Abstract
Using implicit solvent atomistic model and replica exchange molecular dynamics, we study binding of Aβ monomer to zwitterionic dimyristoylphosphatidylcholine (DMPC) lipid monolayer. Our results suggest that Aβ binding to the monolayer is governed primarily by positively charged and aromatic amino acids. Lysine residues tend to interact with surface choline and phosphorous lipid groups, whereas aromatic amino acids penetrate deeper into the monolayer, reaching its hydrophobic core. We show that binding to the DMPC monolayer causes a dramatic conformational transition in Aβ monomer, resulting in chain extension, loss of intrapeptide interactions, and formation of β-structure. This conformational transition is far more significant than that occurring during the initial stages of aggregation in water. We also found that Aβ binding perturbs surface ordering of lipids interacting with Aβ.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is an age-related neurodegenerative disorder, which is linked by genetic and clinical evidence to the production of Aβ peptides during cellular proteolisys and their subsequent aggregation [1, 2]. Although Aβ peptides exist in a variety of alloforms, the most abundant is the 40-mer sequence, Aβ 1−40, which readily forms polymorphic amyloid fibrils depending on preparation conditions [3, 4]. Aβ aggregation leading to fibril assembly proceeds through complex structural transitions initiated with the oligomerization of monomers [5]. Experimental data indicate that Aβ oligomers, even as small as dimers, are the primary cytotoxic species in AD [6, 7].
The precise mechanisms of Aβ cytotoxicity resulting in the neuronal damage and death that is ultimately responsible for AD pathology remain unclear. One plausible mechanism is related to the interaction of Aβ peptides with cellular lipid membranes. In particular, it has been hypothesized that Aβ peptides bind to cellular surfaces with high affinity [8]. A number of experimental studies have indeed offered direct evidence of Aβ association with lipid bilayers [9-11]. These studies have indicated that, depending on Aβ concentration, the peptide may exist on the membrane surface in either monomeric [9] or oligomeric forms [12]. In general, it appears that small oligomers display the highest binding affinity compared to larger aggregates [13, 14]. Experimental data also support the notion that Aβ not only binds to but also penetrates the volume of lipid bilayers, leading to formation of Aβ aggregates in the hydrophobic membrane environment [15–18]. Compelling arguments suggest that membrane-mediated Aβ aggregates are responsible for the appearance of channels and uncontrollable permeation of ions through the membrane [8]. It is likely that multiple pathways of Aβ-membrane interactions exist, including binding of Aβ monomers and their assembly into oligomers on/in the membrane or binding of preformed Aβ aggregates to the membrane. Independent of the specific pathway, the structures of Aβ peptides interacting with the membrane may differ from those in a water environment [19].
Although Aβ interactions with cellular membranes probably lie at the center of AD pathology, there is a lack of corresponding molecular level information. In principle, computer simulations can provide microscopic details of the interactions between Aβ peptides and cellular membranes that are not readily available using other methods [20]. For example, all-atom explicit water molecular dynamics (MD) has been applied recently to examine the adsorption of Aβ monomers on lipid or lipid-like surfaces [21, 22]. MD simulations have suggested that Aβ monomers form mobile small oligomers in the lipid bilayer, which then assemble into larger, channel-forming aggregates, consistent with the atomic force microscopy (AFM) experiments [23]. A simplified representation of lipid bilayer based on a continuum dielectric model has been used to analyze the location and structure of Aβ monomers inserted into the bilayer [24]. This latter study showed that Aβ 1−40 peptides tend to reside at the membrane–water interface. A similar conclusion was reached recently using 200 ns explicit water simulations [25]. However, a number of important questions on Aβ–lipid bilayer interactions remain unanswered. For example, what are the structural changes induced in Aβ monomer upon binding to the membrane? Which amino acids are responsible for binding? Does binding of Aβ monomers to the bilayer affect the structural properties of lipids? Answers to these questions are facilitated by our earlier simulations of Aβ species in aqueous environment [26–28], which can be used as reference in evaluating the changes in Aβ conformations induced by membranes.
The experiments do not implicate a strong propensity of Aβ to insert into zwitterionic bilayers [29], and suggest that, at low concentration (∼150 nM), Aβ resides on the bilayer surface in a monomeric form [9]. Consequently, in this study we use an implicit solvent atomistic model coupled with replica exchange MD to probe the interactions of Aβ monomer with the surface of zwitterionic dimyristoylphosphatidylcholine (DMPC) monolayer. Our results suggest that Aβ binds to the surface of DMPC monolayer forming predominantly interactions with choline and phosphorous groups. We also observed that aromatic amino acids penetrate deeper into the monolayer, reaching its hydrophobic core. The main outcome of our simulations is that binding to the DMPC monolayer causes a dramatic conformational transition in Aβ monomer, resulting in peptide extension, loss of intrapeptide interactions, and formation of β-structure. We also demonstrate that Aβ binding perturbs structural ordering of the lipids interacting with Aβ. The paper concludes with a comparison of our findings with previous studies.
Methods
Molecular dynamics simulations
To simulate interactions of Aβ peptide with the DMPC lipid monolayer (Fig. 1), we used the CHARMM MD program [30], and united atom force field CHARMM19 coupled with the SASA implicit solvent model [31]. The selection of implicit solvent model was dictated by our objective of reducing the computational costs, which would allow us to obtain exhaustive conformational sampling. The latter is extremely challenging when using explicit solvent models. The original forms of the CHARMM19 force field and SASA model were designed for simulations of peptides in water. Their description, applicability and testing can be found in our previous studies [27, 32, 33]. In particular, we have shown that the CHARMM19+SASA force field reproduces the experimental distribution of chemical shifts for Cα and Cβ atoms in Aβ monomers [27, 34]. This force field also correctly predicts the fractions of β-structure in Aβ monomers, dimers, tetramers, and fibrils, and the experimental temperature of dissociation of Aβ fibril [26, 35]. Modifications of the CHARMM19+SASA force field for the simulations of lipids are described in the electronic supplementary material (ESM 1).
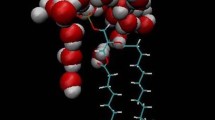
a Sequence of Aβ 10–40 monomer. The N- and C-terminals, Nt and Ct (shown in large letters), span the sequence positions 10–23 and 29–39, respectively. b Snapshot of the simulation system consisting of 81 dimyristoylphosphatidylcholine (DMPC) lipids forming a monolayer and Aβ peptide (purple) bound to the monolayer surface. The lipid choline, phosphorous, glycerol, and fatty acid groups are colored green, blue, red, and grey, respectively. The axis z is normal to the monolayer surface. The layer of constrained hydrocarbons in orange at z= − 22 Å (see Methods) represents the lower boundary of the system. The upper boundary is given by the repulsive wall at z = 80 Å. Phosphorous atoms are distributed around z = 0 Å
In this work we use the N-terminal truncated fragment of the full-length peptide, Aβ 10–40 (Fig. 1a) [28]. Amino truncated Aβ species are naturally occurring and represent a substantial fraction of all Aβ peptides in vivo [36]. Furthermore, experiments [37] and simulations [28] have revealed significant similarities between Aβ 1–40 and Aβ 10–40 oligomers in terms of size distribution and structure. In addition, as recently shown, the N-terminal of Aβ 1–40 monomer does not form extensive long-range interactions or ordered secondary structure [38]. It must be noted that solid-state NMR experiments have revealed ordering of two N-terminal amino acids during fibrillization, suggesting that they play a role in fibril growth [39]. However, the interactions of Aβ with the membranes are likely to differ from the interpeptide interactions responsible for fibril growth and, according to previous computational studies, the N-terminal of the bound Aβ 1–40 monomer is well solvated [24, 40]. These arguments suggest that Aβ 10–40 can be used as an approximate model of the full-length Aβ 1–40 peptide.
The simulation system consisted of Aβ 10–40 monomer interacting with 81 DMPC lipids (Fig. 1b). The selection of DMPC lipids was motivated by their zwitterionic nature, relatively small size, and availability of experimental data. For computational efficiency, we considered a DMPC monolayer (upper leaflet of the bilayer). To mimic the presence of the lower leaflet we placed 324 hydrocarbon atoms constrained harmonically with the constant k = 5 kcal mol−1 Å−1 to the plane z = −22 Å (Fig. 1b). SASA model tends to overestimate the accessible surface areas of atoms in dense hydrophobic medium [31]. According to our test simulations, this effect leads to partial collapse of DMPC bilayers. To resolve this issue and preserve the integrity of the monolayer a soft harmonic constraint with the constant k = 0.6 kcal mol−1 Å−2 was applied to the z-coordinates of phosphorous (P) atoms, forcing them to fluctuate near z = 0 Å. In addition, soft repulsive boundaries along the z-axis were applied to the centers of mass of fatty acid tails −22Å ≲ z ≲ 0Å. Simulations utilized periodic boundary conditions applied in the (x, y) plane. To prevent Aβ from drifting away from the monolayer, a soft repulsive wall was placed at z = 80 Å(Fig. 1b). Therefore, the unit cell dimensions were 70.2 Å × 70.2 Å × 102 Å. A comparison of DMPC monolayer properties with experimental data and the results of other simulations are presented below.
Replica exchange simulations
To perform conformational sampling we used replica exchange molecular dynamics (REMD) [41]. In all, 32 replicas were distributed exponentially in the temperature range from 330 to 560 K. The exchanges were attempted every 20 ps between all neighboring replicas with the average acceptance rate of 26 %. Five REMD trajectories were produced, resulting in the cumulative simulation time of 32 μs. Between replica exchanges the system evolved using NVT underdamped Langevin dynamics with the damping coefficient γ = 0.15 ps−1 and the integration step of 2 fs. Because the initial parts of REMD trajectories are not equilibrated and must be excluded from thermodynamic analysis, the cumulative equilibrium simulation time was reduced to τ sim≈30.7 μs. The REMD trajectories were started with random unbound conformations of Aβ peptide and preformed monolayer. It is important to emphasize that the NVT simulation ensemble and the constraints applied to monolayer atoms preclude full insertion of Aβ monomer into the monolayer. Thus, our REMD simulations are designed to probe binding of Aβ to the monolayer. The convergence of REMD simulations and error analysis are discussed in the electronic supplementary material (ESM 1).
Computation of structural probes
Interactions formed by Aβ peptide and the DMPC monolayer were probed by computing the number of side chain contacts and hydrogen bonds (HBs). A side chain contact occurs if the distance between the centers of mass of side chains is less than 6.5 Å. This cut-off corresponds approximately to the onset of hydration of side chains as the separation distance between them increases. A similar definition of contact has been used for probing interactions between Aβ side chains and monolayer groups. In this case, choline, phosphorous, glycerol, and fatty acid groups were represented by their respective centers of mass. Aβ peptide is assumed bound, if it forms at least one side chain contact with any of the monolayer groups.
The backbone HBs between peptide NH and CO groups were assigned according to Kabsch and Sander [42]. Secondary structure in Aβ peptide was computed using the distribution of (ϕ, ψ) backbone dihedral angles. Specific definitions of β-strand and helix states can be found in our earlier studies [27]. The analysis of conformational ensemble sampled by Aβ peptide was performed using the clustering technique described in ESM 1. The distribution of states produced by REMD was analyzed using multiple histogram method [43], which allowed us to compute thermodynamic averages of various structural quantities. Throughout the paper angular brackets <..> imply thermodynamic averages. Conformational ensemble of Aβ peptide bound to the monolayer was compared with those of Aβ monomer and dimer in water studied by us earlier [26, 27]. To facilitate these comparisons, binding of Aβ peptide to the monolayer was investigated at the temperature 360 K, at which Aβ peptide locks into a fibril-like state during fibril growth in an aqueous environment [32, 35]. A comparison of Aβ binding at 330 K and 360 K is also provided.
Testing the accuracy of the implicit solvent model
To check the quality of DMPC lipid parameterization in the CHARMM19 force field, we performed three test simulations. The first test includes REMD simulations of DMPC monolayer consisting of 36 lipids without the Aβ peptide. The design of these simulations was similar to that for Aβ+monolayer system. We selected a number of DMPC structural quantities, which were reported experimentally, and computed them at 330 K. We found that the monolayer maintains the average surface area per lipid of A l = 60.8 Å2. The experimental value for the DMPC membrane is 60.6 Å2 [44]. From the simulations, the volume per lipid was estimated to be V l = 1,034 Å3, which is close to the experimental value of 1,101 Å3 [45]. We also computed the average monolayer thickness D defined as the average distance between the choline nitrogen atom and the last fatty acid hydrocarbon atom. We found that D = 17 Å, which is similar to the experimental thickness of bilayer leaflet of 18 Å [45].
The conformations of fatty acid tails can be tested by the carbon-deuterium bond order parameter S CD = S mol (3cos2 θ − 1)/2, where θ is the angle between the carbon-deuterium bond and the z-axis and S mol(≈0.5) accounts for anisotropic reorientation of lipids [46]. This order parameter reflects the extent of structural fluctuations in fatty acids and was measured experimentally at 307 K [47]. To compute S CD from the simulation data we used the procedure described previously [46]. Figure 2a demonstrates that the experimental and computed S CD values for individual carbons in the tails are in good agreement.
a Fatty acid order parameters |S CD | for DMPC lipids computed from the replica exchange molecular dynamics (REMD) simulations of monolayer (black line), and measured experimentally for the bilayer (open circles) [47]. b Distributions P(z) of the centers of mass of choline (green), phosphorus (blue), glycerol (red) and fatty acid (grey) groups along the z-axis at 330 K. Solid and dashed lines correspond to the DMPC bilayer simulated with explicit solvent force field and the DMPC monolayer simulated with implicit solvent model, respectively. The midpoint of the distribution of phosphorus atoms is set to z = 0 and the distributions for only one bilayer leaflet are shown. c The free energies of amino acids F(z) along the z-axis: Phe (in blue), Lys (in green). The profiles F(z) are computed for the implicit solvent DMPC model at 330 K using the probability distributions of amino acids along z-axis. Free energy of unbound state (z ≳ 10 Å) is set to zero
To further evaluate the implicit solvent DMPC model, we performed second test simulations in which we used the DMPC bilayer, explicit solvent CHARMM27 force field, NPT ensemble, and periodic boundary conditions. This system contained 72 lipids and 1,848 TIP3P water molecules. Two trajectories with a total simulation time of 100 ns were generated at 330 K. To compare the DMPC monolayer simulated with the implicit solvent model and the DMPC bilayer simulated with explicit solvent force field, we considered the distributions P(z) of the centers of mass of choline, phosphorus, glycerol and fatty acid groups along the z-axis. Figure 2b shows that the centers of these distributions for the two models are in good agreement, although the distributions for the explicit solvent model are somewhat wider.
Tieleman and coworkers [48] have computed the free energies F(z) of amino acid side chains interacting with DOPC bilayer along the z-axis. (DOPC lipids differ from DMPC by a single unsaturated bond in the fatty acid tail.) Their simulations utilized the side chains truncated from their backbones, NPT ensemble at 298 K [49], OPLS-AA force fields for lipids and amino acids, and explicit SPC water model. To compare the results of Tieleman and coworkers with the implicit solvent model developed in this work, we performed the third test simulations, in which we studied the interactions of two amino acids, Lys and Phe, with the monolayer. These amino acids were selected, because they are important for Aβ binding to the monolayer (see Results below). Our simulation system differed from the one designed to probe the monolayer structure (first test simulations) only by the inclusion of a single amino acid (Lys or Phe). To compute the free energies F(z) for Phe and Lys, we used the 640 and 320 ns trajectories generated at 330 K, respectively. Due to convergence concerns we chose the temperature of 330 K, which is slightly higher than that used by Tieleman et al. [48]. The profile of F(z) for Phe shown in Fig. 2c is qualitatively similar to that obtained by Tieleman and coworkers (see Fig. 2c in [48]). Both profiles indicate that Phe penetrates deep into the monolayer (or bilayer) reaching the fatty acid groups (z ≲ −5 Å). The sharp increase in F(z) at z∼ −20 Å for the monolayer, which is absent in the bilayer simulations, is due to the layer of hydrocarbons (Fig. 1b). The free energy of Phe interaction with the monolayer is ∆F = −3.2 kcal/mol, where F b is the minimum F in the bound (or inserted) state (z < 10 Å) and F u is the baseline at z ≳10 Å corresponding to unbound Phe (Fig. 2c). For the monolayer, the free energy of the Phe interaction is ∆F = −3.9 kcal/mol compared to ∆F = −3.1 kcal/mol obtained in the explicit solvent simulations [48].
The profile of F(z) for Lys is qualitatively different from that of Phe (Fig. 2c). A pronounced minimum at z≈3Å reveals the propensity of this amino acid to bind to the surface monolayer groups without penetrating the monolayer volume. For the DOPC bilayer F(z) is similar except for the location of the minimum, which implies somewhat deeper penetration of Lys into the bilayer (up to the glycerol region, see Fig. 5c in [48]). A likely reason for the different locations of F(z) minima is larger partial charges assigned to the glycerol group in the force field of Berger et al. [49] compared to our implicit solvent parameterization. However, both F(z) profiles for the implicit and explicit solvent models do not reveal deep penetration of Lys into the monolayer (bilayer) as seen for Phe. The free energy of Lys interaction with the monolayer is ∆F = −3.2 kcal/mol, whereas ∆F for the DOPC bilayer is −2.9 kcal/mol.
Although our study addresses the thermodynamics of Aβ binding to the monolayer, we add one further line of comparison with experimental data by evaluating kinetic properties. To this end, we performed 80 ns simulations of monolayer without Aβ peptide at constant temperature of 330 K. From these simulations, we determined the lateral diffusion coefficient D xy for short time scale motions to be 5 × 10−7 cm−2 s−1, which is close to the experimental value of 10−7 cm−2 s−1 obtained for DPPC bilayer at 333 K using quasi-elastic neutron scattering [50].
Finally, it is important to assess the impact of the z-constraints imposed on monolayer atoms on Aβ binding. Based on experimental data, the spontaneous curvature of DMPC monolayer is estimated to be from 0.04 to 0.1 nm−1 [51]. The upper boundary in this estimate suggests that the radius of spontaneous curvature R c is about 100 Å. The value of R c for the DMPC bilayer should be about twice larger, i.e., R c~200Å. The size of Aβ monomer bound to the monolayer is <r 1N>≈31Å (see Results below). Because <r 1N> << R c, it appears that Aβ monomer bound to the bilayer experiences relatively small bending of the underlying surface. Note that the R c used represents the lower boundary. Therefore, we believe that the stiffness of the monolayer imposed by the constraints does not significantly perturb the surface adsorption of Aβ monomer.
Taken together, the comparisons of our monolayer model with experimental data and explicit water simulations suggest that the implicit solvent CHARMM19 force field and the DMPC monolayer approximately reproduce the structural properties of the bilayer and the lipid–amino acid interactions.
Results
Aβ monomer binds to DMPC monolayer
Binding of Aβ peptide to the DMPC monolayer was studied by computing the distributions of contacts between Aβ side chains and monolayer groups. At 360 K the probability of Aβ peptide binding to DMPC monolayer P b is≈1.0. Figure 3a shows the numbers of contacts formed by each Aβ residue, <C ml(i)>. On an average, an amino acid from the Nt and Ct terminals forms 2.7 and 2.3 contacts with the monolayer, respectively. In all, there are 79.4 contacts between Aβ and monolayer at 360 K. The plot of <C ml(i)> also reveals considerable variations in the interactions established by each residue. For example, Tyr10, His13, and Lys28 form the largest number of contacts (<C ml(i)>>3.0), whereas Val18 has the smallest number of interaction with the monolayer (<C ml(Val18)> = 1.6). To identify the set of residues responsible for Aβ binding, we assume that it includes the amino acids i, for which \( \left\langle {{C_{ml}}(i)} \right\rangle \geqslant 0.8{\max_i}\left\{ {\left\langle {{C_{ml}}(i)} \right\rangle } \right\} \). Then, the set of binding amino acids includes Tyr10, His13, Lys16, Phe20, and Lys28. Interestingly, four out of five binding residues are located in the Nt terminal indicating that it is the sequence region with the highest affinity for monolayer binding.
a The number of contacts, <C ml(i)>, formed by each Aβ residue i with the DMPC monolayer is shown by the thin black line. We define C ml(i) as the sum of contacts between a given residue i and four lipid structural groups. In the panel, <C ml(i)> is normalized by the largest value. The dotted line at 0.8 marks the threshold value of <C ml(i)> used to identify binding residues Tyr10, His13, Lys16, Phe20, and Lys28. The thick black line shows the normalized difference in the numbers of intrapeptide side chain contacts \( \left\langle {\varDelta C(i)} \right\rangle = \left( {\left\langle {C\left( {i;w} \right)} \right\rangle - \left\langle {C\left( {i;ML} \right)} \right\rangle } \right)/\left\langle {C\left( {i;w} \right)} \right\rangle \) formed by residues i in the monolayer-bound Aβ (ML) relative to those in water (w). The same quantity probing the change in intrapeptide interactions upon dimer formation is represented by the grey dashed line. b The probabilities P(z; i) probe the distribution of amino acids i along the axis z normal to the monolayer surface. The values of P(z; i) are color coded according to the right scale. Several amino acids (Tyr10, Hist13, Phe19, Phe20) are characterized by bimodal P(z; i) distributions implicating their partial insertion into the monolayer. c Probabilities of insertion P ins(i) defined as the probability of occurrence of amino acid i at z < 0, i.e., below the layer of phosphorous atoms
To investigate the distribution of amino acids along the axis z normal to the monolayer surface, we plot in Fig. 3b the probabilities P(z; i) for amino acids i to occur at the point z. This plot reveals uneven distributions of residues along the z axis. Most are characterized by unimodal distributions P(z; i), which peak at z≃4 Å, but there are a few for which P(z; i) is bimodal, featuring a second maximum at z < 0 Å (Fig. 3b). These amino acids partially penetrate the monolayer and interact with the DMPC glycerol groups and fatty acids. To quantify the insertion, we computed the probability of occurrence of amino acid i at z < 0, P ins (i) (Fig. 3c). This plot shows that, on average, the probability of insertion is low (P ins ≈0.13); however, for four residues (Tyr10, His13, Phe19, and Phe20), P ins(i) exceeds 0.2 (the respective probabilities are 0.27, 0.30, 0.22, and 0.26). Further analysis shows that these residues indeed form the largest number of interactions with the glycerol group and fatty acids. For example, for Tyr10, His13, Phe19, and Phe20, the fractions of contacts f c with these core groups out of all interactions with the monolayer are 0.32, 0.30, 0.29, and 0.32. Two other binding amino acids, Lys16 and Lys28, have lower probability of insertion [P ins(i) = 0.16)] and consequently the fractions of contacts f c formed by these residues with glycerol and fatty acid groups are reduced to 0.20 and 0.18. These values are close to the average f c computed for all amino acids (0.20). It also follows from Fig. 3c that the N-terminal has higher average insertion probability [P ins(Nt)≈0.17] than the C-terminal [P ins(Ct)≈0.10].
Conformational ensemble of Aβ bound to the DMPC monolayer
An important question pertains to the effect of Aβ binding to the DMPC monolayer on the peptide structure. To assess structural changes, we used as a reference the conformational ensemble of Aβ monomer in water investigated in our previous studies [26, 27]. First, we analyzed the effect of binding on the distribution of intrapeptide side chain interactions. Figure 3a shows the change in the number of intrapeptide side chain contacts <∆C(i)> formed by each residue i relative to that observed in water. Bound Aβ monomer retains <C(ML)> = 17.5 intrapeptide contacts compared to <C(w)> = 32.2 formed in water, i.e., bound Aβ losses about 45 % of intrapeptide contacts. The disruption of intrapeptide interactions occurs evenly in the Nt and Ct terminals (47 % vs 46 %). Analysis of hydrogen bonds (HBs) also implicates the loss of intrapeptide interactions. For example, the number of intrapeptide HBs decreases from 14.1 in water to just 3.3 in the bound Aβ.
The loss of intrapeptide interactions is consistent with the changes in the peptide end-to-end distance r 1N. Figure 4 compares the probability distributions P(r 1N) for Aβ monomers in water and bound to the monolayer. A shift in P(r 1N) to larger r 1N observed for the bound peptide results in the increase in the average <r 1N> from 18.3 Å in water to 30.7 Å for the bound Aβ (almost 70 % change). Therefore, Aβ peptide upon binding to the monolayer experiences a dramatic extension that should affect its secondary structure. This expectation is confirmed in Fig. 5, which compares the fractions of helix and β-strand structure, <H(i)> and <S(i)>, formed by amino acids i in the peptides bound to the monolayer and in water. The figure implicates unraveling of the helix structure and formation of β-strand. For example, the average fraction of the residues in helical conformation <H> decreases from 0.32 in water to 0.16 in the bound Aβ. Simultaneously, the fraction of β-strand residues <S> increases from 0.24 to 0.37. The changes in <S> and <H> are strongly inversely correlated (the correlation factor is −0.84) suggesting that binding to the monolayer induces helix→strand conversion in the peptide. Interestingly, according to the inset in Fig. 5 this conversion is confined primarily to the Nt terminal, in which <S(Nt)> rises by factor of 2.4 (from 0.17 in water to 0.40 in the bound Aβ), while the helix fraction <H(Nt)> drops more than two-fold (from 0.51 to 0.21). For comparison, the secondary structure changes in the Ct are minor (<S(Ct)> increases from 0.29 to 0.34 and <H(Ct)> decreases from 0.15 to 0.11). Aβ-monolayer interactions appear to be the factor driving this structural transition.
Distributions of secondary structure in Aβ peptide. Upper and lower panels show the fractions of helix and β-strand structure, <H(i)> and <S(i)>, formed by amino acids i in the peptides bound to the monolayer (black) and in water (grey). Inset Change in helix structure per residue, <∆H(i)>, occurring in the bound Aβ relative to that in water. The figure reveals helix→strand transition occurring in the bound Aβ peptide
To map the conformational ensemble of Aβ peptide bound to the monolayer, we used the clustering procedure described in ESM 1. Four major conformational clusters, C1 –C4, were identified, which together comprise 99 % of all structures (Table 1, Fig. 6). The most populated cluster C1 has the largest fraction of residues in β-conformation (S = 0.40) and the lowest helix content (H∼0.1). Consequently, few intrapeptide HBs are observed in this cluster. The contact map in Fig. 6a reveals multiple short range contacts with comparable probabilities (0.2–0.4). The second cluster C2 has a structure generally similar to that of C1. The important distinction of C2 is the formation of the stable contact Gly33–Gly37 with a probability of >0.8 (Fig. 6b). As a result, this cluster has an elevated number of intrapeptide contacts in the Ct and reduced fraction of residues in β-conformation. The structure of cluster C3 is different from that of other clusters. In particular, the distribution of secondary structure along Aβ is highly uneven. In the Nt, helix conformations occur almost twice as frequently than β-strand (0.38 vs 0.23), whereas the opposite trend is seen in the Ct terminal. According to Fig. 6c the formation of helix in the Nt is related to the appearance of the stable contact Val18–Glu22 (its probability >0.7). Consequently, C3 features the largest numbers of intrapeptide HBs (which increase three-fold compared to C1) and contacts. The cluster C4 has a secondary structure similar to that of C1 and C2, with the exception of the conformation of the turn (24–28), which contains a stable contact Gly25–Gly29 (its probability >0.9, Fig. 6d). Cluster C4 has also the smallest radius of gyration (R g = 19 Å). In all clusters, relatively minor variations are observed in the distributions of Aβ-monolayer interactions (Table 1). The Nt terminal always forms the largest number of contacts with the monolayer, whereas the number of Ct-monolayer interactions is reduced by 20–40 %.
Major structural clusters C1–C4 in the conformational ensemble of Aβ peptide bound to the DMPC monolayer. Superposition of Aβ structures from the clusters (thin grey lines) is performed by minimizing their backbone RMSDs. Representative structures for each cluster are shown in aqua. Contact maps visualize the average probabilities P(i, j) of contacts between the side chains i and j in the clusters. Blue spheres in C2–C4 represent the amino acids involved in the formation of the stable contacts that characterize these clusters. C1 has no distinctive stable long-range contacts
Impact of Aβ binding on the structure of DMPC monolayer
Experiments have indicated that the interaction of Aβ peptides with the bilayers may impact lipid structure [17, 52]. To check this possibility, we investigated the structural changes occurring in the monolayer upon Aβ binding. First, we have computed the average surface areas occupied by a lipid near Aβ binding site and in Aβ-free regions. To this end, two-dimensional Delaunay tessellation of the positions of phosphorous (P) atoms was performed and, for each P, a set of triangles was defined using the positions of its nearest neighbors [53]. The area A l per lipid is given approximately by the sum of areas of corresponding triangles divided by a factor of 3. Figure 7a shows the probability distributions P(A l) computed for the lipids that form contacts with the Aβ monomer, and for those that do not interact with the peptide. It can be seen that, for lipids in Aβ-free regions, the maximum in P(A l) is shifted to smaller A l compared to the lipids interacting with Aβ. Indeed, the average A l in the monolayer regions free of Aβ is 59 Å2, whereas for the lipids forming contacts with Aβ it is increased to 66 Å2. This observation suggests that the surface density of lipid molecules is decreased near the sites of Aβ binding.
a Probability distributions P(A l) of the monolayer surface areas per lipid molecule. b Radial pair correlation function g PP(r) measuring the surface density of phosphorous P atoms at the distance r from a given P. In both panels, the plots obtained in the vicinity of the Aβ-monolayer contact and in the peptide-free monolayer regions are in black and grey, respectively. In b, g PP,0 is the phosphorous surface density at r = 40 Å
To gain further insight into the monolayer structure, we computed the two-dimensional radial pair correlation functions g PP(r), which measure the surface density of phosphorous atoms at the distance r from a given P. Similar to the data shown in Fig. 7a, the functions g PP(r) in Fig. 7b are obtained separately for lipids forming contacts with Aβ and for those located in Aβ-free regions. In the latter case, g PP(r) oscillates due to local ordering of phosphorous atoms in well-defined concentric circles. However, the oscillations in g PP(r) are less pronounced around P atoms interacting with Aβ. For example, the amplitude of the peak associated with the nearest neighbor circle is reduced 25 % for lipids forming contacts with the peptide. This finding implies that the local ordering of lipids is compromised by Aβ interactions. We have also computed the carbon-deuterium bond order parameter S CD for lipids interacting with Aβ and located in the Aβ-free regions (data not shown). We found a negligible difference in S CD for these two classes of lipids, suggesting that Aβ binding causes a minor disturbance in the conformations of fatty acids. Taken together, Fig. 7 demonstrates that binding of Aβ peptide to the monolayer causes perturbation in the local lipid surface density.
Discussion
Binding to DMPC monolayer induces structural transition in Aβ peptide and perturbation in lipid packing
Using REMD sampling and the implicit solvent model we have investigated binding of Aβ monomer to DMPC lipid monolayer. We showed that Aβ peptide binds with high affinity to the monolayer and that there are considerable variations in binding propensities among amino acids. For instance, we have classified five amino acids, Tyr10, His13, Lys16, Phe20, and Lys28, as binding. Importantly, of those, four residues are located in the Nt terminal implicating that this sequence region has the highest affinity for binding. Furthermore, Aβ peptide not only binds to the monolayer, but some of its amino acids partially penetrate the monolayer volume. For example, for three binding residues (aromatic Tyr10, His13, and Phe20) the insertion probability P ins (i) is about 0.3 (the respective probabilities are 0.27, 0.30, and 0.26 in Fig. 3c). Further analysis shows that these residues form the largest number of interactions with the glycerol group and fatty acids. Interestingly, two other binding amino acids (polar Lys16 and Lys28) do not penetrate deep into the monolayer, because their insertion probabilities are significantly lower (P ins = 0.16 in Fig. 3c). These results are consistent with the recent free energy computations of amino acid interactions with lipid bilayer in explicit water [48] (see Methods subsection “Testing the accuracy of implicit solvent model”). That study has found that binding thermodynamics strongly favors deep penetration of Phe and Tyr into the bilayer core and the localization of Lys closer to the bilayer surface. Our results are also consistent with the recent investigation of the interactions between Aβ fibril protofilaments and bilayers [54]. In that study, the contacts formed by charged Aβ amino acids were identified as the driving binding factor. Therefore, our results suggest that binding of Aβ to the DMPC monolayer is governed by a mixture of charged and aromatic amino acids. Charged residues tend to interact with the polar surface lipid groups, whereas aromatic amino acids partially penetrate into the monolayer volume.
Our second and more important result is related to the changes in Aβ conformational ensemble occurring in response to binding to the monolayer. We have demonstrated that, upon binding, Aβ peptide loses about half of intrapeptide contacts (Fig. 3a), whereas the number of intrapeptide hydrogen bonds decreases more than four-fold. Simultaneously, the peptide extends dramatically by almost 70 % as measured by the end-to-end distance r 1N (Fig. 4). Loss of intrapeptide interactions and Aβ extension are consistent with the changes in secondary structure observed upon binding to the monolayer. We showed that the average helix content decreases two-fold coupled with the significant increase in the β-strand fraction, especially in the Nt terminal, where it rises by a factor of 2.4 (Fig. 5). Furthermore, cluster analysis showed that, in the most populated cluster C1, the β-strand fraction exceeds the helix fraction by nearly four-fold. These observations suggest that binding to the DMPC monolayer induces a helix→strand transition.
To further substantiate helix→strand transition we computed the autocorrelation function \( s(k) = \overline {\overrightarrow r (i)\overrightarrow r \left( {i + k} \right)} \), where \( \overrightarrow r (i) = {\overrightarrow R_{C\alpha }}\left( {i + 1} \right) - {\overrightarrow R_{C\alpha }}(i) \) is a backbone vector and \( {\overrightarrow R_{C\alpha }}(i) \) is the radius vector of the Cα atom in the residue i. The bar indicates an average over all i and equilibrated Aβ structures. The function s(k) probes the correlations in the chain orientation as a function of the distance k between residues. For the peptide bound to the monolayer s(k) monotonically decreases with k (Fig. 8a) indicating that the correlations in backbone orientation gradually diminish along the chain. This result is expected for the chain molecule forming extended β-like local structure. In contrast, pronounced oscillations in s(k) with the period ∆k≃4 are observed for Aβ monomer in water. This behavior of s(k) is indicative of the helical structure, which implies local ordering of peptide backbone. Thus, Fig. 8a serves as an illustration of the helix→strand structural transition occurring in Aβ due to binding to the monolayer. Our findings are in line with the well known observations that lipid membranes catalyze conformational changes in the bound peptides (e.g., in antimicrobial peptides) [55, 56].
a Function s(k) probes the correlations in the backbone orientation as a function of the number of sequence positions k between two Aβ residues: solid grey, dashed, and black lines are obtained for Aβ monomers in water, in the dimer, and bound to the DMPC monolayer, respectively. Oscillations in s(k) for Aβ monomer or dimer in water reflect the presence of helical structure. A monotonic decrease in s(k) for the bound Aβ points to dramatic structural reorganization involving helix unraveling induced by binding to the monolayer. b Number of Aβ-monolayer contacts <C ml> as a function of secondary structure fraction R, which is either helix R=H (grey line) or β-strand R=S (black line) fraction. The fraction R is considered as a parameter, for which <C ml> is computed. This plot suggests that the increase in β-strand structure coupled with helix melting are associated with the tighter binding to the monolayer
It is also possible to demonstrate directly that the helix→strand transition is indeed associated with interactions with the DMPC monolayer. To this end, we plot in Fig. 8b the number of Aβ-monolayer contacts <C ml> as a function of the helix H and β-strand S fractions. It follows from this figure that the increase in helix content is accompanied by the monotonic decrease in the number of contacts <C ml>. The opposite trend is observed for the β-structure content, which correlates with the tighter binding of Aβ to the monolayer. This conclusion is also consistent with our observation that the most pronounced changes in secondary structure occur in the Nt terminal (Fig. 5), which, according to the cluster analysis, and computations of insertion probabilities (Fig. 3c), has the highest affinity of binding to the monolayer.
It is interesting to compare Aβ conformational changes occurring upon binding to the monolayer with those observed during aggregation [26, 27]. Aβ monomer in the dimer forms <C> = 29.3 intrapeptide contacts, i.e., the loss of intrapeptide contacts is only 8 % compared to the water environment. According to Fig. 3a, the change in the numbers of intrapeptide contacts formed by each residue i in the dimer relative to that in water, <∆C(i)>, is small, and several Aβ sequence positions even gain intrapeptide interactions (<∆C(i)><0). These results are in sharp contrast to those found for the monolayer-bound Aβ, which experiences a dramatic loss of intrapeptide interactions. Consistent with these findings, the autocorrelation function s(k) in Fig. 8a reveals that formation of the dimer is not associated with the radical changes in the local ordering of Aβ backbone compared to the monomer in a water environment. Thus, the conformational changes induced in Aβ by binding to the DMPC monolayer are far more dramatic than those occurring in the initial stages of aggregation.
Our results should be compared with the all-atom explicit water simulations of Aβ 1–40 monomer interacting with DPPC bilayers [40]. Those simulations have shown that Aβ 1–40 inserted in the bilayer adopts a mostly helical structure in the C-terminal end, which persists for up to 90 ns after peptide exit from the bilayer and binding to its surface. In contrast, our simulations indicate that the helix structure content in the C-terminal is low. It is likely that the results of Xu et al. reflect slow structural dynamics of helix melting in the bound Aβ peptides. This interpretation of Xu et al.’s data is supported by experimental observations showing minor helix structure present in Aβ 1–40 peptides bound to zwitterionic DMPC bilayers [15].
Finally, we have found that Aβ adsorption on the DMPC monolayer reduces the surface density of lipid molecules near the sites of Aβ binding. Furthermore, Aβ binding causes disruption in surface packing of lipids as shown in Fig. 7b. Similar results have been reported recently for model phosphatidylcholine bilayers interacting with Aβ 1–40 peptides in explicit water [25]. These observations are consistent with the reduced ordering of lipids near the point of Aβ binding observed experimentally [29]. At the same time, as it follows from the computations of the carbon-deuterium bond order parameter S CD that Aβ binding does not significantly affect the conformations of fatty acids. This result is expected, because in our simulations Aβ monomer as a whole does not penetrate the monolayer volume. If the entire peptide penetrates the monolayer, a disordering effect on the conformations of fatty acids should be expected, as was recently found by Cheng and coworkers [25].
Comparison of Aβ peptide binding to the monolayer at 330 K and 360 K
To facilitate comparison with our previous simulations of Aβ peptides in water, the binding of Aβ to the monolayer was studied at elevated temperature of 360 K. It is important to ascertain that the mechanism of Aβ binding remains valid at temperatures lower than 360 K. To this end, we computed a number of Aβ structural quantities at 330 K and compared them with those at 360 K. Figure S5 shows the numbers of contacts, <C ml(i)> formed by each Aβ residue i with the DMPC monolayer at both temperatures. The figure also compares the numbers of intrapeptide side chain contacts <C i (i)>. It follows from Fig. S5 that both quantities are qualitatively similar at the two temperatures considered. The total number of Aβ-monolayer contacts increases from 79.4 (360 K) to 91.4 (330 K) and, using the same definition as in Fig. 3a, nine amino acids are identified as binding (Tyr10, His13, His14, Lys16, Phe19, Phe20, Asp23, Asn27, Lys28). Thus, as at 360 K, largely the same aromatic and charged residues are classified as binding at 330 K. There is little change in the total number of intrapeptide contacts (17.5 at 360 K vs 17.2 at 330 K), whereas in water at 330 K Aβ monomer forms 33.1 intrapeptide contacts. Therefore, similar to the results at 360 K, binding to the monolayer causes significant loss of intrapeptide interactions at 330 K. Figure S6 presents the probabilities of amino acid insertion into the monolayer at 360 K and 330 K. Although in general P ins(i) increase at lower temperatures, their distribution does not change qualitatively. At 330 K four amino acids with the highest P ins(i) are His13, His14, Phe19, and Phe20, which, apart from His13 also appear among the residues with the largest insertion probability at 360 K. At both temperatures, the N-terminal reveals higher values of P ins(i) than the C-terminal [the average P ins(i) values are 0.27 and 0.16, respectively]. To evaluate the changes in peptide extension, we have computed the distribution of the end-to-end distance r 1N. Figure S7 demonstrates that, at 330 K, Aβ monomer experiences an extension similar to that seen at 360 K as the average <r 1N> increases from 20.1 in water to 30.9 Å in the bound state. Finally, we probed the distributions of secondary structure at 330 K. The average fractions of β-strand and helical structures at 330 K are <S> = 0.35 and <H> = 0.15, whereas in water at the same temperature, <S> = 0.18 and <H> = 0.36. Therefore, as at 360 K, binding of Aβ to the monolayer at 330 K results in a sharp increase in β-content (by two-fold) matched by a considerable loss in helix conformations (more than two-fold drop). Hence, Aβ-monolayer interactions and changes in Aβ conformational ensemble induced by binding are qualitatively similar at 330 K and 360 K.
Structural reorganization in bound Aβ peptide may be generic
Are the conformational changes in Aβ peptide induced by binding to the lipid monolayer generic? To answer this question, we compared our findings with the recent explicit water MD study of short peptides interacting with water–octane interface [57]. In contrast to our work, the latter study used hydrophobic homogeneous octane phase in lieu of the membrane and the peptides composed of repetitive hydrophobic sequences (Gly–Ala and Gly–Val). This choice of simulation system allowed the authors to reveal generic effects likely to be independent of specific sequence or membrane composition. They concluded that the peptide conformational ensemble is affected strongly by binding to the octane surface due to the loss of intrapeptide interactions (such as hydrogen bonds) and chain elongation. Furthermore, surface-bound peptides were found to favor β-strand structures. Their observations are similar to our results, which are obtained for the chemically diverse system involving highly heterogeneous peptide sequence and zwitterionic lipid monolayer. Based on this comparison we argue that the structural reorganization in bound Aβ peptide may indeed represent a generic phenomenon applicable to various sequences and lipid membranes.
Conclusions
In this study, we investigated binding of Aβ monomer to a zwitterionic DMPC lipid monolayer. Our results suggest that Aβ binding to the monolayer is governed primarily by positively charged and aromatic amino acids. Lysine residues tend to interact with surface choline and phosphorous lipid groups, whereas aromatic amino acids penetrate deeper into the monolayer, reaching its hydrophobic core. The main result of our work is that binding to the DMPC monolayer causes a dramatic conformational transition in Aβ monomer, resulting in chain extension, loss of intrapeptide interactions, and formation of β-structure. Interestingly, this conformational transition is far more significant than that occurring during initial stages of aggregation in water. We also found that Aβ binding perturbs surface ordering of lipids interacting with Aβ.
References
Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297:353–356
Shoji M, Golde TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, Cai XD, McKay DM, Tintner R, Frangione B (1992) Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Science 258:126–129
Paravastua AK, Leapman RD, Yaua W-M, Tycko R (2008) Molecular structural basis for polymorphism in Alzheimers β-amyloid fibrils. Proc Natl Acad Sci USA 105:18349–18354
Petkova AT, Yau W-M, Tycko R (2006) Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry 45:498–512
Dobson CM (2003) Protein folding and misfolding. Nature 426:884–890
Haass C, Selkoe DJ (2007) Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimers amyloid β-peptide. Nat Rev Mol Cell Biol 8:101–112
Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA et al (2008) Amyloid-β protein dimers isolated directly from Alzheimers brains impair synaptic plasticity and memory. Nat Med 14:837–842
Arispe N, Diaz JC, Simakova O (2007) Aβ ion channels. prospects for treating Alzheimer’s disease with aβ channel blockers. Biochim Biophys Acta 1768:1952–1965
Nag S, Chen J, Irundayaraj J, Maiti S (2010) Measurement of the attachment and assembly of small amyloid-β oligomers on live cell membranes at physiological concentrations using single-molecule tools. Biophys J 99:1969–1975
Widenbrant MJO, Rajadas J, Sutardja C, Fuller GG (2006) Lipid-induced β-amyloid peptide assemblage fragmentation. Biophys J 91:4071–4080
Yip CM, Darable AA, McLaurin JA (2002) Aβ42-peptide assembly on lipid bilayers. J Mol Biol 318:97–107
Qulst A, Doudevski I, Lin H, Azimova R, Ng D, Franglone B, Kagan B, Ghiso J, Lal R (2005) Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci USA 102:10427–10432
Cizas P, Budvytyte R, Morkuniene R, Moldovan R, Broccio M, Losche M, Niaura G, Valincius G, Borutatite V (2010) Size-dependent neurotoxicity of β-amyloid oligomers. Arch Biochem Biophys 496:84–92
Kremer JJ, Murphy RM (2003) Kinetics of adsorption of β-amyloid peptide aβ(1–40) to lipid bilayers. J Biochem Biophys Methods 57:159–169
Bokvist M, Lindstrom F, Watts A, Grobner G (2004) Two types of Alzheimer’s β-amyloid (1–40) peptide membrane interactions: Aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J Mol Biol 335:1039–1049
Lin H, Bhatia R, Lal R (2001) Amyloid β protein forms ion channels: Implications for Alzheimer disease pathophysiology. FASEB J 15:2433–2444
Nakazawa Y, Suzuki Y, Williamson M, Saito H, Asakura T (2009) The interaction of amyloid aβ(1–40) with lipid bilayers ganglioside as studied by p31 solid state nmr. Chem Phys Lipids 158:54–60
Valincius G, Heinrich F, Budvytyte R, Vanderah DJ, McGillivray DJ, Sokolov Y, Hall JE, Losche M (2008) Soluble amyloid β-oligomers affect dielectric membrane properties by bilayer insertion and domain formation: implications for cell toxicity. Biophys J 95:4845–4861
Kakio A, Yano Y, Takai D, Kuroda Y, Matsumoto O, Kozutsumi Y, Matsuzaki K (2004) Interaction between amyloid β-protein aggregates and membranes. J Peptide Sci 10:612–621
Ma B, Nussinov R (2006) Simulations as analytical tools to understand protein aggregation and predict amyloid conformation. Curr Opin Struct Biol 10:445–452
Davis CH, Berkowitz ML (2009) Structure of the amyloid-β (1–42) monomer absorbed to model phospholipid bilayers: a molecular dynamics study. J Phys Chem B 113:14480–14486
Wang Q, Zhao J, Yu X, Zhao C, Li L, Jeng J (2010) Alzheimer Aβ 1–42 monomer adsorbed on the self-assembled monolayers. Langmuir 26:12722–12732
Janga H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL, Nussinov R, Lal R (2010) Truncated β-amyloid peptide channels provide an alternative mechanism for Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA 107:6538–6543
Miyashita N, Straub JE, Thirumalai D (2009) Structures of β-amyloid peptide 1–40, 1–42, and 1–55-the 672–726 fragment of APP—in a membrane environment with implications for interactions with γ-secretase. J Am Chem Soc 131:17843–17852
Qiu L, Buie C, Reay A, Vaughn MW, Cheng KH (2011) Molecular dynamics simulations reveal the protective role of cholesterol in β-amyloid protein-induced membrane disruptions in neuronal membrane minics. J Phys Chem B 115:9795–9812
Kim S, Takeda T, Klimov DK (2010) Mapping conformational ensembles of aβ oligomers in molecular dynamics simulations. Biophys J 99:1949–1958
Takeda T, Klimov DK (2009) Interpeptide interactions induce helix to strand structural transition in Aβ peptides. Proteins Struct Funct Bioinform 77:1–13
Takeda T, Klimov DK (2009) Probing the effect of amino-terminal truncation for abeta1-40 peptides. J Phys Chem B 113:6692–6702
Ege C, Lee KYC (2004) Insertion of Alzheimer’s Aβ40 peptide into lipid monolayers. Biophys J 87:1732–1740
Brooks BR, Bruccoler RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1982) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem 4:187–217
Ferrara P, Apostolakis J, Caflisch A (2002) Evaluation of a fast implicit solvent model for molecular dynamics simulations. Proteins Struct Funct Bioinform 46:24–33
Takeda T, Klimov DK (2009) Probing energetics of abeta fibril elongation by molecular dynamics simulations. Biophys J 96:4428–4437
Takeda T, Chang WE, Raman EP, Klimov DK (2010) Binding of non-steroidal anti-inflammatory drugs to aβ fibril. Proteins Struct Funct Bioinform 78:2859–2860
Hou L, Shao H, Zhang Y, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon I-JL, Ray DG, Vitek MP et al (2004) Solution NMR studies of the Aβ(1–40) and Aβ(1–42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J Am Chem Soc 126:1992–2005
Takeda T, Klimov DK (2009) Replica exchange simulations of the thermodynamics of Aβ fibril growth. Biophys J 96:442–452
Sergeant N, Bombois S, Ghestem A, Drobecq H, Kostanjevecki V, Missiaen C, Wattez A, David J-P, Vanmechelen E, Sergheraert C, Delacourte A (2003) Truncated beta-amyloid peptide species in pre-clinical alzheimers disease as new targets for the vaccination approach. J Neurochem 85:1581–1591
Bitan G, Vollers SS, Teplow DB (2003) Elucidation of primary structure elements controlling early amyloid β-protein oligomerization. J Biol Chem 278:34882–34889
Vitalis A, Caflisch A (2010) Micelle-like architecture of the monomer ensemble of Alzheimer’s amyloid-β peptide in aqueous solution and its implications for Aβ aggregation. J Mol Biol 403:148–165
Scheidt HA, Morgado I, Rothemund S, Huster D (2012) Dynamics of amyloid β fibrils revealed by solid state NMR. J Biol Chem 287:2017–2021
Xu Y, Shen J, Luo X, Zhu W, Chen K, Ma J, Jiang H (2005) Conformational transition of amyloid β-peptide. Proc Natl Acad Sci USA 102:5403–5407
Sugita Y, Okamoto Y (1999) Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett 114:141–151
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Ferrenberg AM, Swendsen RH (1989) Optimized Monte Carlo data analysis. Phys Rev Lett 63:1195–1198
Kucerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF (2005) Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J 88:2626–2637
Nagle JF, Tristram-Nagle S (2000) Structure of lipid bilayers. Biochim Biophys Acta 1469:159–195
Essex JW, Hann MM, Richards WG (1994) Molecular dynamics simulations of a hydrated phospholipid bilayer. Philos Trans Biol Sci 344:239–260
Aussenac F, Laguerre M, Schmitter J-M, Dufourc EJ (2003) Detailed structure and dynamics of bicelle phospholipids using selectively deuterated and perdeuterated labels. 2 H NMR and molecular mechanics study. Langmuir 19:10468–10479
MacCallum JL, Bennett WFD, Tieleman DP (2008) Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J 94:3393–3404
Berger O, Edholm O, Jahnig F (1997) Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure and constant temperature. Biophys J 72:2002–2013
König S, Pfeiffer W, Bayerl T, Richter D, Sackmann E (1992) Molecular dynamics of lipid bilayers studied by incoherent quasi-elastic neutron scattering. J Phys II 8:1589–1615
Orsi M, Michel J, Essex JW (2010) Coarse-grain modelling of DMPC and DOPC lipid bilayers. J Phys Condens Matter 22:155106
Kremer JJ, Sklansky DJ, Murphy RM (2001) Profile of changes in lipid bilayer structure caused by β-amyloid peptide. Biochemistry 40:8563–8571
Barber CB, Dobkin DP, Huhdanpaa HT (1996) The quickhull algorithm for convex hulls. ACM Trans Math Software 22:469–483
Tofoleanu F, Buchete N-V (2012) Molecular interactions of Alzheimer’s Aβ protofilaments lipid membranes. J Mol Biol 421:572–586. doi 10.1016/j.jmb.2011.12.063
Khandelia H, Langham AA, Kaznessis YN (2006) Driving engineering of novel antimicrobial peptides from simulations of peptide-micelle interactions. Biochim Biophys Acta 1758:1224–1234
White SH, Wimley WC (1998) Hydrophobic interactions of peptides with membrane interfaces. Biochim Biophys Acta 1376:339–352
Nikolic A, Baud S, Rauscher S, Pomes R (2011) Molecular mechanism of β-sheet self-organization at water-hydrophobic interfaces. Proteins Struct Funct Bioinform 79:1–22
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 853 kb)
Rights and permissions
About this article
Cite this article
Kim, S., Klimov, D.K. Binding to the lipid monolayer induces conformational transition in Aβ monomer. J Mol Model 19, 737–750 (2013). https://doi.org/10.1007/s00894-012-1596-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1596-8