Abstract
Glioblastoma (GBM), a malignant form of brain tumor, has a high mortality rate. GRP78, one of the HSP70 protein family members, is overexpressed in GBM. GRP78 is the key chaperone protein involved in the unfolded protein response. Upregulated GRP78 expression in cancer cells inhibits apoptosis and promotes chemoresistance. GRP78 has an ATPase domain, a substrate-binding domain, and a linker region. ATP-competitive inhibitors such as EGCG and OSU-03012 inhibit GRP78 activity and reduce its expression in GBM. However, there is a lack of structural data on the binding modes of these inhibitors to GRP78 ATPase domain. Further, the mode of selectivity of these inhibitors toward GRP78 also is unknown. Toward this end, molecular docking was performed with AutoDock Vina and confirmation obtained by docking using ROSIE. The stability and MM-PBSA binding energy of GRP78-inhibitor complexes as well as energetic contribution of individual residues was analyzed by 50 ns molecular dynamics run with GROMACS. MSA by ClustalW2 identified unique amino acid residues in the ATPase domain of GRP78 which were different from the residues present in other HSP70 proteins. Important and unique amino acid residues of GRP78 such as Ile61, Glu293, Arg297, and Arg367 played a major role in the intermolecular interactions with these inhibitors. The interactions with unique residues of GRP78 as compared with those of HSP70-1A provided the basis for selectivity. It was found that the binding affinity and specificity/selectivity of EGCG toward GRP78 was higher than that toward HSP70-1A, and selectivity was even better than OSU-03012. OSU-03012 was predicted to bind to GRP78. Analyses from MD runs showed tight binding and stability of complexes, and the highest number of hydrogen bonds during the trajectory runs were comparable to those found in the docking studies. Energetic contribution of individual inhibitor-interacting residues showed that energy values of Ile61 and Glu293 were among the most negative. These studies are, to the best of our knowledge, the first studies characterizing EGCG and OSU-03012 interactions with GRP78 on a structural basis and provide a significant insight into their binding modes, selectivity, and structural stability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is the tumor of glial cells in the brain and spinal cord. Thirty percent of the central nervous system tumors are gliomas [1]. Depending on the type of the cell they originate from, gliomas can be of several types: ependymomas which arise from the ependymal cells, astrocytomas originating from the astrocytes (glioblastomas), oligodendrogliomas arising from the oligodendrocytes, brain stem glioma arising in the brain stem region, and mixed gliomas.
About 80 % of gliomas are in malignant form widely known as the glioblastomas (GBMs). Central Brain Tumor Registry of the United States (CBTRUS, 2012) estimates that less than 5 % of the patients survive 5 years post diagnosis and declares it to be the deadliest of all cancers. The exact cause and factors involved in the development of the GBMs are not clearly known, although a few factors have been delineated [2]. No potent and effective cure or treatment for GBMs is known at present. This, coupled with the fact that the blood–brain barrier poses a major hindrance in targeted drug delivery to the brain, makes GBMs less amenable to effective treatment [3].
Glioblastomas generally do not metastasize through blood but spread through the cerebrospinal fluid to the spinal cord. The symptoms usually include nausea, vomiting, seizures, headaches, cranial nerve disorders, weakening of limbs, visual loss, organ failure, and ultimately death.
Studies into the molecular mechanisms of GBMs have established the role of several mutations in tumor suppressor and regulatory genes. Some of these genes and gene products such as ERCC1, ERCC2, XRCC1, isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2), MGMT, p53, GRP78, and predicted CSNK1A1 and Gli2 [4, 5] among others, play a prominent role in GBM development and progression.
Glucose regulated protein 78 (GRP78), a stress-response protein, has been shown to be overexpressed in the endoplasmic reticulum (ER) of GBM cells [6]. It is a stress-induced protein involved in unfolded protein response (UPR) process which corrects protein folding. GRP78, when overexpressed, induces anti-apoptosis and chemo-resistance in the cancerous cells and promotes their survival [7].
Unfolded protein response (UPR) and GRP78 in GBM development
Every cell in our body has the natural ability to adapt to changing environmental conditions and be restored back to its original state. Unfolded protein response (UPR) evolved as a process in minimizing the errors in protein folding integrity. The UPR consists of environmental-stress induced pathways in the endoplasmic reticulum which get triggered by the accumulation of misfolded or unfolded proteins. This misfolding of proteins occurs primarily due to glucose starvation or oxidative stress (hypoxia), thereby, disrupting normal cellular functions.
UPR serves three major purposes: first, it halts normal protein translation in the stressed cell to help it return to its normal functioning; second, it is involved in increase in translation of molecular chaperones and stress-related gene products during stress; and further, if the UPR process fails to correct the misfolded proteins, it degrades them. If the stress continues to persistand normal cellular functions cannot be restored, UPR induces apoptosis in the respective cellafter a certain time period.
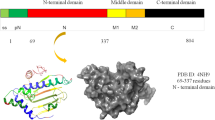
GRP78 is also known as binding immunoglobulin protein (BIP) or heat shock 70 kDa protein 5 (HSPA5). GRP78 is localized in the lumen of endoplasmic reticulum. In humans, it is encoded by HSPA5 gene found on chromosome 9 and has a length of 654 amino acid residues. Belonging to the HSP70 family of molecular chaperone proteins, and activated through UPR process, GRP78 is involved in proper protein folding and assembly, correcting misfolded proteins and targeting them for degradation, ER Ca+2 binding, activating transmembrane ER stress sensors, preventing apoptosis of the affected cell, and is also involved in resolving inflammation by feeding anti-inflammatory signals. When overexpressed, it enables the cancerous cells to survive by inducing anti-apoptotic and chemoresistant properties, leading to cellular proliferation, survival and angiogenesis [8]. The importance of GRP78 stems from the fact that it is thought to be a potential therapeutic target universal to bacterial and viral infections such as Ebola, influenza and hepatitis alongwith brain cancers and GBMs [9].
GRP78 protein structure is composed of three domains out of which two are functional. It has a 44 kDa N-terminal nucleotide binding (ATPase) domain, a 20 kDa polypeptide (substrate) binding domain, and a variable 10 kDa C-terminal tail of unknown function and a linker for connecting the two functional domains. The binding of ATP to the ATPase domain of GRP78 facilitates the binding of the exposed hydrophobic residues of the misfolded or unfolded protein to the substrate binding domain of GRP78. Thereafter, protein disulfide isomerase (PDI) carries on the rearrangement of the protein folds to give the protein its proper conformation [10].
GRP78 & glioblastoma (GBM)
Research over the years has shown that in GBM cells, GRP78 is aberrantly activated or overexpressed. Knockdown experiments of GRP78 in human glioma cell lines have shown decreased GBM cell proliferation [6]. Further, overexpression of GRP78 leads to its binding and inactivation of the UPR genes involved in apoptosis. Continual suppression of the UPR genes by GRP78 induces the cell to become anti-apoptotic and chemo-resistant, decreasing caspase-7 activation and rendering the cells resistant to etoposide- and cisplatin-induced apoptosis [11]. GRP78 is upregulated in hypoxia and glucose starvation, conditions which are present in every tumor environment [12]. So, in order to block the proliferation through apoptosis-induction in GBM cells, GRP78 overexpression needs to be controlled and this could have essential therapeutic benefits. Studies are being conducted to generate suitable inhibitors to overcome the aberrant activation of GRP78 in GBM cells as mentioned below.
Inhibitors against GRP78
Interest in the discovery of therapeutic interventions against GRP78 has led to the design and development of some novel inhibitors [13, 14]. Inhibitors against both the ATPase domain and substrate-binding domain of GRP78 have been explored. GRP78 isoform selectivity among HSP70 protein family is expected to play a crucial role in design and development of these inhibitors.
Among the several inhibitors, (−)-epigallocatechin gallate (EGCG), through its binding to ATPase domain, and OSU-03012, both ATP-competitive inhibitors (Fig. 1), have been shown to inhibit GRP78 activity [9, 15, 16].
EGCG is a flavonoid present in green tea. Recent studies have concluded that EGCG has binding affinity to the nucleotide binding (ATPase) domain of GRP78 and once bound, EGCG inhibits GRP78 association with ATP by competitive inhibition [15, 16]. Thus, it inhibits the ATPase activity of GRP78. Upon EGCG binding to GRP78, the active monomer form of GRP78 gets converted into the inactive dimer and oligomer forms. Further, EGCG binding prevents the formation of the anti-apoptotic GRP78-caspase-7 complex in the endoplasmic reticulum, and thus, helps in induction of apoptosis in the cancer cells.
Sepharose 4B affinity chromatography experiments done with a nested set of overlapping N-terminally (ATPase domain) deleted GRP78 protein showed direct binding of EGCG to the ATPase domain of the GRP78 protein [15]. EGCG was found to induce apoptosis in human glioblastoma cell lines, U-373 MG and U-87 MG [17].
OSU-03012, an ATP-competitive inhibitor, is a derivative of Cox-2 inhibitor celecoxib, and does not inhibit Cox-2. It has anti-cancerous and anti-microbial properties. OSU-03012 treatment in mice decreased the expression of GRP78 and enhanced PERK activity [9]. Knockdown of GRP78 in all cell lines had significantly enhanced OSU-03012 lethality. OSU-03012 did not alter the mRNA stability of GRP78 rather it decreased the protein stability, and treatment with it led to primary GBM cell death in vitro and in vivo. OSU-03012 was shown to inhibit p21-activated kinase 1 (PAK1) activity and compete with ATP binding. In addition, computer modeling predicted a docking site for OSU-03012 in the ATP binding motif of PAK1 [18]. Since it is an ATP-competitive inhibitor, we predict that it might be binding to GRP78 ATPase domain as well. Further, another study [19] has also mentioned that OSU-03012 treatment reduced GRP78 expression in GBM cells and that there is likelihood that OSU-03012 destabilizes GRP78 by binding to it. Like EGCG [20], OSU-03012 readily crosses the blood–brain barrier [19].
There is currently a lack of detailed structural data on the binding of EGCG to GRP78. Comparison studies with EGCG binding and reproduction of similar binding modes and interactions could further help us in our predictions of OSU-03012 binding. Experimental binding data in terms of IC50, Kd or Ki values is also not known at present for both these inhibitors binding to GRP78. Further, exploring the structural determinants of inhibitor selectivity will play an important role in furthering our understanding of selectivity between GRP78 and its HSP70 family members. In the present study, we have attempted to analyze in detail the structural basis, selectivity, and interactions of these inhibitors in the inhibition of GRP78 through molecular docking and molecular dynamics simulations.
Materials and methods
Retrieval of sequences and structures
Full length protein and the ATPase domain sequences of the human GRP78 and other human HSP70 proteins were downloaded from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (http://www.rcsb.org/pdb/home/home.do) in FASTA format. Three dimensional structures of the same were also obtained from PDB. We used the ATP-bound form of GRP78 for our studies as our work involves ATP-competitive inhibitors. For this, we used the structure with PDB ID: 3LDL as there are no multiple crystal structures of ATP-bound forms [13]. 3LDL denotes structure of the ATPase domain of human GRP78, bound to ATP. The PDB IDs for the other human HSP70 proteins, the sequences of which were used for multiple sequence alignment, were: 2E8A, 3LOF, 3GDQ, 3I33, 3FE1, 3FZF, and 4KBO for HSP70-1A, HSP70-1B, HSP70-1L, HSP70-2, HSP70-6, HSP70-8, and HSP70-9, respectively. The three dimensional structure of HSP70-1A with PDB ID 2E8A (resolution 1.7 Å) was used for molecular docking studies. These structures were selected for their resolution, Homo sapiens species origin and for being bound to specific ligands/inhibitors. All the downloaded structures were in .pdb format. Heteroatoms (HETATM) and the water molecules in the PDB files used in docking studies were removed manually and only one chain of a particular PDB structure was used for these proteins.
The three dimensional structures of the two inhibitors, EGCG and OSU-03012, were downloaded from NCBI PubChem database. The downloaded structures were in .sdf format. The .sdf files were converted to .pdb format using OpenBabel (http://openbabel.org/wiki/Main_Page) as AutoDock Vina docking tool requires .pdb format of files for performing docking calculations.
Multiple sequence alignment
Using the FASTA-formatted sequences of full length proteins as well as of their ATPase domains, multiple sequence alignment of GRP78 and other HSP70 proteins, was carried out with the help of ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default parameters and the corresponding results were saved. This alignment was performed to find out the percentage of conserved residues as well as residues unique to GRP78.
Molecular docking with AutoDock Vina
As the studies conducted include ATP-competitive inhibitors binding to ATPase domain, we have used ATPase domains of GRP78 and HSP70-1A proteins. HSP70-1A was chosen as a comparison case, as it is widespread in nature and also exhibits a high level of homology with GRP78. Energy minimizations of the ATPase domains of GRP78 and HSP70-1A were done using Swiss PDB Viewer (DeepView) to lower the molecules’ energy and strain, without causing any significant distortion to the binding pockets.
Molecular docking studies with energy minimized protein models and ATP, ANP, EGCG, and OSU-03012 were done using AutoDock Vina, implemented within the PyRx 0.8 virtual screening tool, using the default parameters. AutoDock Vina is a molecular docking tool, believed to be faster and more accurate than AutoDock, and is used to predict drugs or substrates/ligands binding to proteins. The free-energy scoring function in AutoDock Vina is different from AutoDock 4.2 and combines information from both knowledge-based potentials and empirical scoring functions. Further, it uses Broyden-Fletcher-Goldfarb-Shanno (BFGS) algorithm for local optimization and iterated local search optimizer for global optimization field [21]. ATPase domain of GRP78 (as AutoDock protein) and ATP, ANP, EGCG, and OSU-03012 (as AutoDock ligands) were fed as inputs and the program was run with the default settings. Benchmarking was done through analysis of docked structures of ATPase domain of GRP78 with ATP and ANP (from 3LDL and 3LDO). Docking parameters were selected by comparing these docked structures to the original crystallographic structures obtained from PDB. Using the same default docking parameters, the inhibitors, EGCG and OSU-03012, were docked within the ATPase domain of GRP78.
The result obtained with the lowest binding energy was saved in each case and viewed in Discovery Studio. Another important criterion for selection of the best docked structure was higher number of hydrogen bonds. As there are virtually nil crystallographic structures of EGCG or OSU-03012 bound to any protein with a high homology to GRP78 or other HSP70 proteins, with which docked orientations of our inhibitor molecules could be compared, we relied on the above-mentioned two criteria to select our docked structures.
This whole process was repeated in the case of the ATPase domain of HSP70-1A with ANP, EGCG, and OSU-03012.
Molecular docking with ROSIE
To confirm the docking analysis performed with AutoDock Vina, the docking results were cross-checked using another online docking tool called the Rosetta Online Server That Includes Everyone (ROSIE) (http://rosie.rosettacommons.org/) [22]. ROSIE was selected for cross-checking because it employs a Monte Carlo minimization procedure different from AutoDock Vina. In this minimization procedure, there is simultaneous optimization of the rigid body position as well as orientation of the ligand/small molecule and the protein side-chain conformations. The energy function incorporates van der Waals and electrostatic interaction models, along with a model of implicit solvation and an explicit orientation hydrogen bonding potential. Here, the energy minimized structures of the protein (in .pdb format) and the ligand (in .sdf format) were uploaded and the geometric center (average of X, Y, Z co-ordinates of the ligand atoms) from the original ATP-bound structure of GRP78 was fed into the server and the docking program was run. Benchmarking was also performed prior to using inhibitors for ROSIE run with the same complex structures used in AutoDock Vina.
Molecular dynamics (MD) simulations
The whole simulation process as well as trajectory analyses were done using GROMACS 4.6.3 MD simulation package [23] installed on a Linux cluster with GROMOS96 43a1 force field parameters and SPC water model. The crystal structure of GRP78 protein and docked structures of GRP78 with EGCG and OSU-03012 were fed as starting points. The same simulation procedures were followed for protein and protein-inhibitor complex. Solvation was done in a cubic box using a solute-box distance of 1.0 nm. Ligand topology file was generated using PRODRG2 server and was checked for charges. The total charge of the system was neutralized with 3Na+ counterions. Energy minimization was performed with 1000 steps of steepest descent energy minimization. After applying position restraints on protein and inhibitors, NVT equilibration was done at 300 K and 100 ps of run followed by NPT equilibration of 100 ps with Parrinello-Rahman barostat at reference pressure of 1 bar. After equilibration, production MD run was performed after releasing the solute position restraints for sufficiently long 50 ns run. A constant temperature of 300 K and constant pressure of 1 bar with integration time step of 2 fs was used. All bond lengths were constrained using LINCS algorithm. Particle-mesh Ewald algorithm was used for long range electrostatic interactions, and short-range electrostatics and short-range van der Waals cutoffs were set at 1.4 nm.
Binding energy calculation using MM-PBSA method and energetic contribution of individual residues
Binding energy with MM-PBSA method was calculated using g_mmpbsa tool [24] with default settings. The last 5 ns of production run in the simulation was used and snapshots were extracted every 10 ps and energetic terms calculated. Results are in terms of average and standard deviations for all energetic components. This tool does not calculate entropic terms. It was also used to calculate the energy contribution of individual residues to the overall binding energy with bootstrapping (number of bootstraps = 2000), and results assessed in terms of average energy with standard error.
Visualization and general analysis
The docked structures were visualized with the help of Accelrys’ Discovery Studio 4.0 (Dassault Systemes). Discovery Studio Visualizer is a widely used bioinformatics tool used for molecular visualization and analysis of the three dimensional structures of molecules. Graphic visualization of the three dimensional structures of the docked complexes was performed. Comparison of the conformations and orientations of the ligands in the docked complexes were done. Intermolecular hydrogen bonding patterns and non-bonded contacts were observed in Discovery Studio 4.0 using intermolecular hydrogen bonds and non-bond interactions tabs from Structure menu. Analyses of MD simulation trajectories were done using GROMACS analysis tools, XMGRACE plotting tool, and VMD 1.9.2 [25].
Results and discussion
Multiple sequence alignment
Multiple sequence alignments (MSA) of full length sequences and ATPase domains of GRP78 and other HSP70 proteins (HSP70-1 L, HSP70-1A, HSP70-1B, HSP70-2, HSP70-6, HSP70-8, HSP70-9) were carried out with ClustalW2 tool, to ascertain the conservation of residues and to find the residues unique to GRP78. The unique residues can play a role in specificity and selectivity of inhibitors toward GRP78.
The alignment between full-length sequences indicated that percent sequence identity between sequence of GRP78 with other HSP70 proteins was 64 %.
MSA analysis of the ATPase domains of GRP78 and HSP70-1A protein is shown in Fig. 2. Between GRP78 and other HSP70 proteins, the ligand-binding site residue unique to GRP78 was found to be Ile61 within ∼4.5 Å radius of the ligand (Macias et al. [13] and our study) which was substituted by Thr residue in other HSP70 proteins, with Thr37 in HSP70-1A protein. Further, mention is made of amino acid residues Glu293, Arg297, and Arg367 in GRP78, which are highly conserved in our MSA results. Structurally, however, these are slightly different in position within 4 Å of ligand in GRP78 active site, according to the above mentioned paper, as compared to their counterparts, Glu268, Arg272, and Arg342 in HSP70-1A. These residues, because of difference in position in 3D space, may either interact or not at all, thereby resulting in inhibitor specificity or selectivity.
ATPase domains of GRP78 and HSP70-1A: structural aspects
GRP78 and other HSP70 family members are structurally similar due to the presence of amino acids conserved throughout evolution. GRP78 is thought to be the least conserved member of the HSP70 family. Both GRP78 and other HSP70 proteins possess a nucleotide binding (ATPase) domain and a substrate (protein) binding domain connected via a linker peptide. ATP hydrolysis in both is connected to protein binding and its release from the substrate binding domain. The ATP-bound state has low substrate affinity while the ADP-bound state has high substrate affinity in both cases.
The ATPase domain structures of these isoforms showed that they possess the same secondary structural fold and a highly conserved binding site. The major difference between GRP78 and other HSP70 protein family members was in the residues in the region surrounding the active site. The ATP binding regions of both GRP78 and other HSP70 proteins are deep inside the cavity which makes access to the binding sites difficult. The structural differences in the ATP binding sites of both the proteins are shown in Fig. 3a, b. The ATP binding site of GRP78 appears to be a little broad and open while that of HSP70-1A is narrow and extended. The slight opening in GRP78 observed is due to the rotation of subdomain IIB [26]. The helices and the beta sheets of both proteins are different in orientation at some places, more specifically at the upper right portion with the longer alpha-helix at the opening end of the active site in the figures.
The active site residues of GRP78 bound to ATP comprise: Gly36, Thr37, Thr38, Tyr39, Gly226, Gly227, Gly228, Thr229, Gly255, Glu256, Glu293, Lys296, Arg297, Ser300, Gly363, Gly364, Ser365, Arg367, Ile368, and Asp391. Most of these amino acid residues are hydrogen bonded to the ATP molecule.
The active site residues of HSP70-1A bound to ANP are: Gly12, Thr13, Thr14, Tyr15, Asn57, Ala60, Leu61, Gly201, Gly202, Gly230, Arg258, Glu268, Lys271, Arg272, Ser275, Gly338, Gly339, Ser340, Arg342, Ile343, and Asp366.
The amino acid residues that are different between both the proteins are thought to be of primary importance in imparting differences in binding modes and interactions, thereby, resulting in inhibitor selectivity. From the Macias et al. [13] study and from Fig. 3a and b, one can deduce that these residues are: a) Ile61 of GRP78 located within ∼4.5 Å radius of the ligand in ligand bound structures, not present in HSP70-1A which has Thr37 in the same position of our MSA results. b) Glu293, Arg297, and Arg367 are slightly different in position in GRP78, residing in subdomain IIB [26], as compared to Glu268, Arg272 and Arg342 in HSP70-1A.
Molecular docking
The molecular docking of GRP78 and HSP70-1A with the two inhibitors was done to explore the structural determinants of inhibitor binding and selectivity. The two inhibitors used in this study were EGCG and OSU-03012 (Fig. 1) as no crystal structure data of GRP78 bound to these potent and selective inhibitors is available, their structural details remain to be explored.
The IUPAC name of EGCG is (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-1-benzopyran-3-yl 3,4,5-trihydroxybenzoate with a molecular weight of 458.37 g mol−1. The structure consists of three benzene rings and a cyclohexane ring with an oxygen atom and has many polar hydroxyl group substituents.
OSU-03012 has an IUPAC name of 2-amino-N-[4-[5-phenanthren-2-yl-3-(trifluoromethyl)-pyrazol-1-yl]phenyl]acetamide and has a molecular weight of 460.45 g mol−1. The structure consists of three benzene rings and an imidazole ring and has three fluorides attached to it as polar substituents.
Initially, the docking of ATP and ANP molecules with the ATPase domains of GRP78 and HSP70-1A proteins, respectively, were done with default docking parameters. This provided a benchmark in reproducing the binding mode and interactions observed in the crystallographic structures obtained from PDB and to select the parameters for docking. The docked structures of ATP and ANP with both the proteins were compared with the original crystallographic structures. The same ligand orientations and the same amino acid interactions with the ligand were observed in the docked structures when compared to the crystal structures as seen after structural superimposition of the docked and crystal structures (Fig. 4). This lent further credence that the docking parameters chosen for our work with EGCG and OSU-03012 are reliable. Hence, the default docking parameters of AutoDock Vina were further utilized for docking EGCG and OSU-03012 to the ATPase domains of GRP78 and HSP70-1A.
Docking with EGCG
EGCG bound to GRP78 at the same location where ATP binds, with a binding energy of −8.4 kcal mol−1 for the lowest energy bound structure (Fig. 5), and hence, with comparatively higher affinity than with HSP70-1A (Table 1). Several active site residues formed hydrogen bonds with EGCG, namely, Asp224, Gly227, Gly255, Lys296, Gly364 (2 H-bonds), Ser365, Arg367, and Asp391. Other residues were involved in non-bonded contacts with the inhibitor. It was observed that number of hydrogen bonds were more in the case of GRP78-EGCG complex than with HSP70-1A-EGCG complex, which again shows higher affinity of EGCG to GRP78 and consequently, tighter binding (Table 1).
The lowest energy structure of EGCG obtained after docking to HSP70-1A, had a binding energy of −7.5 kcal mol−1 (Table 1). Five of the active site residues formed hydrogen bonds with EGCG, namely, Asp69, His227, Glu231, Asp232, and Arg261 (Fig. 6). Other residues are involved in non-bonded contacts with the inhibitor. EGCG binds in the same cavity where original ANP was bound to the protein.
EGCG interacted with the unique residue of GRP78, Ile61, and did not interact with its counterpart in the HSP70-1A. The cyclohexane ring of EGCG participates in a hydrophobic interaction with Ile61. Its counterpart, Thr37, in HSP70-1A, has a polar hydroxyl group and so, is unable to form hydrophobic interactions with these moieties. In addition, we also observed residues interacting with EGCG that are different in position between GRP78 and HSP70 due to a shift of the structural part comprising these residues. Glu293 is involved in electrostatic interactions with the benzene ring of EGCG. The carboxyl side chain of Glu293 interacts with the π-electron cloud of the benzene ring through π-anion electrostatic interactions. Its counterpart, Glu268, in HSP70-1A, did not interact at all. Arg297 and Arg367 of GRP78 made hydrophobic bonds with EGCG due to the presence of the guanidium side chain while their counterparts Arg272 and Arg342 in HSP70-1A did not interact. Further, Ermakova et al. [15] suggested that the gallate moiety of EGCG might be critical for interacting with GRP78. We have also found in our studies that the gallate moiety of EGCG participates in the pi-alkyl hydrophobic interactions with Arg297 and Arg367, thus our docking studies provide near-conclusive evidence to the previous studies of Ermakova et al. It is obvious that the positional shift of GRP78 as compared to HSP70-1A, as observed in this and other studies, plays a role in differential interactions with the inhibitor, further lending inhibitor specificity.
Docking with OSU-03012
The next inhibitor, OSU-03012, was also successfully docked into the active site of the two proteins, using the same default docking parameters and the structures with the lowest binding energy were selected. The binding energy value of OSU-03012 complex with GRP78 was −6 kcal mol−1 and with HSP70-1A was −8.4 kcal mol−1. This reflects the tighter binding of OSU-03012 with HSP70-1A. In GRP78, three residues are hydrogen bonded to the inhibitor, which are Glu256 (three H-bonds), Glu293, and Lys296. The other residues interact through non-bonded interactions (Fig. 7).
Among the active site residues of HSP70-1A, Arg36, Ser275, Gly339, and Arg342, are involved in hydrogen bonding with the inhibitor (Fig. 8). The other residues are involved in non-bonded interactions with the inhibitor. The number of hydrogen bonds in GRP78 complexed with OSU-03012 is more than that of HSP70-1A (Table 1).
OSU-03012 like EGCG, is also seen to be involved in interactions with the unique residue of GRP78, Ile61. Thr37 in HSP70-1A, the counterpart of Ile61 of GRP78, does not interact with the inhibitor here. Ile61 has hydrophobic interactions with one of the benzene rings of OSU-03012. OSU-03012 further interacts with Glu293 of GRP78 which is absent in the case of its counterpart Glu268 in HSP70-1A. Glu293 has an electrostatic bond and also a hydrogen bond with one of the benzene ring of OSU-03012. Arg297 in GRP78 makes a hydrophobic bond with OSU-03012 while its counterpart Arg272 in HSP70-1A makes both electrostatic and hydrophobic bonds. Arg367 in GRP78 does not interact while its counterpart, Arg342, in HSP70-1A makes electrostatic bonds with OSU-03012. These differences can also be explained on the basis of positional differences between residues as has been mentioned above.
Our docking results were confirmed through docking of EGCG and OSU-03012 with the ATPase domain of GRP78 using ROSIE online docking tool. The superimposed docked structures of both EGCG and OSU-03012 showed that there was around 85–90 % similarity between the docked poses generated by AutoDock Vina and ROSIE (Fig. 9a, b). The gallate moiety of EGCG appears to be a bit displaced in ROSIE-docked structures, however, what is conclusive is that this does not abolish its important interactions with Arg297 and Arg367, as explained above.
Molecular dynamics simulations
Fifty ns MD simulations were carried out for GRP78 protein alone, GRP78-EGCG and GRP78-OSU03012 complexes. RMSD of final structure relative to energy minimized crystal structure and docked structures was calculated for backbone atoms, in order to monitor structural changes, if any. It is seen from the plot generated (Fig. 10) that the systems attained a stable RMSD value of 0.2–0.25 nm after 30 ns simulation time for both the protein-inhibitor complexes. This means that there are no large conformational changes even when the inhibitor is bound and when the protein is flexible. Both the inhibitors remained bound within the active-site, and their all-atom RMSD values remained constant throughout the entire simulation at 0.1 nm (Fig. 10). RMSF plots vs. protein residue number were also generated in each case (Fig. 11) in order to analyze the mobility of residues before and after the inhibitor is bound. It is observed that compared to protein alone, residues in protein-inhibitor complexes had lower mobility. Most of the active site residues (mostly residues within 200–400 range) of protein-inhibitor complex, had RMSF values less than 0.25 nm in each case, compared to free protein. This shows that the binding of inhibitors reduced the flexibility of active site residues, thus making the complex more stable. The number of inter-molecular hydrogen bonds as a function of time, was calculated by g_hbond utility of Gromacs. In hydrogen bond profiles shown in Fig. 12, EGCG formed 8–9 (highest) hydrogen bonds (one configuration even made ten hydrogen bonds) as compared to OSU-03012 which formed 5–6 (highest) hydrogen bonds during the trajectory period. The highest numbers of hydrogen bonds in MD simulations are comparable to the total number of hydrogen bonds assessed during the docking studies (Tables 2 and 3).
RMSD vs. time plot of the backbone atoms of GRP78 alone (black), GRP78-EGCG complex (red), and GRP78-OSU03012 (green) relative to energy minimized crystal structure and docked structures for 50 ns simulation. RMSD plots of inhibitors (all atoms) relative to initial energy minimized structures are also shown, EGCG (blue) and OSU-03012 (yellow)
RMSF of the whole residues in GRP78 alone (black), GRP78-EGCG complex (red), and GRP78-OSU03012 (green). Residues in complex appear to fluctuate less than free protein. Specifically, the region comprising active site residues, region 200–400 (marked in gray boxes), shows less fluctuation of residues in the case of GRP78-inhibitor complexes as compared to GRP78 alone
Binding energy calculations were done using MM-PBSA method implemented in g_mmpbsa tool. Average values for the last 5 ns of simulations with snapshots generated every 10 ps are reported in Table 4. Results show that EGCG has less negative binding energy than OSU-03012. In EGCG-bound complex, van der Waals energetic term contributes the most toward binding energy while in OSU-03012-bound complex, electrostatic energy term predominates. SASA non-polar solvation energetic terms were more or less similar in both cases, and were favorable to binding while polar solvation energies were unfavorable. Analyses were further done to assess the energetic contribution of individual residues forming hydrogen bonds as well as other non-bonded contacts with the inhibitors. In EGCG-bound complex, Glu293, one of the unique residues and in electrostatic interactions with the inhibitor, was one of the highest contributors toward the binding energy with negative energy values, along with Tyr39 and Gly364. All of the other residues in hydrogen bonded contacts and hydrophobic interactions too contributed to the binding energy except Lys296, Arg297, Arg367, and Asp391. In OSU-03012-bound complex, Glu293 again is one of the most negative energy contributors as well as the residues Glu256 and Asp259. All of the other residues in hydrogen bonded and other non-bonded contacts had negative value of total energy, except Lys296 and Arg297.
EGCG and OSU-03012 binding to GRP78: some mechanistic insights
Differential scanning calorimetry experiments indicate that EGCG binds to unfolded form of GRP78 NBD [16]. These authors mention in their paper, “It is likely that, by analogy with HSP-70 (Liu et al., 2010), an intrinsic mobility of the GRP78 N-terminal domain unfolds it sufficiently to facilitate access of EGCG or HNK under prolonged incubation conditions of a few hours or more”. This is significant in view of the fact that the simulations for EGCG binding to GRP78 from initial time to over 30 ns register an increase of RMSD value from 0.3 to 0.35 nm. After 30 ns, the value of RMSD lowers to 0.2–0.25 nm, in contrast to the free protein which continues to exhibit high RMSD value throughout the simulation course. This may well show unfolding of GRP78 ATPase domain over time, and EGCG binding to an unfolded form. Lowering of RMSD after 30 ns may imply that this unfolded form is being converted into a folded form subsequent to the binding of EGCG. Our MD simulations further predict that OSU-03012 binding seems to convert GRP78 unfolded form immediately to a folded form with RMSD at a constant value of 0.2–0.25 nm throughout. This is possibly due to very strong electrostatic interactions calculated by MM-PBSA method as reported in Table 4. We speculate that the slightly unfolded form of GRP78 may be the native form needed for it to perform its functions. As EGCG binds to the unfolded form but later allows it to fold, and OSU-03012 binding quickly folds the protein leading to a possible dysfunctional state, this may be one of the mechanisms through which the protein may be inhibited by these inhibitors.
As observed from the molecular docking and MD results, both EGCG and OSU-03012 bind selectively and stably to GRP78. Docking studies indicate that EGCG has more negative binding energy in bound form with GRP78 than OSU-03012, but MM-PBSA studies show otherwise. The protein-inhibitor complexes are very stable in water, the mobility of active site residues decreases after the binding of inhibitors; and the systems reach equilibrium conformation after 30 ns as seen from RMSF and RMSD analyses of MD trajectories, respectively. On the basis of our results, we predict EGCG to be a better inhibitor for GRP78 than OSU-03012, due to greater number of hydrogen bonded interactions, more number of unique amino acids involved in the interactions thereby imparting selectivity, as well as its ability to lower the protein flexibility. Even though MM-PBSA predicts OSU-03012 with higher binding energy to be a higher affinity binder than EGCG, a high affinity with the intended target does not always translate into a good inhibitory response in vitro or in vivo, e.g., drug rolipram [27], while a compound with moderate affinity can be a better inhibitor, e.g., drug PU3 [28]. Moreover, we predict that OSU-03012 binds to GRP78 as observed from its binding mode similarity to EGCG binding mode and interactions with unique residues of GRP78 in a manner similar to EGCG. However, it does not appear to be very specific toward GRP78 as observed from its interactions with HSP70-1A residues which are the counterparts of unique residues of GRP78. MD simulations also predict that OSU-03012 binding to GRP78 does not cause large conformational changes as seen from the stable RMSD value after 30 ns throughout the simulation course and RMSF lower than that of free protein in active site regions. Since in GBMs, GRP78 is overexpressed, so, EGCG may predominantly bind to the GRP78 protein and may have a lesser degree of interaction with HSP70-1A. Specific interactions of EGCG and OSU-03012 with the unique residues of GRP78 provide further selectivity and structural basis toward inhibition of GRP78 protein.
Conclusions
GRP78 is overexpressed in GBM and needs to be inhibited or downregulated, keeping in view its conservation in the HSP70 protein family. Despite the discovery of several inhibitors binding to or interacting with GRP78, the structural basis of binding and selectivity has not been elucidated for some of these. The docking studies of the two inhibitors, EGCG and OSU-03012, with the ATPase domains of GRP78 and HSP70-1A as well as molecular dynamics simulations were performed to study their interactions and selectivity. From the results obtained, it can be predicted that EGCG and OSU-03012 have good binding affinity with the ATPase domain of GRP78 as can be observed by the number of hydrogen bonds and other non-bonded interactions and binding energy. The unique residues of GRP78 interacted with both EGCG and OSU-03012, lending a high degree of selectivity over HSP70-1A in the case of EGCG and a lesser degree in the case of OSU-03012. MD simulations predicted tight binding and stability of complexes, and the results complemented some of the results in docking studies. EGCG binds initially to unfolded form of GRP78 which later gets converted into a folded form. OSU-03012 binding appears to immediately fold the protein. EGCG might prove to be a better inhibitor of the ATPase activity of GRP78 due to its specificity and selectivity over HSP70-1A, and greater number of hydrogen bond interactions as well as lowering of the protein’s flexibility. Following the in silico analyses, in vitro and in vivo studies can be done to validate the binding modes of these inhibitors with GRP78 and consequent downregulation of its expression in GBM.
Abbreviations
- GBM:
-
Glioblastoma
- GRP78:
-
Glucose regulated protein 78
- UPR:
-
Unfolded protein response
- HSP70-1A:
-
Heat shock protein 70-1A
- ATP:
-
Adenosine triphosphate
- ANP:
-
Phosphoaminophosphonic acid-adenylate ester
- EGCG:
-
(−)-Epigallocatechin gallate
- PDB:
-
Protein data bank
- SDF:
-
Structure data format
- MSA:
-
Multiple sequence alignment
- MD:
-
Molecular dynamics
- ns:
-
Nanoseconds
- MM-PBSA:
-
Molecular mechanics/Poisson−Boltzmann surface area
References
Goodenberger ML, Jenkins RB et al. (2012) Genetics of adult glioma. Cancer Genet 205(12):613–621
Ostrum QT, Bauchet L, Davis FG et al. (2014) The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol 16(7):896–913
Schwartzbaum JA, Fisher JL, Aldape KD et al (2006) Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2:493–503
Wang P, Dang Q, Zhang C et al (2013) Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 32:3000–3091
Mishra S (2014) CSNK1A1 and Gli2 as novel targets identified through an integrative analysis of gene expression data, protein-protein interaction and pathways networks in glioblastoma tumors: can these two be antagonistic proteins? Cancer Informat 13:93–108
Pyrko P, Schönthall AH, Hofman FM et al (2007) The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res 67:9809–9816
Li J, Lee AS (2006) Induction of GRP78/BiP and its role in cancer. Curr Mol Med 6:45–54
Lee AS (2007) GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res 67(8):3496–3499
Booth L, Cazanave SC, Hamid HA et al (2012) OSU-03012 suppresses GRP78/BiP expression that causes PERK-dependent increases in tumor cell killing. Cancer Biol Ther 13(4):224–236
Mayer M, Kies U, Kammermeier R et al (2000) BiP and PDI cooperate in the oxidative folding of antibodies in vitro. J Biol Chem 275(38):29421–29425
Lee HK, Xiang C, Cazacu S et al (2008) GRP78 is overexpressed in glioblastomas and regulates glioma cell growth and apoptosis. Neurooncology 10:236–243
Huber-Keener KJ, Yang J-M (2011) Impact of metabolic and therapeutic stresses on glioma progression and therapy. In: Chen CC (ed) Advances in the biology, imaging and therapies for glioblastoma. InTech, Rijeka, pp 23–52
Macias AT, Williamson DS, Allen N et al (2011) Adenosine-derived inhibitors of 78 kDa glucose regulated protein (Grp78) ATPase: insights into isoform selectivity. J Med Chem 54(12):4034–4041
Lee JY, Jung KW, Kim Y (2008) Inhibitor design for human heat shock protein 70 ATPase domain by pharmacophore-based in silicoscreening. Bull Korean Chem Soc 29(9):1717–1722
Ermakova SP, Kang SB, Choi BY et al (2006) (−)-Epigallocatechingallate overcomes resistance to etoposide-induced cell death by targeting the molecular chaperone glucose-regulated protein 78. Cancer Res 66(18):9260–9269
Martin S, Lamb HK, Brady C et al (2013) Inducing apoptosis of cancer cells using small-molecule plant compounds that bind to GRP78. Br J Cancer 109:433–443
Yokoyama S, Hirano H, Wakimaru N et al (2001) Inhibitory effect of epigallocatechin-gallate on brain tumor cell lines in vitro. Neuro-Oncology 3:22–28
Porchia LM, Guerra M, Yang YC et al (2007) 2-Amino-N-{4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]-phenyl} acetamide (OSU-03012), a celecoxib derivative, directly targets p21-activated kinase. Mol Pharmacol 72:1124–1131
Booth L, Roberts JL, Tavallai M et al (2015) OSU-03012 and Viagra treatment inhibits the activity of multiple chaperone proteins and disrupts the blood brain barrier: implications for anti-cancer therapies. J Cell Physiol. doi:10.1002/jcp.24977
Smith TJ (2011) Green Tea Polyphenols in drug discovery — a success or failure? Expert Opin Drug Discov 6:589–595
Trott O, Olson AJ (2010) AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31(2):455–461
Lyskov S, Chou FC, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, Das R (2013) Serverification of molecular modeling applications: The Rosetta Online Server That Includes Everyone (ROSIE). PLoS One 8(5), e63906
Pronk S, Pall S, Schulz R et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Kumari R, Kumar R, Open Source Drug Discovery Consortium, Lynn A (2014) g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Humphrey W, Dalke A, Schulten K (1996) VMD — visual molecular dynamics. J Mol Graph 14:33–38
Wisniewska M, Karlberg T, Lehtiö L, Johansson I, Kotenyova T et al (2010) Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B’, and HSPA5/BiP/GRP78. PLoS One 5(1), e8625
Jones NA, Spina D, Page CP (2003) Theophylline and selective phosphodiesterase inhibitors. In: Spina D, Page CP, Metzger WJ, O’Connor BJ (eds) Drugs for the treatment of respiratory diseases. Cambridge University Press, Cambridge, pp 136–171
Chiosis G, Timaul MN, Lucas B, Munster PN, Zheng FF, Sepp-Lorenzino L, Rosen N (2001) A small molecule designed to bind to the adenine nucleotide pocket of Hsp90 causes Her2 degradation and the growth arrest and differentiation of breast cancer cells. Chem Biol 8(3):289–299
Acknowledgments
Part of the computational work (molecular dynamics simulations and MM-PBSA calculations performed by Seema Mishra) reported in this paper was carried out using DBT-funded Bioinformatics Infrastructure Facility at University of Hyderabad.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhattacharjee, R., Devi, A. & Mishra, S. Molecular docking and molecular dynamics studies reveal structural basis of inhibition and selectivity of inhibitors EGCG and OSU-03012 toward glucose regulated protein-78 (GRP78) overexpressed in glioblastoma. J Mol Model 21, 272 (2015). https://doi.org/10.1007/s00894-015-2801-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-015-2801-3