Abstract
We integrated soil models with an established ecosystem process model (SIPNET, simplified photosynthesis and evapotranspiration model) to investigate the influence of soil processes on modelled values of soil CO2 fluxes (R Soil). Model parameters were determined from literature values and a data assimilation routine that used a 7-year record of the net ecosystem exchange of CO2 and environmental variables collected at a high-elevation subalpine forest (the Niwot Ridge AmeriFlux site). These soil models were subsequently evaluated in how they estimated the seasonal contribution of R Soil to total ecosystem respiration (TER) and the seasonal contribution of root respiration (R Root) to R Soil. Additionally, these soil models were compared to data assimilation output of linear models of soil heterotrophic respiration. Explicit modelling of root dynamics led to better agreement with literature values of the contribution of R Soil to TER. Estimates of R Soil/TER when root dynamics were considered ranged from 0.3 to 0.6; without modelling root biomass dynamics these values were 0.1–0.3. Hence, we conclude that modelling of root biomass dynamics is critically important to model the R Soil/TER ratio correctly. When soil heterotrophic respiration was dependent on linear functions of temperature and moisture independent of soil carbon pool size, worse model-data fits were produced. Adding additional complexity to the soil pool marginally improved the model-data fit from the base model, but issues remained. The soil models were not successful in modelling R Root/R Soil. This is partially attributable to estimated turnover parameters of soil carbon pools not agreeing with expected values from literature and being poorly constrained by the parameter estimation routine. We conclude that net ecosystem exchange of CO2 alone cannot constrain specific rhizospheric and microbial components of soil respiration. Reasons for this include inability of the data assimilation routine to constrain soil parameters using ecosystem CO2 flux measurements and not considering the effect of other resource limitations (for example, nitrogen) on the microbe biomass. Future data assimilation studies with these models should include ecosystem-scale measurements of R Soil in the parameter estimation routine and experimentally determine soil model parameters not constrained by the parameter estimation routine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soils are important in the terrestrial carbon cycle for their role in the cycling and storage of carbon (Jenkinson and Rayner 1977; Schimel and others 1994; Jobbagy and Jackson 2000; Trumbore 2000; Raich and others 2002). Soil processes such as decomposition are strongly influenced by soil microbial communities (Fierer and others 2003; Lipson and Schmidt 2004; Crawford and others 2005; Monson and others 2006b; Göttlicher and others 2006). These microbial communities have been found to be quite heterogenous exhibiting both temporal (Lipson and Schmidt 2004) and spatial variation (Fierer and others 2003) in species assemblage. Temperature and other environmental factors such as moisture strongly influence these microbial communities and their associated CO2 fluxes from soil (Davidson and Janssens 2006). Current projections of increased surface temperature and changes in moisture (Alley and others 2007) will likely affect soil microbial interactions, ultimately changing the efflux of CO2 from soils and producing feedbacks in the terrestrial carbon cycle (Schimel and Gulledge 1998).
From an ecosystem perspective, the CO2 produced from respiration by soil organisms and its subsequent diffusion from the soil is a large component in the overall net ecosystem CO2 exchange (NEE, a complete list of symbols used to refer to fluxes is given in Table 1) (Goulden and others 1996; Lavigne and others 1997; Janssens and others 2001; Griffis and others 2004; Davidson and others 2006b; Monson and others 2006a, b; Chapin and others 2006). Measurements of NEE can complement manipulative experiments and elucidate how biological processes contribute to terrestrial ecosystem CO2 exchange. For example, recent studies at a high-elevation subalpine forest (the Niwot Ridge AmeriFlux site) demonstrated that winter soil respiration (R Soil) contributes to 35–48% of total ecosystem respiration (TER), with a large proportion of this respiration from microbial biomass (Monson and others 2006a, b). Additionally, seasonal variation in observed soil respiration fluxes is coincident with seasonal variation in microbial community composition (Lipson and others 2000; Lipson and Schmidt 2004; Monson and others 2006b).
Long-term records of NEE are a good candidate to investigate how environmental variation affects soil fluxes. Plot-level experimentation has shown differential responses of autotrophic and heterotrophic respiration to environmental drivers such as temperature, moisture, and substrate supply (Tang and others 2005b; Borken and others 2006; Hartley and others 2006; Scott-Denton and others 2006). As NEE is an aggregate measurement of ecosystem processes, long-term records of NEE can potentially be used to independently determine autotrophic and heterotrophic ecosystem respiration.
Modelling is an approach well-suited to exploring how soil carbon processes affect NEE, as direct soil measurements can induce a disturbance to the soil matrix and potentially bias results (Ryan and Law 2005). Recent reviews of soil measurements and soil modelling have emphasized the need for greater focus on understanding the short-term controls of soil respiration and coupling of belowground processes with aboveground processes (for example, photosynthesis) (Smith and others 1998; Fitter and others 2005; Ryan and Law 2005; Trumbore 2006; Davidson and others 2006a).
Plot-level measurements of R Soil are part of the standard measurements at many FLUXNET sites (http://www.fluxnet.ornl.gov/fluxnet). At the Niwot Ridge AmeriFlux site, Scott-Denton and others (2003) showed that the rhizospheric component (roots plus nearby microbes) is a significant contributor to R Soil. A subsequent study showed that autotrophic (root) and heterotrophic (microbial) respiration responded differently to environmental variation (Scott-Denton and others 2006), as summertime decreases in R Soil resulted from lower heterotrophic, rather than autotrophic, contributions to R Soil. The SIPNET ecosystem model (simplified photosynthesis and evapotranspiration model) is a useful tool at FLUXNET sites to decompose NEE into its component fluxes of photosynthesis (GEE) and TER (Braswell and others 2005; Sacks and others 2006, 2007). The current version of SIPNET does not explicitly model the contribution of roots or microbes to R Soil.
Model-data fusion or data assimilation techniques to extract information from models and observations (Raupach and others 2005) is a strategy to understand soil processes by directly using long-term records of NEE. One application of model-data fusion in the environmental science community is to extract meaningful information about ecosystem processes (such as process-level parameters) from the inherent stochasticity in environmental observations (Braswell and others 2005; Clark 2005; Knorr and Kattge 2005; Raupach and others 2005; Williams and others 2005; Xu and others 2006; Sacks and others 2007). Model-data fusion can determine which parameters are well-constrained by the existing data. Identifying the poorly constrained parameters thereby focuses future measurements on the processes in most need of further research. Comparing different models of soil respiration with the same data assimilation framework assists in model selection and makes identification of poorly constrained parameters and processes more robust.
The objective of this study was to model at the ecosystem level the rhizospheric and heterotrophic contributions of R Soil with SIPNET. To constrain the model, we used a multi-year dataset that consists of net CO2 fluxes made at the Niwot Ridge AmeriFlux site in conjunction with a model-data fusion approach to estimate model parameters. Our results were evaluated by (a) model predictions of measured ecosystem fluxes, (b) comparisons of the modelled contributions of R Soil to TER, root respiration (R Root) to R Soil, and (c) literature comparisons of these quantities and estimated parameters.
Materials and Methods
Site Description
Measurements for this study were made at the Niwot Ridge AmeriFlux site, a subalpine forest at 3050 m elevation west of Boulder, Colorado (40°1’58”N; 105°32’46”W). The three dominant conifer species at Niwot Ridge include subalpine fir (Abies lasiocarpa), Engelmann spruce (Picea engelmannii), and lodgepole pine (Pinus contorta). Mean annual precipitation averages 800 mm and the mean annual temperature is 1.5°C (Monson and others 2002). The site has been extensively studied; for further details, see Bowling and others (2001, 2005), Monson and others (2002, 2006a, b), Scott-Denton and others (2003, 2006), Turnipseed and others (2003, 2004), Yi and others (2005), and Sacks and others (2007).
Net ecosystem exchange (NEE) was measured via eddy covariance. Details about the eddy covariance and meteorological measurements at Niwot Ridge can be found in Monson and others (2002). From November 1998 through the present, half-hourly fluxes of CO2 along with corresponding climate data have been measured at this site. For this study, we used half-hourly flux and meteorological data from 1 November 1998 to 31 December 2005 aggregated into a twice-daily timestep. A net CO2 flux measurement determines NEE, which is equal to the sum of photosynthesis (GEE) and total ecosystem respiration (TER). Sign conventions in the micrometeorological literature (and here) typically define all nonradiative CO2 fluxes as positive when directed to the atmosphere, so the GEE flux is negative and the TER flux is positive. Gaps in the half-hourly flux data arose from instrument malfunction or periods of atmospheric stability which can underestimate the flux measurement. These half-hourly gaps were then filled with nonlinear regression or functional fits with environmental variables such as incoming radiation, air temperature, or soil temperature (Monson and others 2002). If more than 50% of the half-hourly flux data for a given twice-daily timestep was gap-filled, this timestep was excluded from the data assimilation routine. Sacks and others (2006) further described the gap-filling and data processing methods used.
The Niwot Ridge forest is aggrading carbon, with cumulative annual NEE ranging from −60 to −80 g C m−2 (Monson and others 2002). These values for cumulative NEE are lower than other forest ecosystems and are attributable to the fact that this is a high-elevation site with extreme climate conditions (Monson and others 2002).
In addition to NEE, six additional climate variables measured at the site were used in the model: air temperature, soil temperature, precipitation, flux density of photosynthetically active radiation, relative humidity, and wind speed. The model was run on a twice-daily time step. The exact length of each day or night time step was determined from the day of year and latitude. For this study CO2 fluxes are reported as g C m−2 day−1.
SIPNET Ecosystem Model
The basic model formulation of SIPNET has been described in previous papers (Braswell and others 2005; Sacks and others 2006, 2007). SIPNET is a simplified version of the PnET family of models (Aber and Federer 1992; Aber and others 1996). The base model for SIPNET has three vegetation carbon pools (wood, leaves, and soil) and includes a model for soil moisture. The soil moisture model was developed by Sacks and others (2006) and is described in detail in that study. The initial conditions and fluxes are characterized by parameters listed in Table 2. Because Niwot Ridge is a coniferous forest, the model assumes an evergreen phenology. Biomass is added at a rate proportional to the net primary productivity (photosynthesis less autotrophic respiration, NPP). Photosynthesis adds biomass to the wood carbon pool and is the only way that carbon can be added to the ecosystem. Allocation to other carbon pools (such as leaves) decreases the wood carbon pool.
For this study we modified the soil respiration calculations to model winter CO2 fluxes more accurately. Previous studies (Braswell and others 2005; Sacks and others 2006, 2007) reduced soil respiration at all times of the year by a factor proportional to soil wetness. For this study, we applied this modification only when the soil temperature was greater than 0°C.
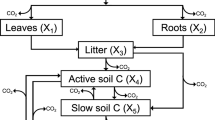
The most significant changes to SIPNET are explicit modelling of root carbon dynamics and modelling the influence of soil microbes on the soil carbon pool. Model parameters are determined from literature values or estimated with the model-data fusion routine described in section “Parameter Estimation Routine”. The various model structures are described in detail below and are conceptually shown in Figure 1. Table 2 lists the fixed and estimated parameters for each model modification.
Base Model
This model is the same one used in (Sacks and others 2006, 2007) and is shown in Figure 1A. No explicit modelling of root or soil microbial dynamics occurs. The only allocation to new biomass is to the leaf carbon pool. Wood and leaf respiratory losses are modelled with the following equation:
where R X is the actual respiration rate from pool X (g C m−2 day−1), K X a base rate (day−1), C X the amount of carbon in a given pool (g C m−2). Needle and wood respiration use the same Q 10 value. For needle respiration, the base respiration rate is adjusted by a factor of \(Q_{10}^{-T_{{\text{Opt}}}/10},\) where T Opt is an optimum temperature for photosynthesis (Aber and Federer 1992; Aber and others 1996). Soil heterotrophic respiration is treated as in equation (1), except that soil temperature is used instead of air temperature and respiration is reduced by a factor proportional to the fractional soil wetness when soil temperatures are above zero:
where R Soil is the actual respiration rate (g C m−2 day−1), K S a base respiration rate (day−1), C S the amount of soil carbon (g C m−2), W S the soil water amount (cm water equivalent), and W S,C is the soil water holding capacity (cm water equivalent). With this formulation of R Soil, heterotrophic respiration (R H) and autotrophic soil respiration are not distinguished.
Roots Model
This model expands on the Base model and is shown in Figure 1B. The wood carbon pool is split into (a) aboveground biomass, (b) fine roots, and (c) coarse roots. Allocation among the four carbon pools (wood, leaves, fine roots, coarse roots) occurs at a rate proportional to the mean NPP over the past 5 days as described in equation (3):
To determine appropriate values for the percentage of NPP allocated to coarse and fine roots (αCR and αFR, respectively), we assume that total belowground carbon allocation (TBCA) is approximately twice litterfall input I L (Raich and Nadelhoffer 1989). Assuming that TBCA is equal to (αCR + αFR)NPP, the sum of αCR and αFR can be found if litterfall inputs are known:
Litterfall rates (I L) and NPP derived from the Base model provide an estimate for αCR + αFR in equation (4). Examining the frequency distribution of αCR + αFR showed that the most frequent value of αCR + αFR is approximately 0.4 (results not shown). Assuming that this belowground allocation is split equally between fine and coarse roots (McDowell and others 2001; Joslin and others 2006), this yields values of αCR and αFR to be 0.2.
Fine and coarse root respiratory losses (R Fine-Root and R Coarse-Root, respectively) are modelled with equation (1) using soil temperature for the Q10 functional response. Heterotrophic respiration R H is modelled with equation (2). Overall soil respiration R Soil equals the sum of R Fine-Root, R Coarse-Root, and R H.
In addition to respiration, root carbon losses also occur through exudation into the soil. Root exudation occurs at a rate proportional (β X ) to the mean photosynthesis over the past 5 days:
Quality Model
This model is shown in Figure 1C and expands the Roots model by structuring the soil carbon pool into a discrete number of soil pools that theoretically represent a continuum between more labile (high substrate quality, easily decomposed) to more recalcitrant (low substrate quality, less easily decomposed) pools of carbon. For this study, the number of soil carbon pools was fixed at three. These pools are parameterized by a variable q representing the “quality” of a particular pool (Ågren and Bosatta 1987, 1996; Bosatta and Ågren 1991). For this application, the variable q takes on values between zero and one (Bosatta and Ågren 1985). Higher values of q represent more labile soil carbon and lower values more recalcitrant soil carbon.
Variation in the dynamics of each soil pool occurs by associating inputs such as litter with different quality pools. For this study, leaf litter and root exudates enter the highest quality pool, and wood litter enters the second highest quality pool. Associating litter or exudates with a particular soil quality pool potentially leads to different sizes of each of the respective soil pools. As respiration is dependent on pool size, we expect variation in the amount of carbon respired across these soil pools.
In addition to litter inputs and respiration losses, decomposition influences soil pool carbon content in the Quality model. This model assumes that in the process of decomposition the quality (q) of carbon that had been decomposed decreases and enters another soil quality pool. Soil carbon of quality q i is incorporated into microbial biomass at the following ingestion rate:
where ε is the efficiency in converting carbon into biomass (no units), C S,i the soil carbon in quality pool i (g C m−2), μmax the specific microbial ingestion rate (h−1) and \(\tilde{C}_{{\text{B}},0}\) is the average microbial biomass density of carbon (g C m−2). Growth respiration (R Growth) is given by equation (7):
This growth respiration is associated with respiration from the growth of new microbial biomass. Heterotrophic respiration, or maintenance costs from existing soil microbes, is dependent on soil carbon of quality q i . This respiration is characterized with equation (8), where K H is a base respiration rate (day−1):
Overall soil respiration consists of root respiration (R Fine-Root and R Coarse-Root), growth respiration (R Growth) and the heterotrophic respiration (R H,i ) across all different soil carbon quality pools.
Linear Soil Respiration Models
Each model variant (Base, Roots, and Quality) adds an additional level of structure to soil carbon. We investigated if this additional complexity leads to an over-specification of soil processes by substituting linear relationships describing soil heterotrophic respiration in the Base and Roots models. Rather than having soil heterotrophic respiration dependent on soil carbon pool size (equation (2)), for this model variant soil heterotrophic respiration was modelled by the following function:
where “max” represents “maximum of” and a 0 and a 1 represent the parameters characterizing the assumed linear relationship between heterotrophic respiration and soil temperature. A linear function in equation (9) was chosen over more complicated functions to avoid overfitting the data. Equation (9) was substituted for equation (2) in the Base and Roots models, respectively. When equation (9) is used, no explicit modelling of soil carbon dynamics occurs. We refer to a Base and Roots model run using equation (9) for R H as “Base-Linear” and “Roots-Linear”, respectively.
Parameter Estimation Routine
The parameter optimization method used in this study was a variation of the Metropolis algorithm developed by Metropolis and others (1953). A similar parameter estimation routine was used in Braswell and others (2005) and Sacks and others (2006, 2007); here we only describe the relevant details.
Each given model (Base, Roots, Quality, Base-Linear, Roots-Linear) has a set of parameters used to characterize the model (Table 2). Each model has two types of parameters: fixed and estimated. Fixed parameter values are derived from literature or from unpublished data collected at the Niwot Ridge site. Estimated parameters are found from the Metropolis algorithm. Each estimated parameter is given a range of allowable values; usually this range is selected from literature or from conventional knowledge. The probability distribution over the range of every parameter was assumed to be uniform. The parameters were estimated using twice-daily NEE data in the parameter estimation routine.
The parameter estimation routine proceeds by exploring the parameter space to find the parameter set that maximizes the likelihood L:
where n was the number of data points, x i and η i are the measured and modelled data and σ is the standard deviation of the data about the model. Values of x i are twice-daily measurements of NEE. This likelihood function assumes that all errors are Gaussian distributed and that the standard deviation σ followed a uniform distribution. As in Braswell and others (2005) and Sacks and others (2006), for this study σ is estimated by finding the value σe that maximizes the likelihood:
Braswell and others (2005) used synthetic data sets with different values of σ and found that σe did, in fact, seem to reproduce the σ used to generate a synthetic data set.
At each time step, the current parameter set generates estimates of GEE, aboveground leaf respiration (R L) and wood respiration (R W), and R Soil. These fluxes then determine modelled values of NEE via equation (12), which is then used for η i in equation (10):
Both measured and modelled values of NEE characterize the likelihood (equation (10)). The Metropolis algorithm then proceeds to find the best parameter set that maximizes the likelihood. In the implementation of the Metropolis algorithm we use the log-likelihood because it is mathematically and computationally easier to determine.
The parameter optimization proceeds by randomly selecting a particular parameter, changing its value by a random amount, and evaluating the log-likelihood with this proposed (new) parameter set. If this proposed parameter set increases the log-likelihood, then this parameter set is accepted. If the proposed parameter set did not increase the log-likelihood, it is still accepted with a probability equal to the difference of the log-likelihoods. After a suitable spin-up period, the collection of accepted parameter sets characterizes the joint posterior probability distribution of the parameters (Hurtt and Armstrong 1996; Braswell and others 2005). For a particular parameter, statistics from the frequency distribution of accepted values (typically 150,000 values) determines final parameter statistics.
Past studies with SIPNET (Braswell and others 2005; Sacks and others 2006, 2007) estimated parameters by using the entire available record of flux measurements in the optimization. This approach makes it difficult to evaluate model performance because parameters have already been optimized to match measured data. As a result, for this study we partition the data into two periods: the first 3 years of flux measurements from Niwot Ridge (November 1, 1998 to December 31, 2001) are used in the parameter estimation routine. This set of flux data will be referred to as the “optimization period.” The remainder of the unused data (January 1, 2002 to December 31, 2005) is subsequently used to evaluate the different models. This set of data will be referred to as the “corroboration period.” Mean cumulative annual NEE from 1999 to 2001 (optimization data) was −73 g C m−2, ranging from −49 to −89 g C m−2. For the corroboration period (2002–2005), mean cumulative annual NEE was −60 g C m−2, ranging from −21 to −88 g C m−2. When the year of lowest net CO2 uptake (−21 g C m−2 during 2002) is excluded from the corroboration period, mean cumulative annual NEE was −72 g C m−2 and ranged from −62 to −88 g C m−2.
Results
The parameter set that yielded the highest log-likelihood in the parameter estimation for each model variation (Base, Roots, Quality) is shown in Table 3. In addition, the mean and standard deviation generated from the set of accepted parameter values are reported. Three types of behavior characterize the posterior distributions. Well-constrained parameters are ones where the best and mean values are similar and the standard deviation is typically small (for example, A Max, T Min, K). An edge-hitting parameter is one where the best or mean value is near the edge of its allowed range (for example, K H). Finally, a noninformative parameter is one where the best and mean values differed significantly and the parameter standard deviation is quite large (for example, δW, δCR). A noninformative parameter suggests the NEE data used in the parameter optimization could not constrain that particular parameter. Some parameters have remarkable consistency among the Base, Roots, and Quality models (for example, A Max and T Min), whereas others have considerable variation (for example, δCR and Q10CR).
Table 4 compares the log-likelihood for each of the models using the best parameter set retrieved from the parameter estimation. A higher log likelihood (closer to zero) indicates the model has a better fit with the data. The Roots and Quality models increased the overall log-likelihood from the Base model with the maximum improvement (highest log-likelihood) in the Roots model. With each model refinement (Roots and Quality models), additional parameters are introduced. Introducing these parameters to the model increases the degrees of freedom and may lead to a higher log-likelihood by overfitting the data. Hence, one must ask if the increased log-likelihood associated with the Roots and Quality models truly represents a better model or is an overfit of the data. The Bayesian Information Criterion (BIC) (Schwartz 1978) assesses this by introducing a penalty for each additional parameter:
where n is the number of data points used in the optimization, M is the number of estimated parameters, and LL is the log-likelihood. A lower value of BIC indicates greater support for the model from the data (Kendall and Ord 1990). Values of the BIC in Table 4 indicate that the Roots model had the greatest support from the data.
Figure 2 compares measured NEE to modelled NEE for the Quality model using the best (for example, highest likelihood) parameter set and data from the corroboration period. Similar results for the other models were obtained and hence are not shown. Figure 2 distinguishes between winter and summer time periods. For each year, summer was determined by the zero crossing of daily integrated NEE, indicating an ecosystem transition from a net source of CO2 to a net sink of CO2.
During the winter, the Quality model predicts more net CO2 uptake (more negative NEE) than observed. During the summer this pattern is reversed, with the model predicting less net uptake (less negative NEE, Figure 2B). Similar results to the patterns in Figure 2 were found in Sacks and others (2006, 2007).
Figure 3 shows estimated values of NEE, GEE and TER for each of the model variants. Clearly, all models are able to generate consistent estimates of GEE and TER. Figure 4A shows the twice daily values of measured and modelled NEE for the Roots model. Examination of the difference between measured and modelled cumulative NEE (Figure 4B) shows marked differences between the Base, Roots, and Quality models. Although all models correctly show that the ecosystem is a net sink of CO2, they differ in their predictions of the size of this sink. The Base model eventually predicts less cumulative net uptake than the Quality or Roots models. All models predict similar levels of photosynthesis but have different respiration rates (Figure 3). Similar levels of photosynthesis are a consequence of generating similar values for photosynthetic parameters in the estimation routine (Table 3). If data were being overfitted, there is no prior reason to expect this robustness in the photosynthetic parameters. Additionally, from Figure 4B the Roots model underestimates respiration, whereas the Base and Quality models overestimate respiration. No model could capture the decreases in net CO2 uptake caused by drought during the summer of 2002 (Figure 3E).
Comparisons of measured and modelled cumulative NEE. Panel (A) shows comparison among twice-daily values of measured NEE and modelled NEE for the Roots model. Similar results for the other model variants were obtained. Panel (B) shows the difference between measured and modelled values of cumulative NEE for each of the model variants. Positive values indicate that the model is producing more negative values of NEE than measurements. Gray-shaded panels in both plots represent the optimization period of fluxes used to estimate model parameters.
Figure 5 shows values of R Soil (Figure 5A–C), R Root (Figure 5D, E) and R H (Figure 5F, G) for each of the model variants. Note that the Base model does not model root dynamics, hence R Root and R H are subsumed into R Soil. Estimates of R Root for the Roots and Quality models all reached a maximum during summer months. This summer peak in root respiration agrees with seasonal trends shown in Bond-Lamberty and others (2004b). Model estimates of R H show summer-time decreases in the Roots and Quality models. These decreases are attributed to decreases in soil moisture, which reduce the amount of respiration. Suppression of heterotrophic, but not root, respiration during periods of soil moisture limitations is consistent with observations reported by Scott-Denton and others (2006).
Figure 6 shows the distribution of values of R Soil/TER and R Root/R Soil for each of the model variants. The Roots and Quality models have seasonal variation in the contribution of R Soil to TER and R Root to R Soil, with wintertime values having R Soil/TER closer to unity. This seasonal variation arises from the assumption that foliar respiration only occurs when the air temperature is above a certain threshold, T S, which for the Base, Roots, and Quality models was estimated to be 0.06°C; thus, in the parameter estimation routine all ecosystem respiration is forced toward R Root and R Soil during the winter.
Discussion
Overall Model Comparisons
The models produce reasonable values of NEE that corresponded well with measurements (Figure 4A). Overall, these soil carbon models did not detract from the ability of SIPNET to partition NEE into GEE and TER (Figure 3).
Variation in the long-term modelled NEE (Figure 4B) between the Base, Roots, and Quality models is strongly dependent on the assumptions governing each model and the values of the parameters estimated from the data assimilation routine. As estimated photosynthetic parameters between the models were quite robust, variation in long-term modelled NEE is a consequence of the lack of robustness in parameters describing turnover and respiration rates between the Base, Roots, and Quality models. Higher values of base respiratory rate parameters (for example, K H, K CR, K FR) would increase overall respiration rates. A higher pool turnover rate would reduce the amount of carbon in a particular pool. As respiration rates are assumed to be proportional to pool size, variation in turnover rate parameters indirectly affect respiratory fluxes and ultimately modelled NEE values.
The model with the lowest BIC is the Roots model (Table 4). The penalty for each additional parameter (the value of ln(n) in equation (13)) is approximately 8. Arguably the lower BIC from the Roots model is a marginal improvement over the Base and Quality models (approximately 1% from the Base model). The Roots and Quality models have at least 6 more additional parameters than the Base model. If some of these additional parameters could be determined from direct experimentation, this would reduce the value of the BIC for the Roots and Quality and make the BIC comparison more robust.
Estimation of Soil Respiration Fluxes
A given model structure can have significant effects on modelled soil respiration fluxes (Figure 5 and Table 3). Past studies with SIPNET have been able to successfully partition NEE into GEE and TER (Braswell and others 2005; Sacks and others 2006, 2007), but had less success in partitioning TER into its autotrophic and heterotrophic components. Inclusion of root carbon pools, and structuring the soil carbon through a soil quality approach show improvements in the ability to match measured CO2 fluxes with the Roots and Quality models having higher log-likelihoods than the Base model (Table 4).
The estimated contribution of soil respiration to total ecosystem respiration (R Soil/TER) predicted by the various models presented in this study can be compared to previously published studies (Table 5). Mean values as an average across the corroboration period are reported, however, we note that seasonal variation in soil respiration fluxes was found in the Roots and Quality models (Figure 6). Model predicted values of R Soil/TER for the Roots and Quality models all fall within ranges of published studies at Niwot Ridge (Monson and others 2006a) or other literature studies (Lavigne and others 1997; Law and others 1999; Janssens and others 2001; Davidson and others 2006b). Note that the Base model significantly underestimates R Soil/TER. The mean values reported in Table 5 may be biased toward wintertime values, which were generally close to unity for the Roots and Quality models (Figure 6B, C). We argue these modelled values are overestimated; Monson and others (2006a) estimated wintertime values of R Soil/TER at Niwot Ridge to be 0.35–0.48. As discussed in section “Results”, these large values of wintertime R Soil/TER reflect the model assumption that foliar respiration is zero below a temperature threshold.
Modelled values of the contribution of R Root to R Soil are approximately 0.8, with considerable variation (Table 5). These values are significantly higher than those reported from girdling studies at Niwot Ridge (0.4–0.5, Scott-Denton and others (2006)) and in literature. A recent meta-analysis by Subke and others (2006) showed that the contribution of heterotrophic respiration to soil respiration ranged between 0.4–0.7, implying that the contribution of R Root to R Soil is approximately 0.3–0.6. Variation in this ratio may depend on forest age (Bond-Lamberty and others 2004b), time of year (Bond-Lamberty and others 2004b), plant phenology (Davidson and others 2006b), litter inputs (Dehlin and others 2006; Cizneros-Dozal and others 2007), nutrient cycling (Brooks and others 2004), or ecosystem type (Subke and others 2006). Methodological differences may have an effect on experimental estimates of R Root and by association R Root/R Soil; these are discussed further in Hanson and others (2000), Hendricks and others (2006), Jassal and Black (2006), and Subke and others (2006).
The Roots and Quality models led to better agreement from the Base model in determining the contribution of R Soil to TER (Table 5). In spite of these encouraging results, additional work is needed to further partition R Soil into its constituent components R Root and R H. Improvements could be made to the model structure by modelling physical and biological effects on winter soil respiration fluxes (Massman and others 1997; Brooks and others 2004; Hubbard and others 2005; Monson and others 2006b). Incorporating these changes to SIPNET could improve the ability to estimate R Root/R Soil.
The use of linear functions to describe soil heterotrophic respiration (for example, equation (9)) contrasts with more mechanistic descriptions of soil heterotrophic respiration (for example, equations (2) and (8)). Describing soil respiration with a linear functions provides a worse model-data fit (higher BIC, Table 4) and unrealistic estimates of heterotrophic respiration than with mechanistic approaches (results not shown). Often a fitted linear model defines the best a process model can expect to do, suggesting the Base-Linear and Roots-Linear models omit crucial processes. Previous SIPNET studies (for example, Sacks and others (2006, 2007)) and the results from the Roots model suggest that these processes are most likely time-scales and a degree of uncoupling between soil and aboveground processes.
Evaluation of Allocation Parameters and Estimated Turnover Rates
Biomass turnover rate parameters estimated from the various models can be compared to turnover rates from published studies (Table 6). As discussed in section “Results”, leaf turnover rate (δL) is a parameter well-constrained from the data assimilation. When these values are converted to litterfall rates, they compare favorably with literature values. Needle turnover rates in the range of 0.050–0.090 y−1 correspond to leaf litterfall rates of 100–180 g C m−2 y−1, assuming leaf biomass is approximately 900 g C m−2 and the fractional carbon content of leaves is 0.45. For our simulations, the final values of leaf biomass across all model variants range from 900 to 950 g C m−2 (results not shown). Annual litterfall input at Niwot Ridge has been estimated to be 184 g C m−1 y−1 (Sacks and others 2007). These litterfall rates are certainly within the range of values from a variety of ecosystems reported in Davidson and others (2002). Laiho and Prescott (1999) reported annual litterfall input for A. lasiocarpa and P. englemanii forests to be 200 and 240 g C m−2 y−1, respectively. Assuming that half of the litter is foliage, this would correspond to leaf litter input rates of 100–120 g C m−2 y−1.
The wood turnover rate parameter (δW) is not well-constrained for any of the model variants. Assuming wood biomass is 5500 g C m−2 and a fractional carbon content of 0.5 (Laiho and Prescott 1999), then wood turnover rates of 0.01–0.04 y−1 correspond to wood litterfall rates of 110–440 g C m−2 y−1. Woody litterfall input ranges derived from reported values in Laiho and Prescott (2004) were 100–120 g C m−2 y−1; these measured values are near the low end of the modelled ranges. Hence, we can conclude that estimated values of δW for the Roots and Quality models may be too high. With these high values of δW, simulations of the Quality model predicted a much lower final pool size for wood biomass (<1000 g C m−2) then would be expected in the context of the above studies.
Estimated fine and coarse root turnover rates have considerable variation among each of the model variants, suggesting that NEE data alone could not adequately constrain these parameters. We would not have any prior reason to suspect that half-daily NEE data could constrain these parameters, as wood and roots require several months to years to decompose. To obtain a good constraint for these parameters stable isotope or radiocarbon approaches (Gaudinksi and others 2001) would be needed. While there is considerable variation in the root turnover parameters, published literature values have considerable variation as well. Gill and Jackson (2000) reviewed 190 published studies and reported root turnover rates in temperate coniferous forests to range from 0.1–0.8 y−1. This variation in turnover rates may arise from methodological biases used to determine these rates (Hendricks and others 2006; Subke and others 2006).
The considerable variation in the estimated turnover parameters may reflect the simplistic way SIPNET treats turnover of particular carbon pools. Turnover rate parameters represent not only loss from a particular carbon pool as litter, but also subsequent decay and release of that carbon into the soil carbon pool. Decay and subsequent release into the soil typically occurs at a much slower rate than litterfall, which would decrease the value of a particular turnover parameter. Reported decay values in coniferous forests are much lower than input rates (Johnson and Greene 1991; Laiho and Prescott 2004).
This study assumes a constant allocation to roots, leaves, and wood. This constant allocation assumption has broad support in the literature (Raich and Nadelhoffer 1989; Bond-Lamberty and others 2004a; Hendricks and others 2006; Subke and others 2006). We derived values of αCR and αFR from an assumed relationship between TBCA and litterfall rate (Raich and Nadelhoffer 1989). This relationship has been found to be highly uncertain, and perhaps larger, for forests not at steady state (Davidson and others 2002). Niwot Ridge is recovering from early twentieth-century logging (Monson and others 2002); hence applying the relationship from Raich and Nadelhoffer (1989) may underestimate αCR and αFR.
We chose a 5-day time period between photosynthesis and structural allocation of carbon. This argues that there is a 5-day lag between recently assimilated carbon to when it could potentially be respired. Past studies using (a) stable isotope tracers (Ekblad and Högberg 2001; Bowling and others 2002; Knohl and others 2005; Schaeffer and others In press), (b) tree girdling to remove photosynthate supply (Högberg and others 2001), (c) radiocarbon tracing techniques (Carbone and others 2007), or (d) soil CO2 measurements combined with measurements of NEE (Tang and others 2005a) have estimated this lag to vary from 1 to 10 days. Hence, our choice of a 5-day lag is appropriate but highly uncertain.
This study assumed that carbon exudates from the rhizosphere into the soil are proportional to mean GEE over the past 5 days. The 5-day lag and proportionality constants (βFR and βCR) are conservative estimates from field studies (Jakobsen and Rosendahl 1990; Rangel-Castro and others 2005; Christensen and others 2007; Kaštovská and Šantr

čkova 2007); these values are highly uncertain and are strongly dependent on soil substrate quality and microbiota (Kaštovská and Šantr

čkova 2007).
Evaluation of Quality model
The Quality model presented here is based on a discrete version (Bosatta and Ågren 1985) of a continuous time, continuous quality model (Ågren and Bosatta 1987, 1996; Bosatta and Ågren 1991). Additional complexities for the Quality model could be incorporated. First, previous modelling studies have argued that μmax and ε should be an increasing function of quality (Ågren and Bosatta 1987, 1996; Bosatta and Ågren 1991, 1999). Second, temperature may have an additional effect on soil parameters (Holland and others 2000; Tjoelker and others 2001; Wythers and others 2005; Davidson and Janssens 2006). Inclusion of temperature dependence in the ingestion rate (μmax), efficiency (ε), or Q10 values could generate different results than the ones presented here. Initial data assimilation tests showed that there was not enough information in the flux data to include these additional complexities.
For this study, litter and root exudates are quality dependent, leading to potential differences in the respiration from these different soil quality pools. The assumption of quality-dependent litter is supported in a recent study by Dehlin and others (2006), which found that microbial communities responded differently to mixtures of different substrates than when grown with individual substrates alone. Additionally, for this study, soil is always degraded to a lower quality with no delay between subsequent ingestion and release back to the soil. Subsequent degradation in soil quality may be valid for application of the Quality model on longer timescales (decades to centuries, Ågren and Bosatta (1987)), but may not be appropriate for SIPNET on a twice-daily timestep.
One of the challenges of the soil quality model is parameterizing measurements of soil quality mathematically. The most general definition of quality is the accessibility of a substrate for decomposition (Ågren and Bosatta 1996; Bosatta and Ågren 1999). Alternatively, soil quality may be explicitly parameterized by physical parameters (water content, density), chemical factors (pH, total organic carbon), biological activity (microbial activity, presence of pathogens) (Burns and others 2006), or radiocarbon content (Trumbore 2000). Future studies should explicitly link these additional definitions of soil quality with measurements (for example, soil temperature and moisture).
The microbiota play a critical role and their dynamics reflect community processes, nutrient limitations, and specific substrate effects (Brooks and others 1996; Monson and others 2006b; Scott-Denton and others 2006; Lipson and others In review). None of these influences are explicit to this model. Future model refinements are needed to address these issues.
Future Work
Many estimated parameters from the Roots and Quality models (for example, turnover parameters) were not as well constrained as other parameters, (for example, A max, T min, T Opt). Site-specific determination of these unconstrained parameters can improve upon the results here by reducing the number of degrees of freedom, potentially improving model-data fits. Repeating this analysis with the inclusion of additional data streams in the data assimilation (such as measurements of R Soil appropriately scaled to the ecosystem) might better constrain some of the model parameters. Future studies should also attempt to incorporate in the data optimization more discrete measurements such as litterfall measurements or needle biomass surveys. Such measurements would complement NEE measurements by providing a stronger direct constraint on pool size. Equation (10) would then contain additional terms describing these constraints. The combination of time series data (NEE measurements) with discrete data (R Soil measurements) in the likelihood function will require careful consideration.
Fundamental to the approach of SIPNET is the introduction of additional complexity only when needed to avoid overfitting data. The Roots and Quality models increase confidence in the ability of SIPNET to characterize soil respiration at the ecosystem scale when compared to the Base SIPNET model or linear models of soil respiration. However, additional modifications such as site-specific determination of soil pool turnover parameters and consideration of multiple resource limitations are needed. Yet we stress that this conclusion could not have been reached if a more complicated model was initially used. Furthermore, ecosystem-scale R Soil measurements would provide additional comparisons to potentially strengthen our conclusions. This could be achieved with plot-level measurements of R Soil, however care must be exercised when scaling these plot-level R Soil values to the ecosystem (Lavigne and others 1997; Dore and others 2003).
Conclusions
This study modified an established process-based ecosystem model and evaluated contrasting models of soil carbon processes. Comparisons of the original model to model refinements found no noticeable difference in model predictions of GEE and TER. However, these model refinements strongly diverged from the original model in their estimates of soil respiration fluxes.
Results from this study strongly conclude that the NEE flux alone is not a strong constraint on soil fluxes. Modifications to the basic model structure of SIPNET by adding explicit dynamics to the root and heterotrophic components helped, but introduced additional complexities.
This study found that mechanistic representations of soil heterotrophic respiration are better than using a fitted linear model. This is strong evidence that the structure of the soil ecosystem (roots, soil organic matter quality) have direct and first-order effects on fluxes that can not be captured in a parametric zeroth or simple first order regression model.
Therefore, extracting information about the environmental controls on the fluxes requires (a) the correct model structure (which will be even somewhat more complex than presented here) and (b) additional data constraining soil processes such as microbial growth efficiency or net and gross nitrogen mineralization.
References
Aber JD, Federer A (1992) A generalized, lumped-parameter model of photosynthesis, evapotranspiration and net primary production in temperate and boreal forest ecosystems. Oecologia 92:463–474.
Aber JD, Reich PB, Goulden ML (1996) Extrapolating leaf CO2 exchange to the canopy: a generalized model of forest photosynthesis compared with measurements by eddy correlation. Oecologia 106:257–265.
Ågren G, Bosatta E (1987) Theoretical analysis of the long-term dynamics of carbon and nitrogen in soils. Ecology 68(5):1181–1189.
Ågren GI, Bosatta E. 1996. Theoretical ecosystem ecology: understanding element cycles. Cambridge University Press
Alley R, Berntsen T, Bindoff NL, Chen Z, Chidthaisong A, Friedlingstein P, Gregory J, Hegerl G, Heimann M, Hewitson B, Hoskins B, Joos F, Jouzel J, Kattsov V, Lohmann U, Manning M, Matsuno T, Molina M, Nicholls N, Overpeck J, Qin D, Raga G, Ramaswamy V, Ren J, Rusticucci M, Solomon S, Somerville R, Stocker TF, Stoot P, Stouffer RJ, Whetton P, Wood RA, Wratt D. 2007. Climate change 2007: the physical science basis, summary for policymakers, Intergovernmental Panel Climate Change
Arthur MA, Fahey TJ (1992) Biomass and nutrients in an Englemann spruce-subalpine fir forest in north central Colorado: pools, annual production, and internal cycling. Canadian Journal of Forest Research 22:315–325.
Bhupinderpal-Singh, Nordgren A, Löfvenius MO, Högberg MN, Mellander PE, Högberg P (2003) Tree root and soil heterotrophic respiration as revealed by girdling of boreal scots pine forest: extending observations beyond the first year. Plant Cell Environment 26:1287–1296.
Bond-Lamberty B, Wang C, Gower ST. 2004a. A global relationship between the heterotrophic and autotrophic components of soil respiration? Global Change Biology 10:1756–1766 doi:10.1111/j.1365-2486.2004.00816.x
Bond-Lamberty B, Wang C, Gower ST (2004b) Contribution of root respiration to soil surface CO2 flux in a boreal black spruce chronosequence. Tree Physiology 24:1387–1395.
Borken W, Savage K, Davidson EA, Trumbore SE. 2006. Effects of experimental drought on soil respiration and radiocarbon efflux from a temperate forest soil. Global Change Biology 12:177–193 doi:10.1111/j.1365-2486.2005.01058.x
Bosatta E, Ågren GI (1985) Theoretical analysis of decomposition of heterogeneous substrates. Soil Biology and Biochemistry 17(5):601–610.
Bosatta E, Ågren GI (1991) Dynamics of carbon and nitrogen in the organic matter of the soil: a generic theory. The American Naturalist 138(1):227–245.
Bosatta E, Ågren GI (1999) Soil organic matter quality interpreted thermodynamically. Soil Biology and Biochemistry 31:1889–1891.
Bowling DR, Tans PP, Monson RK. 2001. Partitioning net ecosystem carbon exchange with isotopic fluxes of CO2. Global Change Biology 7:127–145.
Bowling DR, McDowell NG, Bond BJ, Law BE, Ehleringer JR. 2002. 13C content of ecosystem respiration is linked to precipitation and vapor pressure deficit. Oecologia 131:113–124.
Bowling DR, Burns SP, Conway TJ, Monson RK, White JWC. 2005. Extensive observations of CO2 carbon isotope content in and above a high-elevation subalpine forest. Global Biogeochemical Cycles 19:GB3023 doi:10.1029/2004GB002394
Braswell BH, Sacks WJ, Linder E, Schimel DS. 2005. Estimating diurnal to annual ecosystem parameters by synthesis of a carbon flux model with eddy covariance net ecosystem exchange observations. Global Change Biology 11:335–355 doi:10.1111/j.1365-2486.2005.00897.x.
Brooks PD, Williams MW, Schmidt SK. 1996. Microbial activity under alpine snowpacks, Niwot Ridge, Colorado. Biogeochemistry 32:93–113.
Brooks PD, McKnight D, Elder K. 2004. Carbon limitation of soil respiration under winter snowpacks: potential feedbacks between growing season and winter carbon fluxes. Global Change Biology 11:231–238 doi:10.1111/j.1365-2486.2004.00877.x
Burns RG, Nannipieri P, Benedetti A, Hopkins DW. 2006. Defining soil quality. In: Bloem J, Hopkins DW, Benedetti A, Eds. Microbiological methods for assessing soil quality. CABI Publishing. pp 15–22
Carbone MS, Czimczik CI, McDuffee KE, Trumbore SE. 2007. Allocation and residence time of photosynthetic products in a boreal forest using a low-level 14C pulse-chase labeling technique. Global Change Biology 13:466–477. doi: 10.1111/j.1365-2486.2006.01300.x
Chapin FS, Woodwell GM, Randerson JT, Rastetter EB, Lovett GM, Baldocchi DD, Clark DA, Harmon ME, Schimel DS, Valentini R, Wirth C, Aber JD, Cole JJ, Goulden ML, Harden JW, Heimann M, Howarth RW, Matson PA, McGuire AD, Melillo JM, Mooney HA, Neff JC, Houghton RA, Pace ML, Ryan MG, Running SW, Sala OE, Schlesinger WH, Schulze ED. 2006. Reconciling carbon-cycle concepts, terminology, and methods. Ecosystems 9:1041–1050 doi:10.1007/s10021-005-0105-7
Christensen S, Bjørnlund L, Vestergård M. 2007. Decomposer biomass in the rhizosphere to assess rhizodeposition. Oikos 116:65–74 doi:10.1111/j.2006.0030-1299.15178.x
Cizneros-Dozal LM, Trumbore SE, Hanson PJ. 2007. Effect of moisture on leaf litter decomposition and its contribution to soil respiration in a temperate forest, J Geophys Res. 112:GB01013 doi:10.1029/2006JG000197
Clark JS. 2007. Models for ecological data: an introduction. Princeton University Press
Crawford JW, Harris JA, Ritz K, Young IM. 2005. Towards an evolutionary ecology of life in soil. TRENDS in Ecology and Evolution 20:81–87 doi:10.1016/j.tree.2004.11.014
Davidson EA, Janssens IA. 2006. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440:165–173 doi:10.1038/nature04514
Davidson EA, Savage K, Bolstad P, Clark DA, Curtis PS, Ellsworth DS, Hanson PJ, Law BE, Luo Y, Pregitzer KS, Randolph JC, Zak D. 2002. Belowground carbon allocation in forests estimated from litterfall and IRGA-based soil respiration measurements. Agricultural and Forest Meteorology 113:39–51.
Davidson EA, Janssens IA, Luo Y. 2006a. On the variability of respiration in terrestrial ecosystems: moving beyond Q10. Global Change Biology 12:154–164 doi:10.1111/j.1365-2486.2005.01065.x
Davidson EA, Richardson AD, Savage KE, Hollinger DY. 2006b. A distinct seasonal pattern of the ratio of soil respiration to total ecosystem respiration in a spruce-dominated forest. Global Change Biology 12:230–239 doi:10.1111/j.1365-2486.2005.01062.x
Dehlin H, Nilsson Charlotte MC, Wardle DA. 2006. Aboveground and belowground responses to quality and heterogeneity of organic inputs to the boreal forest. Oecologia 150(1):108–118. doi: 10.1007/s00442-006-0501-5
Dore S, Hymus GJ, Johnson DP, Hinkle CR, Valentini R, Drake BG. 2003. Cross validation of open-top chamber and eddy covariance measurements of ecosystem CO2 exchange in a Florida scrub-oak ecosystem. Global Change Biology 9:84–95. doi:10.1007/s004420100667
Ekblad A, Högberg P. 2001. Natural abundance of 13C in CO2 respired from forest soils reveals speed of link between tree photosynthesis and root respiration. Oecologia 127:305–308. doi:10.1007/s004420100667
Fierer N, Schimel JP, Holden PA. 2003. Variations in microbial community composition through two soil depth profiles. Soil Biology and Biochemistry 35:167–176.
Fitter AH, Gilligan CA, Hollingworth K, Kleczkowski A, Twyman RM, Pitchford JW, Members of the NERC Soil Biodiversity Programme. 2005. Biodiversity and ecosystem function in soil. Funct Ecol 19:369–77. doi 10.1111/j.1365-2435.2005.00969.x
Gaudinksi JB, Trumbore SE, Davidson EA, Cook AC, Markewitz D, Richter DD. 2001. The age of fine-root carbon in three forests of the eastern United States measured by radiocarbon. Oecologia 129:420–429 doi:10.1007/s004420100746
Gill RA, Jackson RB (2000) Global patterns of root turnover for terrestrial ecosystems. New Phytologist 147:13–31.
Göttlicher SG, Steinmann K, Betson NR, Högberg P. 2006. The dependence of soil microbial activity on recent photosynthate from trees. Plant Soil 287:85–94 doi:10.1007/s11104-006-0062-8
Goulden ML, Munger JW, Fan SM, Daube BC, Wofsy SC (1996) Measurements of carbon sequestration by long-term eddy covariance: methods and a critical evaluation of accuracy. Global Change Biology 2:169–182.
Griffis TJ, Black TA, Gaumont-Guay D, Drewitt GB, Nesic Z , Barr AG, Morgenstern K, Kljun N. 2004. Seasonal variation and partitioning of ecosystem respiration in a southern boreal aspen forest. Agricultural and Forest Meteorology 125:207–223.
Hanson PJ, Edwards NT, Garten CT, Andrews JA. 2000. Separating root and soil microbial contributions to soil respiration: a review of methods and observations. Biogeochemistry 48:115–146.
Hartley IP, Armstrong AF, Murthy R, Barron-Gafford G, Ineson P, Atkin OK. 2006. The dependence of respiration on photosynthetic substrate supply and temperature: integrating leaf, soil, and ecosystem measurements. Global Change Biology 12:1954–1968 doi:10.1111/j.1365-2486.2006.01214.x
Hendricks JJ, Hendrick RL, Wilson CA, Mitchell RT, Pecot SD, Guo D (2006) Assessing the patterns and controls of fine root dynamics: an empirical test and methodological review. Journal of Ecology 94:40–57.
Högberg P, Nordgren A, Buchmann N, Taylor AFS, Ekblad A, Högberg MN, Nyberg G, Ottosson-Löfvenius M, Read DJ. 2001. Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature 411:789–792.
Holland EA, Neff JC, Townsend AR, McKeown B. 2000. Uncertainties in the temperature sensitivity of decomposition in tropical and subtropical ecosystems: implications for models. Global Biogeochemical Cycles 14(4):1137–1151.
Hubbard RM, Ryan MG, Elder K, Rhoades CC. 2005. Seasonal patterns in soil surface CO2 flux under snow cover in 50 and 300 year old subalpine forests. Biogeochemistry 73:93–107 doi:10.1007/s10533-004-1990-0
Hurtt GC, Armstrong RA. 1996. A pelagic ecosystem model calibrated with BATS data. Deep-Sea Research Part II-Topical Studies in Oceanography 43:653–683.
Jakobsen I, Rosendahl L (1990) Carbon flow into soil and external hyphae from roots of mycorrhizal cucumber plants. New Phytologist 115:77–83.
Janssens IA, Lankreijer H, Matteucci G, Kowalski AS, Buchmann N, Epron D, Pilegaard K, Kutsch W, Longdoz B, Gruenwald T, Montagnani L, Dore S, Rebmann C, Moors EJ, Grelle A, Rannik U, Morgenstern K, Oltchev S, Clement R, Gudmundsson J, Minerbi S, Berbigier P, Ibrom A, Moncrieff J, Aubinet M, Bernhofer C, Jensen NO, Vesala T, Granier A, Schulze ED, Lindroth A, Dolman AJ, Jarvis PG, Ceulemans R, Valentini R. 2001. Productivity overshadows temperature in determining soil and ecosystem respiration across European forests. Global Change Biology 7:269–278.
Jassal RS, Black TA. 2006. Estimating heterotrophic and autotrophic soil respiration using small-area trenched plot technique: theory and practice. Agricultural and Forest Meteorology 140:193–202.
Jenkinson DS, Rayner DH. 1977. The turnover of soil organic matter in some of the Rothamsted classical experiments. Soil Science 123:298–305.
Jobbagy EG, Jackson RB. 2000. The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecological Applications 10(2):423–436.
Johnson EA, Greene DF (1991) A method for studying dead bole dynamics in Pinus contorta var. latifola- Picea engelmannii forests. Journal of Vegetation Science 2:523–530.
Joslin JD, Gaudinski JB, Torn MS, Riley WJ, Hanson PJ. 2006. Fine-root turnover patterns and their relationship to root diameter and soil depth in a 14C-labeled hardwood forest. New Phytologist 172:523–535 doi:10.1111/j.1469-8137.2006.01847.x
Kaštovská E, Šantr
 kova H. 2007. Fate and dynamics of recently fixed C in pasture plant-soil system under field conditions. Plant and Soil 300(1–2): 61–69. doi 10.1007/s11104-007-9388-0
kova H. 2007. Fate and dynamics of recently fixed C in pasture plant-soil system under field conditions. Plant and Soil 300(1–2): 61–69. doi 10.1007/s11104-007-9388-0
Kendall MG, Ord JK. 1990. Time Series. Oxford University Press, New York.
Knohl A, Werner RA, Brand WA, Buchmann N. 2005. Short-term variations in δ13C of ecosystem respiration reveals link between assimilation and respiration in a deciduous forest. Oecologia 140:70–82. doi: 10.1007/s00442-004-1702-4
Knorr W, Kattge J. 2005. Inversion of terrestrial ecosystem model parameter values against eddy covariance measurements by Monte Carlo sampling. Global Change Biology 11:1333–1351 doi:10.1111/j.1365-2486.2005.00977.x
Laiho R, Prescott CE. 1999. The contribution of coarse woody debris to carbon, nitrogen, and phosphorus cycles in three Rocky Mountain coniferous forests. Canadian Journal of Forest Research 29: 1592–1603 doi:10.1139/X03-241
Laiho R, Prescott CE. 2004. Decay and nutrient dynamics of coarse woody debris in northern coniferous forests: a synthesis. Canadian Journal of Forest Research 34:763–777 doi:10.1139/X03-241
Lavigne MB, Ryan MG, Anderson DE, Baldocchi DD, Crill PM, Fitzjarrald DR, Goulden ML, Gower ST, Massheder JM, McCaughey JH, Striegl MRRG. 1997. Comparing nocturnal eddy covariance measurements to estimates of ecosystem respiration made by scaling chamber measurements at six coniferous boreal sites. J Geophys Res 102(D24):28977–28985
Law BE, Ryan MG, Anthoni PM. 1999. Seasonal and annual respiration of a ponderosa pine ecosystem. Global Change Biology 5:169–182.
Lipson DA, Schmidt SK. 2004. Seasonal changes in an alpine soil bacterial community in the Colorado Rocky Mountains. Applied and environmental microbiology 70(5):2867–2879 doi:10.1128/AEM.70.5.2867-2879.2004
Lipson DA, Schmidt SK, Monson RK. 2000. Carbon availability and temperature control the post-snowmelt decline in alpine soil microbial biomass. Soil Biology and Biochemistry 32:441–448.
Lipson DA, Monson RK, Schmidt SK, Weintraub M. In review. The trade-off between growth rate and yield in microbial communities and its consequences for soil respiration
Massman WJ, Sommerfeld RA, Mosier AR, Zeller KF, Hehn TJ, Rochelle SG. 1997. A model investigation of turbulence-driven pressure-pumping effects of the rate of diffusion of CO2, N2O, and CH4 through layered snowpacks. J Geophys Res 102(D15):18,851–18,863.
McDowell NG, Balster NJ, Marshall JD (2001) Belowground carbon allocation of Rocky Mountain Douglas-fir. Canadian Journal of Forest Research 31:1425–1436.
Metropolis N, Rosenbluth AW, Rosenbluth MN (1953) Equations of state calculations by fast computing machines. Journal of Chemical Physics 21:1087–1092.
Monson RK, Turnipseed AA, Sparks JP, Harley PC, Scott Denton LE, Sparks K, Huxman TE (2002) Carbon sequestration in a high-elevation, subalpine forest. Global Change Biology 8:459–478.
Monson RK, Burns SP, Williams MW, Delany AC, Weintraub M, Lipson DA. 2006a. The contribution of beneath-snow soil respiration to total ecosystem respiration in a high-elevation subalpine forest. Global Biogeochemical Cycles 20:GB3030 doi:10.1029/2005GB002684
Monson RK, Lipson DL, Burns SP, Turnipseed AA, Delany AC, Williams MW, Schmidt SK (2006b) Winter forest soil respiration controlled by climate and microbial community composition. Nature 439:711–714 doi:10.1038/nature04555
Raich JW, Nadelhoffer KJ (1989) Belowground carbon allocation in forest ecosystems: global trends. Ecology 70(5):1346–1354.
Raich JW, Potter CS, Bhagawati D. 2002. Interannual variability in global soil respiration, 1980–94. Global Change Biology 8:800–812.
Rangel-Castro JI, Prosser JI, Ostle N, Scrimgeour CM, K. Killham, Meharg AA. 2005. Flux and turnover of fixed carbon in soil microbial biomass of limed and unlimed plots of an upland grassland ecosystem. Enivornmental Microbiology 7:544–552 doi:10.1111/j.1462-2920.2005.00722.x
Raupach MR, Rayner PJ, Barrett DJ, DeFries RS, Heimann M, Ojima DS, Quegan S, Schmullius CC. 2005. Model-data synthesis in terrestrial carbon observation: methods, data requirements and data uncertainty specifications. Global Change Biology 11:378–397 doi:10.1111/j.1365-2486.2005.00917.x
Ryan MG, Law BE (2005), Interpreting, measuring, and modeling soil respiration. Biogeochemistry 73:3–27 doi:10.1007/s10533-004-5167-7
Sacks WJ, Schimel DS, Monson RK, Braswell BH. 2006. Model-data synthesis of diurnal and seasonal CO2 fluxes at Niwot Ridge, Colorado. Global Change Biology 12:240–259 doi:10.1111/j.1365-2486.2005.01059.x
Sacks WJ, Schimel DS, Monson RK. 2007. Coupling between carbon cycling and climate in a high-elevation, subalpine forest: a model-data fusion analysis. Oecologia 151:54–68 doi:10.1007/s00442-006-0565-2
Schaeffer SS, Anderson DE, Burns SP, Monson RK, Sun J, and Bowling DR. 2007. Canopy structure and atmospheric flows in relation to the δ13C of respired CO2 in a subalpine coniferous forest, Agric Forest Meteorol (in press). doi:10.1016/j.agrformet.2007.11.003
Schimel DS, Brawell BH, Holland EA, McKeown R, Ojima DS, Painter TH, Parton W, Townsend AR. 1994. Climatic, edaphic, and biotic controls over storage and turnover of carbon in soils. Global Biogeochemical Cycles 8(3):279–293.
Schimel JP, Gulledge J. 1998. Microbial community structure and global trace gases. Global Change Biology 4:745–748.
Schwartz G. 1978. Estimating the dimensions of a model. Annals of Statistics 6(2):461–464.
Scott-Denton LE, Sparks KL, Monson RK. 2003. Spatial and temporal controls of soil respiration rate in a high-elevation, subalpine forest. Soil Biology & Biochemistry 35:525–534.
Scott-Denton LE, Rosenstiel TN, Monson RK. 2006. Differential controls by climate and substrate over the heterotrophic and rhizospheric components of soil respiration. Global Change Biology 12:205–216 doi:10.1111/j.1365-2486.2005.01064.x
Sellers PJ, Randall DA, Collatz GJ, Berry JA, Field CB, Dazlich DA, Zhang C, Collelo GD, Bounoua L. 1996. A revised land surface parameterization (SiB2) for atmospheric GCMs, 1: Model formulation. J Climate 9:676–705.
Smith P, Andrén O, Brussaard L, Dangerfield M, Ekschmitt K, Lavelle P, Tate K. 1998. Soil biota and global change at the ecosystem level: describing soil biota in mathematical models. Global Change Biology 4:773–784.
Subke J-A, Inglima I, Cotrufo F. 2006. Trends and methodological impacts in soil CO2 efflux partitioning: a metaanalytical review. Global Change Biology 12:921–943 doi:10.1111/j.1365-2486.2006.01117.x
Tang J, Baldocchi DD, Xu L. 2005a. Tree photosynthesis modulates soil respiration on a diurnal time scale. Global Change Biology 11:1298–1304 doi:10.1111/j.1365-2486.2005.00978.x
Tang J, Misson L, Gershenson A, Cheng W, Goldstein AH. 2005b. Continuous measurements of soil respiration with and without roots in a ponderosa pine plantation in the Sierra Nevada Mountains. Agricultural and Forest Meteorology 132:212–227.
Tjoelker MG, Oleksyn J, Reich PB. 2001. Modelling respiration of vegetation: evidence for a general temperature-dependent Q10. Global Change Biology 7:223–230.
Trumbore S. 2000. Age of soil organic matter and soil respiration: radiocarbon constraints on belowground C dynamics. Ecological Applications 10(2):399–411.
Trumbore S. 2006. Carbon respired by terrestrial ecosystems - recent progress and challenges. Global Change Biology 12:141–153 doi:10.1111/j.1365-2486.2005.01067.x
Turnipseed AA, Anderson DE, Blanken PD, Baugha WM, Monson RK. 2003. Airflows and turbulent flux measurements in mountainous terrain Part 1. Canopy and local effects. Agricultural and Forest Meteorology 119:1–21.
Turnipseed AA, Anderson DE, Burns SP, Blanken PD, Monson RK. 2004. Airflows and turbulent flux measurements in mountainous terrain Part 2. Mesoscale effects. Agricultural and Forest Meteorology 125:187–205.
Wang C, Yang J. 2007. Rhizospheric and heterotrophic components of soil respiration in six Chinese temperate forests. Global Change Biology 13:123–131 doi:10.1111/j.1365-2486.2006.01291.x
Williams M, Schwarz PA, Law BE, Irvine J, Kurpius MR. 2005. An improved analysis of forest carbon dynamics using data assimilation. Global Change Biology 11:89–105 doi:10.1111/j.1365-2486.2004.00891.x
Wythers KR, Reich PB, Tjoelker MG, Bolstad PB. 2005. Foliar respiration acclimation to temperature and temperature variable q10 alter ecosystem carbon balance. Global Change Biology 11: 435–449 doi:10.1111/j.1365-2486.2005.00922.x
Xu T, White L, Hui D, Luo Y. 2006. Probabilistic inversion of a terrestrial ecosystem model: Analysis of uncertainty in parameter estimation and model prediction. Global Biogeochemical Cycles 20:GB2007 doi:10.1029/2005GB002468
Yi C, Monson RK, Zhai Z, Anderson DE, Lamb B, Allwine G, Turnipseed AA, Burns SP. 2005. Modeling and measuring the nocturnal drainage flow in a high-elevation, subalpine forest with complex terrain. J Geophys Res 110:D22303 doi:10.1029/2005JD006282
Acknowledgments
JMZ would like to thank Frederick Adler and Aaron McDonald for helpful discussions. The authors thank Steve Aulenbach for computer support. JMZ was funded as a fellow in the U.S. Department of Energy, Global Change Education Program, administered by the Oak Ridge Institute for Science and Education. WJS was supported by a National Science Foundation Graduate Research Fellowship. Additional funding was provided by a grant to DRB from the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-04ER63904, as part of the North American Carbon Program. Partial support for these studies was provided by funds from the Western Office of the National Institute for Climate Change Research (NICCR) under and agreement with the U.S. Department of Energy (BER Program). The authors would like to thank three anonymous reviewers for improving previous versions of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zobitz, J.M., Moore, D.J.P., Sacks, W.J. et al. Integration of Process-based Soil Respiration Models with Whole-Ecosystem CO2 Measurements. Ecosystems 11, 250–269 (2008). https://doi.org/10.1007/s10021-007-9120-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10021-007-9120-1









 kova H. 2007. Fate and dynamics of recently fixed C in pasture plant-soil system under field conditions. Plant and Soil 300(1–2): 61–69. doi
kova H. 2007. Fate and dynamics of recently fixed C in pasture plant-soil system under field conditions. Plant and Soil 300(1–2): 61–69. doi