser MA, Scott WK, Martin ER, Booze MW, Qin XJ, Walter JW, Nance MA, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner CB, Jankovic J, Goetz CG, Small GW, Mastaglia F, Haines JL, Pericak-Vance MA, Vance JA (2004) Apolipoprotein E controls the risk and age at onset of Parkinson Disease. Neurology 62:2005–2009
Mui S, Rebeck GW, McKenna-Yasek D, Hyman BT, Brown RH Jr (1995) Apolipoprotein E epsilon 4 allele is not associated with earlier age at onset in amyotrophic lateral sclerosis. Ann Neurol 38:460–463
Siddique T, Pericak-Vance MA, Caliendo J, Hong ST, Hung WY, Kaplan J, McKenna-Yasek D, Rimmler JB, Sapp P, Saunders AM, Scott WK, Siddique N, Haines JL, Brown RH (1998) Lack of association between apolipoprotein E (APOE) genotype and sporadic amyotrophic lateral sclerosis (ALS). Neurogenetics 1:213–216
Bachus R, Bader S, Gessner R, Ludolph AC (1997) Lack of association of apolipoprotein E epsilon 4 allele with bulbar-onset motor neuron disease. Ann Neurol 41:417
Moulard B, Sefiani A, Laamri A, Malafosse A, Camu W (1996) Apolipoprotein E genotyping in sporadic amyotrophic lateral sclerosis: evidence for a major influence on the clinical presentation and prognosis. J Neurol Sci 139 [Suppl]:34–37
Drory VE, Birnbaum M, Korczyn AD, Chapman J (2001) Association of APOE epsilon4 allele with survival in amyotrophic lateral sclerosis. J Neurol Sci 190:17–20
Al-Chalabi A, Enayat ZE, Bakker MC, Sham PC, Ball DM, Shaw CE, Lloyd CM, et al (1996) Association of apolipoprotein E ε4 allele with bulbar-onset motor neuron disease. Lancet 347:159–160
Smith RG, Haverkamp LJ, Case S, Appel V, Appel SH (1996) Apolipoprotein E epsilon 4 in bulbar-onset motor neuron disease. Lancet 348:334–335
Wilbourn AJ (1998) Clinical neurophysiology in the diagnosis of amyotrophic lateral sclerosis: the Lambert and the El Escorial criteria. J Neurol Sci 160 [Suppl 1]:S25–S29
Vance JM (1998) The collection of biological samples for DNA analysis. In: Haines JL, Pericak-Vance MA (eds) Approaches to gene mapping in complex human diseases. Wiley-Liss, New York
Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ (1993) Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 43:1467–1472
Rimmler JB, Haynes CS, McDowell JG, Stajich JE, Adams CS, Slotterbeck BD, Rogala AR, West SG, Gilbert JR, Hauser ER, Vance JM, Pericak-Vance MA (1999) DataTracker: comprehensive software for data quality control protocols in complex disease studies. Am J Hum Genet 65 [Suppl]:A442
Haynes C, Speer MC, Peedin M, Roses AD, Haines JL, Vance JM, Pericak-Vance MA (1995) PEDIGENE: a comprehensive data management system to facilitate efficient and rapid disease gene mapping. Am J Hum Genet 57:A193
O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266
Zaykin D, Zhivotovsky L, Weir BS (1995) Exact tests for association between alleles at arbitrary numbers of loci. Genetica 96:169–178
Allison DB (1997) Transmission-disequilibrium tests for quantitative traits. Am J Hum Genet 60:676–690
Abecasis GR, Cardon LR, Cookson WO (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292
Rabinowitz D (1997) A transmission disequilibrium test for quantitative trait loci. Hum Hered 47:342–350
Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154
Martin ER, Bass MP, Kaplan NL (2001) Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Gen 68:1065–1067
Martin ER, Bass MP, Pericak-Vance MA, Hauser ER (2003) Genotype-based associtation test for general pedigrees: the genotype-PDT. Genet Epidemiol 25:203–213
Schmidt S, Barcellos LF, DeSombre K, Rimmler JB, Lincoln RR, Bucher P, Saunders AM, Lai E, Martin ER, Vance JM, Oksenberg JR, Hauser SL, Pericak-Vance MA, Haines JL, Multiple Sclerosis GG (2002) Association of polymorphisms in the apolipoprotein e region with susceptibility to and progression of multiple sclerosis. Am J Hum Genet 70:708–717
Li CC (1972) Populations subdivision with respect to multiple alleles. Ann Hum Genet 33:23–29
Acknowledgements
We would like to thank the families and patients with ALS for their participation in this study. Without their help this work would not have been possible. This research was supported by the National Institute of Neurological Disorders and Stroke (NS 37912, NS021442), Les Turner ALS Foundation, Vena E. Schaff ALS Research Fund, Harold Post Research Professorship, Herbert and Florence C. Wenske Foundation, Ralph and Marian Falk Medical Research Trust, Abbott Labs Duane, and Susan Burnham Professorship. R.H.’s laboratory also receives support for this work from the NINDS, the NIA, project ALS, the Angel Fund, the ALS Association, and the Al-Athel ALS Research Foundation. P.S. is supported through the laboratory of H.R. Horvitz, an investigator of the Howard Medical Institute, Department of Biology, Massachusetts Institute of Technology, Cambridge, Mass.
Author information
Authors and Affiliations
Corresponding author
Additional information
R.H. Brown Jr and T. Siddique contributed equally to this project.
Rights and permissions
About this article
Cite this article
Li, YJ., Pericak-Vance, M.A., Haines, J.L. et al. Apolipoprotein E is associated with age at onset of amyotrophic lateral sclerosis. Neurogenetics 5, 209–213 (2004). https://doi.org/10.1007/s10048-004-0193-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-004-0193-0