Abstract
BMS-488043, like its predecessor BMS-378806, is a small molecule that can block the interactions between gp120 and CD4, and has shown good clinical efficacy. However, the crystal structure of drug-gp120 complexes or the full-length gp120 free of bound ligand is unpublished until now. Docking combined with molecular dynamics simulation is used to investigate the binding mode between BMS-488043 and gp120. On the basis of the analysis of the simulated results, the plausible binding mode is acquired, such as the changes of binding mode in the trajectory and the calculated binding free energy. Subsequently, a number of residues which make contacts with the small molecule are studied by binding free energy decomposition to understand the mutation experiments, such as Trp427, Ser375, and Thr257 residues with the help of the acquired binding mode above. Especially, the importance of the hydrophobic groove formed by residues Ile371 and Gly472 which bind BMS-488043 is elaborated, which has not been explored much. In addition, theoretical investigations on the dynamics behavior of the gp120 associated with BMS-488043 enhanced binding are performed; the results indicate that the BMS-488043 may be more deeply inserted into the Phe43 cavity compared with the previous binding mode acquired by docking.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
HIV-1, an incurable disease discovered more than 25 years, has infected about 60 million individuals, with about 20 million killed [1]. The intricate and multi-step HIV attacking process involves attaching to host cells, binding CD4 and coreceptor, and fusing membrane. All of these stages are only regulated by the viral envelope (Env) proteins, an exterior glycoprotein gp120 and a transmembrane domain gp41, both of which are processed from the gp160 precursor and projected from the membrane of the virion [2]. Sequence comparisons reveal that the gp120 glycoprotein is composed of five conserved domains (C1 to C5) [3] and five variables (V1 to V5). Dramatic conformational changes [4, 5] in the gp120 glycoprotein can be observed during the initial attachments of gp120 binding to CD4 on the target cell surface, including two sets of β-sheets, the exposure of V1/V2 and V3 loop structures, and the orientation changes of gp120; the changes make two sets of β-sheets spatially separated in unbound gp120, and bring together upon CD4 wedging into the bridging sheet which consists of a four-stranded β-sheet minidomain [4, 5]. Also, they expose the V1/V2 and V3 loop structures, and then change the orientation of gp120 so that the bridging sheet and the V3 loop are directed toward the host cell membrane. These changes allow gp120 to interact efficiently with one of the chemokine receptors, CCR5, and CXCR4, which serve as obligated co-receptors for HIV-1 [6–10] that enable the fusion of the viral and the host membranes. Figure 1 displays the gp120 bound with CD4 structure.
BMS-488043 (BMS043) (Fig. 2) and its predecessor BMS-378806 (BMS806) [11–13], which inhibit the binding interactions of CD4 and gp120, are low-molecular-weight attaching inhibitors of HIV-1 entry. With improved pharmacokinetic properties, BMS043 demonstrates antiviral efficacy and a favorable safety profile in HIV-infected subjects [14, 15], which prohibits HIV-1’s infection via envelope conformational alterations and primarily interfering with cellular CD4 binding. BMS806 binds to gp120 with a small binding enthalpy change in a mostly entropy-driven process [16, 17]. The precise gp120 binding site of the two inhibitors is unclear. However, with the combination of biochemical and genetic approaches, the potential binding site is mapped to a specific area within the CD4 binding pocket (Phe43 cavity) of the gp120 envelope protein [11, 18].
In order to understand the effective mechanism of the Phe43 cavity as the binding site of BMS806, the binding modes between the BMS molecules and gp120 have been investigated by theoretical methods [19, 20]. A docked binding mode proposed by Kong has been studied by molecular dynamics simulation [19], but the short time scale (2500 ps) of production simulation is not long enough to obtain sufficient sampling, and Teixeira gives a similar binding mode between BMS806 and gp120 only [20]; furthermore, the crystal structure of unliganded gp120 is not solved, the structure of gp120 in the CD4 bound state in the docking experiments of the two studies above is used as a receptor, while the previous research works [4, 5, 11, 17] have shown that the tertiary structures of unliganded gp120 and BMS-gp120 complex are not the same as the structure of CD4-gp120 complex. Consequently, in order to evaluate the stability and plausibility of the docked binding modes and investigate the dynamical behaviors of gp120 upon BMS binding, longer time scales for simulations are needed. Recently, the intermediate structure of gp120 predicted by Da and colleagues [21] is used as a receptor in docking studies, a special binding mode [22], in which the heterocyclic ring of BMS043 is docked into a cleft between LB and β3-β5 loops, is proposed to explain the prevention of the bridging sheet formation and interference of the gp41 exposure, but the accuracy of the intermediate structure of gp120 needs to be verified by experiments. In this paper, the structure of gp120 in the CD4 bound state is used as the receptor and the Phe43 cavity is thought to be the target of BMS043. In order to evaluate the plausibility of the binding modes, the 36,200 ps molecular dynamics simulations are performed, we focus on the explanation for the mutation experiments to the residues in the Phe43 cavity and the dynamics behavior of gp120 upon BMS043 binding.
Materials and methods
Structural model preparations
The calculations are based on the X-ray structure of gp120 in the bound state with CD4 and 17b obtained from the Brookhaven Protein Data Bank (PDB) with the PDB codes 1G9N [23]. The coordinates of the clinical primary YU2 isolate substrate-free gp120 core and the CD4-gp120 complex are used as the MD initial structure, the variable loops V1/V2 and V3 as well as the N- and C-termini are truncated in the crystal structure, which are regions extending away from the main body of the gp120 envelope glycoprotein. The inhibitor BMS043 structure is obtained from the PubChem Substance Database and the coordinates of the small molecule are converted into PDB format by the GlycoBioChem PRODRG2 Server [24].
Automated docking
The gp120 structures are taken every 2000 ps of the trajectory of CD4-gp120 complex, the X-ray structure of gp120 in the CD4 bound state are used as receptors in the docking process, and the parameters sets for dynamics simulation of CD4-gp120 complex in AMBER10 are used for the following simulations of the three binding modes; the root-mean-square deviations from the average structure of gp120 in the trajectory are 1.58 (starting structure), 1.23 (2,200 ps), 1.03 (4,200 ps), 1.27 (6,200 ps), and 1.15 Å (8200 ps), five docking experiments are carried out totally; the obtained results show that almost all the clusters’ benzene rings are inserted into the Phe43 cavity with only a few clusters’ heterocyclic rings being inserted into the cavity. In order to sample sufficiently, the typical docking results are used as the starting structures for the following AMBER10 MD simulations [25, 26] and are rescored by using MM/GBSA [27, 28] when they arrive at equilibrium states.
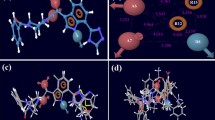
The structures of gp120 glycoprotein are used as the starting structure for AutoDock 4.2 [29] where the AutoDockTools package is used to generate all the necessary input files and the docking grids. The Lamarckian genetic algorithm (LGA) is employed to search for the optimal conformation. The atomic affinity grids on the active site are created by using AutoGrid [29]. Each grid map consists of a 50 × 40 × 40 grid point with 0.375 Å in each dimension. The center of the grid is set to the position of the neighborhood of the Phe43 cavity by the average coordinates of Cα atoms of Trp427 and Gly473. The global optimization is performed with a population of 50 randomly positioned individuals, with a maximum of 2.5 × 106 energy evaluations and a maximum of 2.7 × 105 generations. The rest of the parameters are set as default values. At the end of a docking experiment, docking solutions with a ligand all-atom root mean square deviation (RMSD) within 2 Å of each other are clustered together by using the rms_analysis utility. Three reasonable clusters of them are selected as representative binding modes based on whether the ligand is bound inside a pocket of the receptor: mode I, mode II, and mode III (benzene rings in mode I and mode II penetrated into the Phe43 cavity, heterocyclic ring in mode III inserted into the Phe43 cavity). Figure 3 shows their binding modes, and the heterocyclic rings in mode I and mode II have different orientations.
Three binding modes (a Mode I. b Mode II. c Mode III) were extracted from the results of the docking experiments, respectively; and three binding modes (a’ Mode I. b’ Mode II. c’ Mode III) at 30 ns of simulations are shown, respectively. The molecular ribbon diagram of the YU2 isolate gp120 core in complex with BMS043 is shown above. The ligand (BMS043) and nearby residues were represented by a stick and colored by heteroatom. The figures created by Chimera
MD simulation
The protonation state of the protein is adjusted to mimic physiological pH conditions. All Lys residues are positively charged and His residues are modeled as neutral by protonating the Nε2 atom. The rest of the ionizable residues are set to pH = 7 protonation state. Additionally, the name of entire cysteine residues is changed into CYX because all the cysteine residues in gp120 core are connected by disulfide bonds which are connected together during AMBER MD simulations [23], and the monosaccharides’ names can be recognized by AMBER software. Hydrogen atoms are added by using the LeaP module in AMBER automatically. All MD simulations presented in this work are performed by using the AMBER10 suite of programs [25, 26]. The AMBER ff03 force field [30] and the GLYCAM06 parameter sets [31] are used as the parameters for glycoproteins, ions, and water molecules; whereas we use the general AMBER force field (GAFF) [32] to describe the ligands. Partial charges and force field parameters for the inhibitors are generated automatically by using the Antechamber program [33] in AMBER10. Atomic charges are derived from the AM1-BCC charge method [34, 35]. An appropriate number of Cl- counterions are added to neutralize the system and the molecules are solvated in a box of water by using the TIP3P water model [36] with at least 10.0 Å of water around every atom of the solute. A 10.0 Å cutoff is applied for the evaluation of the non-bonded interactions. Particle mesh Ewald [37] summation is used to calculate the long-range electrostatic interactions during minimization and molecular dynamics.
All structures are minimized by using the sander module in AMBER10. The minimization procedure is split into two stages. First the solvent is allowed to relax while the protein atoms are restrained to their original position with a force constant of 500 kcal mol−1 Å−2. Afterward, the entire system is relaxed and minimized without any restraints. The first stage is performed by using the steepest descent minimization of 2500 steps, followed by a conjugated gradient minimization of 2500 steps, while the second stage consists of 4000 steps of steepest descent, follows by 6000 steps of conjugated gradient minimization.
The MD simulations are carried out by utilizing the periodic boundary condition. The SHAKE algorithm [38] is applied to constrain all bonds involving hydrogen, and the time step of all MD simulations is 0.002 ps. Atomic coordinates are collected at intervals of 1.0 ps to analyze the detailed structures. Initial velocities are assigned from a Maxwellian distribution at the initial temperature. Langevin dynamics with a collision frequency of 2.0 ps−1 are employed to increase the temperature of the system gradually from 0 to 300 K in 20 ps and followed by 200 ps equilibration to remove steric clashes. Subsequently, 36,000 ps (800 ps for CD4-gp120 complex) MD simulations with standard NPT conditions are performed for data collection.
Free energy calculations and decomposition
The docking pose from each cluster after molecular dynamics simulations is rescored by using MM/GBSA in AMBER10 [27, 28], and the modified GB model [39, 40] is used. In order to calculate the averaged binding free energy, 200 snapshots are extracted from the trajectory every 10 ps from 28,000 to 30,000 ps. The averaged binding free energy is calculated by estimating the free energy change associated with the sum of molecular mechanical energies in vacuo plus the solvation free energy and then subtracting the product of entropy change and absolute temperature. The formula is used as the following:
where ∆E MM can be further divided into the electrostatic, van dev Waals, and internal energies contributions, and can be estimated through molecular mechanics method. ∆G sol includes the polar solvation free energy calculated with the generalized Born (GB) approximation model [41–43] and the non-polar part obtained as a function of the solvent-accessible surface area (SASA) [44]. The last term in the above equation (T∆S) is the solute entropy which is usually estimated by normal mode analysis method [45]. However the normal mode calculation to estimate the entropy contribution is somewhat problematic and time-consuming. Furthermore, differences in the entropy effect can be omitted since the two systems are so similar, thus the contribution of entropy to binding free energy is not explicitly taken into account in this work.
The methodology of binding energy decomposition is used to describe the energy contribution of each residue from the association of the receptor with the ligand in three parts: the molecular internal energy in vacuo, the polar contribution of solvation free energy, and the nonpolar solvation part. In addition, all the energies are decomposed into backbone and side-chain atoms, and the key residue contributions to the binding are analyzed through energy decomposition.
Results and discussion
In order to evaluate the plausibility of the binding modes (mode I, mode II, and mode III) predicted by AutoDock, the molecular dynamics simulations are performed; the stability of BMS043 and gp120 complex and changes of the three binding modes along their MD trajectories are analyzed; furthermore, the statistical data of hydrogen bonds and binding free energies are discussed. Afterward, the binding free energy decomposition of the most plausible binding mode is performed for further understanding the interactional mechanism of BMS043 with GP120 compared with the mutation experiments. Lastly, the dynamics behaviors of gp120 associated with BMS043 binding are investigated.
Stability of the trajectories during simulations
The root-mean-square deviation (RMSD) from the X-ray structure provides a direct measurement of the structural drift from the initial coordinates as well as the atomic fluctuation over the course of a MD simulation. As mentioned above, upon engagement of CD4, the V1/V2 and V3 loop structures of gp120 undergo dramatic conformational shifts and two sets of β-sheets are brought together forming the bridging sheet [4, 5], while the formation of a gp120-inhibitor complex involves relatively small conformational changes, in other words, BMS043 prevents conformational changes in gp120 upon CD4 engagement [17, 46]. Therefore, some structural domains of gp120-inhibitor complex have several changes relative to the structure of CD4-gp120 complex, especially for the two sets of β-sheets. The available structure used in the simulations derives from the X-ray structure of gp120 bound with CD4 and 17b; when the inhibitor binds to gp120 in the CD4 bound state, theoretically the structure of the gp120-inhibitor complex will reverse into a different tertiary structural shape that may be more like unliganded gp120; hence, it is quite difficult to get a very stable phase of the gp120 and BMS043 complex. In this article, the emphases are not focused on the equilibrium structure of the bridging sheet that has large fluctuations during simulations, thus, we calculate the RMSD values of main chain with respect to the starting structure of mode I, mode II, and mode III (Fig. 4), in which the residues 119–203 and 422–435 of the bridging sheet (34 positions) are not involved in the RMSD calculations.
After a period of 15,000 and 26,200 ps simulations for mode I and mode III, both the gp120 protein and the small molecule tend to reach the stable state. The average RMSD values of backbone atoms of gp120 protein and the small molecule in the mode I complex are 1.74 and 1.66 Å at the 15,000–36,000 ps intervals. The converged RMSD values of mode III complex are also observed in the 26,200–36,000 ps intervals with average RMSD values of 2.50 and 2.16 Å for the protein and small molecule, respectively. However, the two curves of RMSD values for mode II complex still do not converge after 36,000 ps simulation. Especially, the RMSD curve of the small molecule has a dramatically increase at 8000 ps in the trajectory, which is actually abnormal, this indicates that the binding mode of the mode II makes the protein unstable and unsuitable.
Changes of binding mode along the MD trajectories
Indeed, substitutions of Thr257 and Ser375 on the wall of the Phe43 cavity with larger amino acid residues, like T257R or S375W, are made in previous experiments [18]. These mutations decrease the BMS806 affinity but stabilize the CD4-gp120 complex by filling the Phe43 cavity with Arg and Trp long side chains [18]. It suggests that BMS806 is deeply inserted into this cavity.
Figure 3a’, b’ and c’ show the binding modes of ligand and gp120 at 30,000 ps of simulations. In order to examine three binding modes in dynamics trajectories, one compares the binding modes above with the starting structures, and finds that the ligand in the mode II diffuses away from the Phe43 cavity after 26,000 ps simulation, confirming that the binding mode of mode II is very unstable again, the possibility of binding mode II should be obviated. While it is observed from Fig. 3a’ and c’ that the ligand in mode I and mode III is inserted into gp120 binding pocket more deeply and mode I or mode III are comparatively a more appropriate binding mode. Additionally, whatever the binding mode is, the benzene ring of mode I or the heterocyclic ring of mode III is placed in the groove formed by the residues Ile371 in the α3-helix and the Gly472 after 30,000 ps simulations, indicating that this groove is very important for the ligand binding.
Hydrogen bonds analysis between gp120 and BMS043
In order to understand dynamical flexibilities of gp120 and the interactions between protein and ligand, hydrogen bonds for mode I and mode III modes are analyzed. The geometric criterion for the formation of hydrogen-bond is common with a H-acceptor distance less than 3.5 Å and the donor-hydrogen-acceptor angle larger than 120°. However, no strong hydrogen bond is found in mode I and mode II. A weak hydrogen bond between the ND2 atom of Asn425 and the OAC atom of BMS043 in mode I has an occurrence of 35.35 %; while in mode III there are three weak hydrogen bonds, N-H(Gly473)…OAB(BMS043), ND2-HD22(Asn425)…OAD(BMS043) and OH-HH(Tyr384)…N-7(BMS043), whose occupancy rates are 49.45 %, 33.94 %, and 27.70 %, respectively; generally, it reflects the fact that the interacting strength between protein and ligand in mode III is stronger than that in mode I complex. We do not find the hydrogen bond between the OG atom of Ser375 and the N-7 atom of BMS043 which exists in the complex of gp120 and BMS806 [19]. The result may be attributed to the fact that the limit volume of the binding cavity is hard to accommodate the larger group of the ligand of the two methoxyl groups on the heterocyclic ring. As a consequence, the steric clashes are increased correspondingly, which make the distance between the OG atom of Ser375 and the N-7 atom of BMS043 longer than that in the previously studied system. In the present model the calculated distance between the two atoms in mode III at 30,000 ps is about 6.55 Å, which is far beyond the necessary range of hydrogen bond.
Binding free energy calculations
Although previous research work suggests that the Phe43 cavity is conserved [23], the ligand can induce and trigger changes of gp120 to accommodate ligand. The docking experiments do not sufficiently incorporate the protein flexibility coupled to ligand binding, while molecular dynamics simulation in explicit water allows the motion of gp120 and ligand, this superiority makes them suit each other better, or fall apart if the binding mode is not appropriate. Therefore, it is more accurate to predict the binding mode by rescoring the typical docking pose after molecular dynamics simulations by using MM/GBSA method [27, 28] than by using the scoring function in AutoDock.
In order to analyze the binding free energies, a total of 200 snapshots are taken at a time interval of 10 ps from 28,000 to 30,000 ns of the MD trajectories. The calculated binding free energies are averaged from these 200 snapshots. Figure 5 shows the binding free energies and energy components, i.e., electrostatic, van der Waals, nonpolar solvation, and polar solvation components. The van der Waals and electrostatic terms provide the major favorable contributions to the inhibitor binding, whereas polar solvation terms are a disadvantageous ingredient. Nonpolar solvation terms, which correspond to the burial of SASA upon binding, contribute in a slightly favorable way. The benzene ring or heterocyclic ring in the ligand being inserted into the Phe43 cavity results in different effects on the separate energy components. As can be seen from Fig. 5, the nonpolar solvation terms provide slightly favorable contributions to the binding of inhibitor to gp120 similarly in both binding modes, but when the heterocyclic ring is inserted into the Phe43 cavity (mode III), the electrostatic and van der Waals terms have larger contributions, while the polar solvation terms have larger positive contributions in comparison with that of the benzene ring in the Phe43 cavity (mode I). However, the total binding free energy between BMS043 and gp120 in mode III is −27.39 kcal mol−1 which is bigger than that in mode I (−23.21 kcal mol−1). Consequently, mode III is a better binding mode than mode I energetically, which is in good agreement with the analysis of hydrogen bonds and the calculated results by Kong and co-workers [19].
Binding free energy components (kcal mol−1) of BMS043 to gp120 calculated by the MM/GBSA method. Components are as follow: 1 electrostatic, 2 van der Waals, 3 nonpolar solvation, 4 polar solvation, 5 total binding free energy. The standard deviations of mode I and mode III are 2.37 and 3.23 kcal mol−1, respectively
Binding energy decomposition
In order to obtain a detailed insight into the protein-ligand binding process, the binding free energy of mode III is decomposed into per-atom contributions which can be summed over atom groups to obtain different energy contributions arising from residues backbone and side-chain by using MM/GBSA method. The sum of the electrostatic term in the internal energy (∆G TELE ) and the electrostatic term in the solvation energy (∆G TGB ) constitutes the electrostatic component of the protein-ligand binding free energy, one can see from Table 1 that this quantity is a positive number (unfavorable) for most of the residues we have studied. Apparently, it is because the favorable electrostatic interactions between the protein and ligand do not fully counteract the unfavorable electrostatic interactions between solute molecules (the protein and ligand) and solvent molecules. Similarly, the sum of the van der Waals term in the internal energy (∆G TVDW ) and the nonpolar term in the solvation energy (∆G TGBSUR ) constitutes the nonpolar component of the protein-ligand binding free energy, this quantity is a negative number (favorable) in all cases. This analysis suggests that the nonpolar interaction is the driving force for the binding of this inhibitor to gp120.
In the previous studies [11, 18], many mutation experiments have been conducted to investigate the effect of sensitivity to BMS; the research shows that W427V completely abates the effect of BMS043 while W427F mutant does not affect the binding affinity of BMS806 [18]. As can be seen from Table 1, the interaction of Trp427 with BMS043 is much stronger in comparison with that of the other residues; it contributes −2.24 kcal mol−1 to the binding affinity, in which VDW interaction is the dominating force. From their relative positions, it can be found that this large VDW contribution is mainly originated from the interaction between heterocyclic ring of BMS043 and methylene of Trp427, the methylene plays an important role in the binding process. BMS806 can bind to W427F mutant in the mutant experiment [18], it is possible that the residues tryptophan and phenylalanine have similar structure of side chain and both contain a methylene which may be critical for gp120 binding BMS043, the VDW interaction is not affected when Trp427 mutated to Phe427. However for W427V, a hydrogen atom on the methylene is replaced by a methyl group that can compress the limited volume of Phe43 cavity and squeeze BMS043 out of the cavity; therefore, W427V has completely negated effects for BMS043 binding.
Ser375 and Thr257, another two important residues, are situated at the bottom of the Phe43 cavity formed by the outer domain (Fig. 6) and contribute −0.24 and −0.78 kcal mol−1 to the binding affinity, respectively. By comparison of the side chain and backbone contributions in Thr257, the binding free energy contributions mainly come from VDW interactions of the side chain of Thr257 with heterocyclic ring of BMS043, while the backbone contribution is very small; one can see from Fig. 6 that the methyl group of Thr257 is underneath heterocyclic ring of BMS043, indicating that the major contribution of VDW interaction comes from the methyl group of Thr257, this finding can be used to explain why T257A can slightly affect the sensitivity of ligand binding and T257G can affect the ligand binding more seriously [18]. For S375A mutation, the sensitivity of the small molecule binding is almost not affected [18]. From calculated results involving Ser375, one can find that the Ser375 side chain or backbone provide very slight contributions, thus S375A will not affect the ability of the small molecule binding. Previous experimental studies [47, 48] indicate that the S375W and T257R diminish the size of the Phe43 cavity and stabilize the conformation closer to the CD4-bound state even in the absence of ligands, but these mutations largely negate BMS806 binding. As seen from Fig. 6, one finds that the methoxyl group at C-9 on the heterocyclic ring of BMS043 is just near the Ser375 and Thr257 residues. The distance between oxygen atom of Ser375 side chain and the carbon atom of the methoxyl group of BMS043 as well as the distance between the carbon atom of methyl group of Thr257 and the carbon atom of the methoxyl group of BMS043 are less than 3.5 Å. Large side chain mutations in these two residues located inside the cavity may make steric hindrance for the binding.
The structure-BMS043 of mode III at 30 ns of simulation surrounded by the vicinal residues. The backbone of gp120 represented by ribbon and colored as tan. BMS043 and wire representation of the vicinal residues (the residues in 4.0 Å of BMS043 were selected) represented by stick and colored as heteroatom
Figure 6 shows that the phenyl of BMS043 is accommodated by the groove formed by the residues Ile371, Gly367, and Gly472 which form hydrophobic interactions with the phenyl group of ligand.
As seen from Table 1, one can find that the total binding free energy value of the contribution of Ile371 in the groove, which mainly originates from the van der Waals interaction between the side chain (the isobutyl group) of Ile371 and the benzene ring and heterocyclic ring of the ligand, is very large (−2.55 kcal mol−1). Mutation experiments [11] show that I371F mutations do not negate BMS806 binding and reduce the BMS806 affinity by half, the reason can be that Phe has a longer functional group on the side chain compared with the isobutyl group of Ile and may prevent the benzene ring of BMS806 from interacting with other hydrophobic residues, such as Gly472 and Gly367. However, the hydrophobic property of the groove is not changed generally; thus, the BMS806 can still be bound to I371F mutant.
Similar to the residue Ile371, the other important residue Gly472 has a large contribution as well; the favorable interaction mostly comes from the VDW interactions between the backbone of Gly472 and the benzene ring of BMS043. Gly367 and Asp368 are also involved in this groove, the hydrophobic residue Gly367 is favorable to the ligand binding by contacting with the phenyl of BMS043. However, for the acidic amino acid Asp368, it forms an important electrostatic interaction with Arg59 in CD4 of CD4-gp120 complex and the interaction is critical for CD4 binding. However, the hydrophilic side chain of Asp368 is adverse for interacting with hydrophobic benzene ring of BMS043, which contributes the unfavorable values (0.23 kcal mol−1). Thus, if this residue is mutated to Gly, or a polar substituent is added to the phenyl ring, and or the phenyl moiety is replaced by heterocyclic ring to make contact with the acidic amino acid, the binding ability between the protein and BMS043 would become better. In order to demonstrate whether this binding mode is effective or not, more mutation experiments to the hydrophobic groove should be done.
As for mode III shown in Table 1, other residues in the binding mode, such as Ser256, Phe382, Tyr384, Asn425, Met426, Gly473, Asp474, Met475, and Arg476, have contributions to binding energies with BMS043. As shown in Fig. 6, the 3-methoxyl groups on the heterocyclic ring have strong VDW interactions with Asn425 and Met426, while Phe382, Tyr384, and Ser256 are deeply inserted into the cavity and interact with the heterocyclic ring. Furthermore, the sulfur atom of Met475 can strongly interact with the N-H atom of BMS043, and enhance the interactions between them. In addition, Gly473 and Asp474 residues, which are positioned in the vicinity of the methyl-piperazine, contribute to the binding affinity between inhibitor and gp120.
Changes in the conformation of gp120 during the simulations
In order to investigate the motion of gp120 associated with the BMS043 binding, the root-mean-squared fluctuation (RMSF) of the Cα atoms from the backbone of gp120 core over the whole MD trajectories is calculated, which reflects the mobility of a certain residue around its average position (Fig. 7a), the average structure of mode III of the equilibrium state and its overlaps with the starting structure are exhibited in Fig. 7b.
a The Cα root mean square fluctuations (RMSF) of gp120 in complex with BMS043, gp120 bound with CD4, and CD-free gp120 during the entire MD simulation represented in black, red, and blue line, respectively. b Overlay of the average structure of BMS-gp120 complex in mode III (sky blue) from the last 8 ns of the MD trajectories with the crystal structure of gp120 (tan)
As seen from the overall curves of RMSF for three structure states (Fig. 7a), the dynamics behavior of gp120 bound with CD4 is the most stable, some domains of gp120 in the CD4 bound state become unstable when CD4 is removed, and the dynamics behavior of the complex with the Phe43 cavity binding BMS043 is enhanced. However, in total the fluctuation trends of gp120 in three conditions (three structure states) are similar, for example, almost all of the residues which have large fluctuations are located either at the N- and C-termini or in loops/links regions and α2-helix (335–352) keeps a good rigidity in the three structural states. The N- and C-termini will oscillate all the time when the inner domain being linked with gp41 is truncated, consequently, they undoubtedly have large RMSF values; the residues 206–215 and 218–247 in the inner domain and the residues 263–268, 278–282, 351–358, 389–413, 435–445, and 457–465 in the outer domain have comparably large fluctuations in the BMS043-gp120 complex, most of which are located far away from the binding cavity. The large fluctuation can also be found in the domains of CD4-free gp120, thus the large mobility of these domains may not be associated with the Phe43 cavity binding BMS043. As far as the stable α2 structure is concerned, Tan and Rader [49] have pointed out that this domain is a rigid core regardless of strain or binding CD4 or other mimics, though BMS043 is not a CD4 mimics, this core in our system is still rigid upon BMS043 binding, reflecting that the universe character in the complex of CD4 or its mimics and gp120 is also applicable for the BMS043-gp120 complex.
A lot of dynamics simulations of glycoprotein have been studied [21, 22, 48–55], the focus here is placed on the influence to behavior of the Phe43 cavity and the hydrophobic groove in mode III upon BMS043 binding. In comparison the average RMSD values of the residues locate in the Phe43 cavity of mode III with that of CD4-bound gp120, one can find that the residues 377–382 have lager flexibilities, it is mentioned above that the heterocyclic ring of BMS043 is inserted into the Phe43 cavity more deeply in contrast with the starting structure, therefore, the enhanced dynamics behavior may be attributed to the regulation of Phe382 for accommodating BMS043. In addition, the dynamics behaviors of the hydrophobic groove consisting of the residues 472–473 and 367–371 in mode III have also enhanced. In order to stabilize the α3-helix (368–371), strong electrostatic interactions are formed between Asp368 and Arg56 in CD4 of the CD4-gp120 complex, α3-helix is no longer restrained when CD4 is truncated. Hence, Asp368 and Pro369 in CD4-free gp120 have large average RMSDs of 1.25 and 1.02 Å as compared with the values of respective 0.56 and 0.77 Å in the CD4-gp120 complex. While the residues in mode III become more flexible (1.30 and 1.36 Å for averaged RMSDs of Asp368 and Pro369, respectively), indicating that the dynamics behavior of α3-helix is further enhanced upon BMS043 binding. Our results are consistent with the observation of Shrivastava and LaLonde [50] that the dynamical behavior of the α3-helix is enhanced in gp120 bound with NBD-556. As seen from the average structure of mode III superimposed with the starting structure (Fig. 7b), the α3-helix has moved toward the outer domain compared with the original position in the starting structure, consequently, the entrance size of Phe43 cavity is increased, which is more beneficial for the ligand to be inserted into the cavity, it suggests that the BMS043 may be more deeply inserted into this cavity of BMS043-gp120 structure compared with the position of BMS043 in mode III and the previous binding mode by docking [19, 20]. Recently, the crystal structure of unliganded gp120 being solved by Kwon (coree gp120) [56] shows that the unliganded gp120 assumes the CD4-bound conformation because the conformational equilibrium of unliganded gp120 favors the CD4-bound state when the V1/V2 and V3 loops are truncated, while the full-length wild type and the more truncated coremin gp120 (10 residues in the V3 loop are not included in the conformation, which are included in the conformation of coree gp120) used in our simulations are more likely to assume the conformation before the inducement of CD4 [56]. The prediction about α3-helix in the unliganded gp120 and BMS-gp120 complex in this paper is based on the mobility of the coremin gp120. Although Kwon [56] solved the crystal structure of the NBD-556-gp120 complex which also assumes the CD4-bound conformation, BMS043 is more likely to be bound to the conformation close to the full-length unliganded gp120 [56]. Thus, the mobility α3-helix in the BMS043-gp120 complex is reasonable, and our studies do not contradict that of Kwon.
In terms of the behaviors of residues Gly472 and Gly473 in the hydrophobic groove, they also have enhanced mobility, which may be attributed to the hydrophobic interaction with phenyl ring of BMS043 and the inherent flexibilities of link region (links α5 and β24). In addition, it can be seen from Fig. 7a that the bridging sheet has very large fluctuations and the β2/3 (119–203) have more fluctuations than β20/21 (422–435) which are consistent with the previous studies [55].
Conclusions
In order to get the plausible binding mode of BMS043 to gp120, the structure of gp120 core is extracted from the crystal structure (1G9N), and the protein-ligand docking is conducted by AutoDock 4.2, three binding modes of the small molecule to the receptor are obtained. The small molecule in the three cases is located in the Phe43 cavity. The dynamical behavior of the three binding modes of the complex is investigated by explicit water, unrestrained molecular dynamics (MD) simulations. According to the changes of binding mode and the previous experimental results [18] which indicated the small molecule is deeply inserted in the Phe43 cavity. It is found from our simulated results that the BMS043 in the Phe43 cavity in mode I or mode III are more deeply inserted into the cavity partly and the other portion of MBS043 outside of the Phe43 cavity ultimately formed hydrophobic interactions with the groove composed of Gly367, Ile371, and Gly472. From the obtained calculated results of hydrogen bonds analysis and the binding free energies of mode I and mode III by using MM/GBSA method, we find that binding mode III is the more plausible binding mode. Afterward, binding free energy decomposition of mode III is performed for further understanding the interactional mechanism of BMS043, the calculated results of the residues in the Phe43 cavity are in good agreement with those of the mutation experiments. It is proposed that more mutation experiments to the hydrophobic groove composed of Gly367, Ile371, and Gly472 should be done to determine its importance to drug design.
On the basis of the BMS043-gp120 structure in mode III, the dynamics behavior of gp120 associated with BMS043 binding is investigated, the overall mobility of gp120 is enhanced, and α3 keeps a good rigidity during the whole simulation of BMS043-gp120 complex, which is consistent with previous studies [49]. As seen from the RMSF of residues in the Phe43 cavity and the superimposition of the average structure of mode III with the starting structure, it is found that the dynamics behaviors of the residues 377–382 in mode III are enhanced, which may be attributed to the deeper insertion of BMS043 into the Phe43 cavity, and that the mobility of α3 toward outer domain increases the entrance size of the Phe43 cavity. This can explain the plausibility of the model of BMS043-gp120 complex proposed by Da [22] in which the BMS043 is located in the bottom of the Phe43 cavity. It is believed that our studies will be helpful for further understanding the binding mode of BMS043-gp120 and the drug design.
References
UNAIDS (2009) 2008 Report on the global AIDS epidemic. World Health Organization
Tilton JC, Doms RW (2010) Entry inhibitors in the treatment of HIV-1 infection. Antivir Res 85(1):91–100
Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, Parks ES, Parks WP, Josephs SF, Gallo RC (1986) Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45(5):637–648
Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC (2005) Structure of an unliganded simian immunodeficiency virus gp 120 core. Nature 433(7028):834–841
Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA (1998) Structure of an HIV gp 120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody NATURE-LONDON-:648–659
Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W (1996) The [beta]-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85(7):1135–1148
Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P, Marmon S, Sutton R, Hill M, Davis C, Peiper S, Schall T, Littman D, Landau N (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature 381(6584):661–666
Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW (1996) A dual-tropic primary HIV-1 isolate that uses fusin and the [beta]-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85(7):1149–1158
Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP (1996) HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381(6584):667–673
Feng Y, Broder CC, Kennedy PE, Berger EA (1996) HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272(5263):872–877
Guo Q, Ho HT, Dicker I, Fan L, Zhou N, Friborg J, Wang T, McAuliffe BV, Wang HH, Rose RE (2003) Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J Virol 77(19):10528–10536
Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HGH, Rose R, Yamanaka G (2003) A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci U S A 100(19):11013–11018
Wang T, Zhang Z, Wallace OB, Deshpande M, Fang H, Yang Z, Zadjura LM, Tweedie DL, Huang S, Zhao F (2003) Discovery of 4-benzoyl-1-[(4-methoxy-1H-pyrrolo [2, 3-b] pyridin-3-yl) oxoacetyl]-2-(R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J Med Chem 46(20):4236–4239
Hanna G, Lalezari J, Hellinger J, Wohl D, Masterson T, Fiske W, Kadow J, Lin P, Giordano M, Colonno R (2004a) Antiviral activity, safety, and tolerability of a novel, oral small-molecule HIV-1 attachment inhibitor, BMS-488043, in HIV-1-infected subjects. In: p abstract no. 141
Hanna G, Yan J, Fiske W, Masterson T, Zhang D, Grasela D (2004b) Safety, tolerability, and pharmacokinetics of a novel, small-molecule HIV-1 attachment inhibitor, BMS-488043, after single and multiple oral doses in healthy subjects. In: p abstract no. 535
Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB III, Sodroski J, Freire E (2006) Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry 45(36):10973–10980
Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S (2004) Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci USA 101(14):5036–5041
Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB III, Sodroski JG (2004) Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol 78(7):3742–3752
Kong R, Tan JJ, Ma XH, Chen WZ, Wang CX (2006) Prediction of the binding mode between BMS-378806 and HIV-1 gp120 by docking and molecular dynamics simulation. Biochim Biophys Acta (BBA)-Proteins Proteomics 1764(4):766–772
Teixeira C, Serradji N, Maurel F, Barbault F (2009) Docking and 3D-QSAR studies of BMS-806 analogs as HIV-1 gp120 entry inhibitors. Eur J Med Chem 44(9):3524–3532
Da LT, Quan JM, Wu YD (2009) Understanding of the bridging sheet formation of HIV-1 glycoprotein gp120. J Phys Chem B 113(43):14536–14543
Da LT, Quan JM, Wu YD (2011) Understanding the binding mode and function of BMS-488043 against HIV-1 viral entry. Proteins: Struct Funct Bioinforma 79(6):1810–1819
Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA (2000) Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8(12):1329–1339
Schuttelkopf AW, Van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D: Biol Crystallogr 60(8):1355–1363
Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26(16):1668–1688
Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1–3):1–41
Gohlke H, Kiel C, Case DA (2003) Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J Mol Biol 330(4):891–913
Zoete V, Meuwly M, Karplus M (2005) Study of the insulin dimerization: Binding free energy calculations and per©\residue free energy decomposition. Proteins: Struct Funct Bioinforma 61(1):79–93
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24(16):1999–2012
Kirschner K, Yongye A, Tschampel S, Gonzalez-Outeirino J, Daniels C, Foley B, Woods R (2008) GLYCAM06: a generalizable biomolecular force field. Carbohydrates J Comput Chem 29(4):622–655
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174
Okur A, Wickstrom L, Layten M, Geney R, Song K, Hornak V, Simmerling C (2006) Improved efficiency of replica exchange simulations through use of a hybrid explicit/implicit solvation model. J Chem Theory Comput 2(2):420–433
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) Fast, efficient generation of high©\quality atomic Charges. AM1©\BCC model: I. Method. J Comput Chem 21(2):132–146
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high©\quality atomic charges. AM1©\BCC model II. Parameterization and validation. J Comput Chem 23(16):1623–1641
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Onufriev A, Bashford D, David A (2000) Modification of the generalized Born model suitable for macromolecules. J Phys Chem B 104(15):3712–3720
Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large–scale conformational changes with a modified generalized born model. Proteins: Struct Funct Bioinforma 55(2):383–394
Bashford D, Case DA (2000) Generalized Born models of macromolecular solvation effects. Annu Rev Phys Chem 51(1):129–152
Simonson T (2001) Macromolecular electrostatics: continuum models and their growing pains. Curr Opin Struct Biol 11(2):243–252
Tsui V, Case DA (2000) Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers 56(4):275–291
Sitkoff D, Sharp KA, Honig B (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem 98(7):1978–1988
Kottalam J, Case D (1990) Langevin modes of macromolecules: applications to crambin and DNA hexamers Biopolymers 29 (10©\11):1409–1421
Ho HT, Fan L, Nowicka-Sans B, McAuliffe B, Li CB, Yamanaka G, Zhou N, Fang H, Dicker I, Dalterio R (2006) Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J Virol 80(8):4017–4025
Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J (2002) Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol 76(19):9888–9899
Pan Y, Ma B, Keskin O, Nussinov R (2004) Characterization of the conformational state and flexibility of HIV-1 glycoprotein gp120 core domain. J Biol Chem 279(29):30523
Tan H, Rader A (2009) Identification of putative, stable binding regions through flexibility analysis of HIV–1 gp120. Proteins: Struct Funct Bioinforma 74(4):881–894
Shrivastava I, LaLonde JM (2011) Enhanced dynamics of HIV gp120 glycoprotein by small molecule binding. Biochemistry 50(19):4173–4183
Shrivastava I, LaLonde JM (2010) Fluctuation dynamics analysis of gp120 envelope protein reveals a topologically based communication network. Proteins: Struct Funct Bioinforma 78(14):2935–2949
Liu SQ, Liu SX, Fu YX (2008) Molecular motions of human HIV-1 gp120 envelope glycoproteins. J Mol Model 14(9):857–870
Hsu STD, Bonvin AMJJ (2004) Atomic insight into the CD4 binding–induced conformational changes in HIV–1 gp120. Proteins: Struct Funct Bioinforma 55(3):582–593
Hsu STD, Peter C, Van Gunsteren WF, Bonvin AMJJ (2005) Entropy calculation of HIV-1 Env gp120, its receptor CD4, and their complex: an analysis of configurational entropy changes upon complexation. Biophys J 88(1):15–24
Pan Y, Ma B, Nussinov R (2005) CD4 binding partially locks the bridging sheet in gp120 but leaves the [beta] 2/3 strands flexible. J Mol Biol 350(3):514–527
Do Kwon Y, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T (2012) Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci 109(15):5663–5668
Acknowledgments
This work is supported by Natural Scientific Fund of China (11179035) and 973 fund of Chinese Ministry of Science and Technology (2010CB934504).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, L., Chen, H., Zhao, RN. et al. The investigations on HIV-1 gp120 bound with BMS-488043 by using docking and molecular dynamics simulations. J Mol Model 19, 905–917 (2013). https://doi.org/10.1007/s00894-012-1619-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1619-5