Abstract
Isoproturon is the only herbicide that can control Phalaris minor, a competitive weed of wheat that developed resistance in 1992. Resistance against isoproturon was reported to be due to a mutation in the psbA gene that encodes the isoproturon-binding D1 protein. Previously in our laboratory, a triazole derivative of isoproturon (TDI) was synthesized and found to be active against both susceptible and resistant biotypes at 0.5 kg/ha but has shown poor specificity. In the present study, both susceptible D1(S), resistant D1(R) and D2 proteins of the PS-II reaction center of P. minor have been modeled and simulated, selecting the crystal structure of PS-II from Thermosynechococcus elongatus (2AXT.pdb) as template. Loop regions were refined, and the complete reaction center D1/D2 was simulated with GROMACS in lipid (1-palmitoyl-2-oleoylglycero-3-phosphoglycerol, POPG) environment along with ligands and cofactor. Both S and R models were energy minimized using steepest decent equilibrated with isotropic pressure coupling and temperature coupling using a Berendsen protocol, and subjected to 1,000 ps of MD simulation. As a result of MD simulation, the best model obtained in lipid environment had five chlorophylls, two plastoquinones, two phenophytins and a bicarbonate ion along with cofactor Fe and oxygen evolving center (OEC). The triazole derivative of isoproturon was used as lead molecule for docking. The best worked out conformation of TDI was chosen for receptor-based de novo ligand design. In silico designed molecules were screened and, as a result, only those molecules that show higher docking and binding energies in comparison to isoproturon and its triazole derivative were proposed for synthesis in order to get more potent, non-resistant and more selective TDI analogs.

Pharmacophore of modeled and MD simulated (D1/D2) PS-II reaction center of Phalaris minor has been simulated for de novo synthesis of a TDI (triazole derivative of isoproturon)-based molecule. De novo synthesized molecules were redocked and reranked as more potent and selective herbicides for P. minor. The figure shows the modeled and MD simulated structure of the PS-II reaction center (D1(R)/D2) of P. minor (left), and the docked conformation of TDI in the binding cavity of D1(R) protein (right)
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phalaris minor is a major weed of wheat that has become a serious threat to wheat production in India since the 1970s onwards [1]. Isoproturon—the first and only herbicide recommended for this weed—worked well for nearly 20 years. Due to its rigorous and continuous use, P. minor has developed resistance to isoproturon, as reported by Malik [2] and Walia [3]. To overcome the problem of isoproturon resistance, alternative herbicides were introduced onto the market in 1997–1998; viz., fenoxaprop, clodinafop, tralkoxydim and sulfosulfuron [4]. Some of these alternative herbicides have also developed multiple resistance to their modes of action [5]. Therefore, it is of the utmost important to keep changing and introducing new herbicides at regular intervals, i.e., before the development of resistance.
In our earlier work, we reported polymorphism in both biotypes of P. minor [6]. Later, three point mutations (G24S, V29A and F97L) in the D1(R) protein of the resistant biotype were reported by Tripathi et al. [7], clearly indicating that the P. minor populations have developed resistance to isoproturon because of alteration in the target site [7]. Isoproturon is a known inhibitor of PS-II and, like other urea herbicides, it binds to the to the D1 protein of the PS-II reaction center and inhibits electron transport by acting as a non-reducible analog of plastoquinone QB. To overcome the resistance in P. minor we have synthesized and tested 17 isoproturon analogs based on the pharmacophoric features of resistant D1(R) protein. Of these, a triazole derivative was found to be active at 0.5 kg/ha in both biotypes but had poor selectivity between wheat and weeds. This triazole derivative of isoproturon (TDI) causes undesirable growth retardation in wheat plants for a short period of time as compared to controls [8]. Singh et al. considered only protein D1, where a few residues at the N and C terminals of D1 were truncated that might play some structural role in the interaction between ligands and cofactor and D2 protein. In the present study, the whole reaction center D1/D2 of PS-II of P. minor has been modeled and energy minimized, followed by molecular dynamics (MD) simulation in a lipid biphasic system with ligands and cofactors. The constructed model was used in molecular docking and receptor based de novo drug design studies considering TDI as a lead molecule in order to synthesize more potent and selective analogs.
Materials and methods
Functional annotation of psbA
Amino acid sequence information of both susceptible [gi|31506000|gb|AAP47827.1| PsbA] and resistant D1(R) proteins [gi|30413115|gb|AAP33145.1| PsbA] of P. minor were taken from the NCBI database (http://www.ncbi.lm.nih.gov). IProClass functional annotation was performed to determine the family of the protein [9].
Phylogenetic analysis
Phylogenetic linkage analysis for the D1 protein of PS-II reaction center of Tritium aestivum, P. minor (both of same family) and T. elongatus (highest BLAST-p score) was performed using clustalW (http://www.genome.jp/tools/clustalw/).
D1 protein homology modeling
Template selection
BLAST-p (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) with protein data bank (PDB) for both D1 and D2 proteins was performed to find the nearest homologue.
Target-template alignment
The clustalW server (http://www.genome.jp/tools/clustalw/) was used for sequence alignment with the BLOSUM scoring matrix method using gap penalty 10 and gap extension 0.1. Sequence alignments for both susceptible and resistant D1(S) and D1(R) as well as for psbD sequence were generated.
Modeling
The sequence of the complete CDS was constructed with the aid of phylogenetic analysis. Sequences were aligned based on the sequence of theThermosynechococcus elongatus a/d-chain (Protein Data Bank code 2AXT) [10], and built based on the template 3D structures using Prime (Prime, v2.2, Schrodinger, LLC, New York, NY). Nearby ligands and cofactor were included in the model. Model of susceptible D1(S) protein, resistant D1(R) protein and D2 protein were constructed from the structure of bacterial reaction center.
Energy minimization
After model building of all proteins; the loop regions of susceptible D1(S), resistant D1(R) and D2 proteins were refined in Swiss PDB Viewer (SPDBV). Individual loops were minimized using 500 steps of steepest descent then subsequently by 100 steps of the conjugate gradient method. Harmonic constraints was used for loop energy minimization in SPDBV [http://spdbv.vital-it.ch/index.html]. The modeled D1 and D2 structures were finally energy minimized with Schrodinger Macromodel module (Macromodel, version 9.8, Schrodinger) using MMFFs force field in a water environment until converging at a termination gradient of 0.05 kJ mol−1 Å−1. The H-bonds were fixed using the SHAKE algorithm during MD.
Evaluation of structural model
Stability of the homology model was validated by the Structural Analysis and Verification Server (http://nihserver.mbi.ucla.edu/SAVES/), which has inbuilt tools such as PROCHECK, [11] WHAT_CHECK, WHATIF, PROSA, ERRAT, PROVE and VERIFY-3D.
Establishment of the reaction center
The D1/D2 reaction center was constructed with one bicarbonate, one β-carotene, five chlorophyll, two phenophytin and two plastoquinone ligands. Cofactors: iron and the oxygen evolving center (OEC) were retained along with D1/D2 protein. Two reaction centers, S and R, of P. minor were used for further MD simulation analysis [12].
MD simulation
Complete models of PS-II both resistant and susceptible, along with cofactors and ligands were simulated with GROMACS 4.0.7 in lipid environment of 1-palmitoyl-2-oleoylglycero-3-phosphoglycerol (POPG) [13]. Then itp files of ligands and cofactors were generated using the Dundee server [http://davapc1.bioch.dundee.ac.uk/prodrg/]. To maintain the compatibility with ligands and cofactors, the ffgmx force field parameter was used in GROMACS [14]. The simulation started with preprocessing of the D1/D2 protein reaction center with ffgmx force field parameter. Thereafter, ligands pdb were processed in the dundee server and added to protein.pdb files to generate a complete protein-ligand file [http://davapc1.bioch.dundee.ac.uk/prodrg/]. Four parameter files were used as input for MD simulations; energy minimization (em.mdp), temperature equilibration (nvt.mdp), pressure equilibration (npt.mdp) and MD (md.mdp) files [15] [http://cluster.earlham.edu].
Simulation in lipid environment
Lipid PDB was obtained from lipid book data bank (http://lipidbook.bioch.ox.ac.uk/lipid/show/id/2.html). Models were centered in a box of dimensions 5.90790:5.89520:7.54620. The solvent box started with a pre-equilibrated box of 128 POPG and 3,527 water molecules.
Co-ordinates of the PS-II reaction center complex and equilibrated solvent box were merged using the command genbox, which removes water molecules automatically where their Van der Waals radii would overlap with the protein structure. The whole system was then solvated with water, resulting in box of 5.90790:5.89520:7.54620 dimension with 128 POPG and 3,982 water molecules. All water present in the hydrophobic core was removed manually. To neutralize the system, water molecules were replaced with 138 Na+ and 10 Cl− ions to neutralize the overall system charge and provide an additional salt concentration. After ions were added, 3,962 water molecules remained in the system. The particle mesh Ewald (PME) method was used for calculation of coulombic interactions.
Additionally, sodium and chloride ions were added because explicit considerations of ions have beneficial effects on secondary structure stability [16]. The system was minimized for 20 ps using the steepest decent/conjugate gradient method, then equilibrated for pressure with isotropic pressure coupling (Tp = 1.0 ps) to 1 bar and temperature coupling (Tt = 0.1 ps) to 310 K, using a Berendsen protocol; the MD simulation was finally carried out for 1,000 ps using the parameters listed in Table S1.
Simulation of the binding site
CASTp program was used for cavity area and volume simulation in both susceptible and resistant biotype of PS-II reaction center [17]. Cavities that possess mutated residues were considered for molecular docking.
Docking
Autodock 3.0 [18] was used for docking of herbicide (isoproturon) with the reaction center proteins. TDI was also docked at the binding site, considering it as a lead molecule in order to enhance specificity for weeds (Fig. 1).
The structures of susceptible and resistant D1/D2 PS-II reaction centers were set up for docking with isoproturon and TDI. Amino acids at positions 62 and 67 were considered as the center of the grid map in both reaction centers of PS-II. The dimensions of the grids for susceptible reaction centers D1(S) /D2 proteins of PS-II were 36:36:36 Å, with the center coordinates (31.5, 26.5, and 64.0) close to the G62 and V67 of D1 protein. Similar parameters were used for resistant reaction center D1(R)/D2; the grid center was close to residues S62 and A67. Auto Dock3 with a genetic search algorithm was chosen for all dockings. The optimized Auto Dock run parameters were set for both isoproturon and TDI. Both molecules were preprocessed for docking with the help of autotors (root, and no. of rotatable bonds). Maximum number of energy evaluations, number of generation in the genetic algorithm (GA) and number of GA per run were set to 2,500,000; 2,700,000 and 50 respectively.
Receptor based pharmacophore and de novo drug design
The POCKET module of Ligbuilder [19] was used for receptor-based pharmacophore simulation for the best docked conformation of TDI. By running the POCKET program, the binding pocket from the 3D structure of PS-II models (represented in PDB format) was analyzed, and the information necessary to run the GROW module was prepared. The key interaction sites within the binding pocket were also derived. Fragments from TDI were used as seeds in the GROW module to construct the ligand molecules for the target protein by applying the growing strategy. Three subsets of TDI (1H-1,2,4-triazole; propan-2-ylbenzene; (1R)-2-amino-1-(dimethylamino)-1-(methylamino)ethanol) were used as seeds. The GROW module was used with population size of 3,000; a generation limit of 20; and a maximum output of 200 as a parameter. All the resultant molecules were collected in ligand_collection_file (ligbuilder lig file). The PROCESS module was used to extract the grown molecules from the Grow module with default chemical criteria except mol. weight, which was set to 400 Da. A total of 200 molecules was filtered out according to their binding affinities to the target protein.
Virtual screening of de novo simulated molecules
Molecules simulated by LigBuilder using a de novo receptor-based drug design approach were docked by the molegro virtual docker to rerank them in terms of docking affinity [20]. The binding site for the modeled D1/D2 reaction center was defined with a cavity volume of 30.208 Å and surface area of 116.4 Å. The center of the binding site was 31.38:33.02:69.14 in the X, Y and Z dimensions, covering an area with a radius of 12 Å. Grid resolution was set to 0.3 Å. Docking wizard parameters were as follows: number of runs 10; max iteration 1,500; max population size 50, using Mol Dock SE with maximum poses returned to one.
Results and discussion
Annotations of psbA CDS and phylogenetic analysis
Functional annotation has shown that the D1 protein belongs to superfamily Photo_RC_L/M (SSF81483) and family Photo_RC (PF00124). The print database identifies it as reaction center protein (PR00256) with five conserved motifs. Prosite accession no. (PS00244) has the signature for photosynthetic reaction center [NQH]-x(4)-P-x-H-x(2)-[SAG]-x(11)-[SAGC]-x-H-[SAG](2). INTERPRO, BLOCK ID CATH and SCOP ID are IPR000484, IPB000484, 1PRC and 2RCR, respectively. Functional annotation of the sequence has also shown that D1 proteins of P. minor, both susceptible and resistant is a psbA fragment of 263 amino acid residues, which is truncated at both N- and C-terminals and has a molecular weight of 28.538 kDa.
Phylogenetic analysis
PDB BLAST of the psbA amnio acid sequence of the D1 protein of P. minor has shown that it is closest to the wheat family while that of the psbD amino acid sequence of D2 protein has shown that it is closest to 2AXT of T. elongatus with highest score. To construct a complete CDS, phylogenetic analysis was performed. Pairwise analyses of sequences shows that, among the sequences analyzed, T. aestivum and P. minor have the highest alignment score. The phylogram was constructed by the ‘Neighbor-Joining using % identity’ method. The phylogram shows that P. minor is closer to T. aestivum (wheat) than to T. elongatus.
Based on this finding, they D1 protein of P.minor was aligned with refseq of T. aestivum (32 kDa) D1 protein (>gi|13928185|dbj |BAB47014.1|) to determine residues of the C and N terminii of the D1 protein. Alignment showed 38 residues (MTAILERRESTSLWGR FCNWITSTENRLYIGWFGVLMI) truncated at the N-terminus and 52 residues truncated at C-terminus (FNQSVVDSQGRVINTWADIINRANLGMEVMHERNAHNFPLDLAAVEVPS) of both D1 proteins. These truncated sequences were concatenated to both S and R D1 proteins for modeling, complete sequence were used for MD simulation.
Based on the phylogenetic relationships, the wheat reference sequence (|13928190|dbj|BAB47019.1| of PS-II D2 protein [Triticum aestivum]) from the NCBI reference sequence database was used for homology modeling and for construction of the PS-II reaction center (Fig. S1).
Homology modeling
Susceptible D1(S) protein and resistant D1(R) protein produced significant alignment with A-chain of PS-II of T. elongatus (pdb|2AXT|A, pdb|2AXT|AA), and D2 protein shows excellent alignment with D-chain of PS-II of T. elongatus (pdb|2AXT|DD) (Table S2). The 3.0 Å resolution structure of the bacterial reaction center was obtained from PDB. The crystal structure of T. elongatus PS-II model consists of 38 polypeptides along with 139 ligands and 10 cofactors [10]. Considering the sequence homology, for psbA sequence (D1) and for psbD (D2) were taken as templates [10].
D1 protein homology modeling
Modeling was performed by prime (Prime, v2.2, Schrodinger, LLC), ligands and cofactors were introduced into the model from the template, which was close to the reaction center. Incorporated molecules were one β-carotene, one bicarbonate ion, one phenophytin-a, two chlorophyll-a and two plastoquinones, along with cofactors iron and OEC. Modeled proteins were energy minimized (Fig. 2).
Modeled structure of a susceptible D1 protein; b resistant D1 protein; side chains of Ser62, Ala67, and Leu135 are shown in the figure. Structures are shown in ribbon drawing and ligands in stick form. Diagrams were generated using Yasara [http://www.yasara.org/]
D2 protein homology modeling
D2 protein was modeled using prime (Prime, v2.2, Schrodinger, LLC) with ligands; β-carotene (1), bicarbonate ion (1), chlorophyll-a (4), phenophytin-a (2), plastoquinone (2) and cofactor; iron (Fig. 3).
Validation
The results were validated at using the NIH server (http://nihserver mbi.ucla.edu/SAVS/ http://nihserver.mbi.ucla.edu/SAVES/}. Individual proteins modeled with overall quality factors of 91.584, 94.118, and 89.024 for susceptible D1(S) proteins, resistant D1(R) protein and D2 protein, respectively. Residues were checked for stereochemical parameters for both S and R biotypes and all the residues were in the allowed region of ϕ and φ angles, althouth the D2 protein has 1.7% generously allowed region (Table S3).
PROSA (https://prosa.services.came.sbg.ac.at/prosa.php) was used to check 3D models of protein structure for potential errors. The Z-score indicates overall quality and measures the deviation of the total energy of the structure with respect to an energy distribution derived from random conformations. A negative PROSA score indicates the correctness of the modeled protein. The PROSA profiles calculated for the modeled susceptible D1(S) protein and D2 protein were similar to the template A-chain and D-chain, respectively. The overall quality of the target D1 proteins of both S and R models was reflected in the Z scores of −2.75 and −2.40, respectively, which are comparable to the template Z-score −3.13. The Z-scores of the modeled D2 protein and template D-chain are very close, at −0.15 and −0.25, respectively. The PROSA potential energy curve of the modeled proteins were very similar to their respective templates.
Construction of PS-II reaction center
Two models of the reaction center—one for resistant biotype and another for susceptible biotype—were constructed. The ligands and cofactor common to D1 and D2 protein were removed. The final PS-II reaction center has D1(S/R) protein , D2 protein, ligands (one β-carotene, one bicarbonate ion, five chlorophyll-a, two phenophytin-a, two plastoquinone) and cofactors (iron and OEC).
MD simulation
MD simulation was performed for both susceptible and resistant D1(S/R)/D2 PS-II reaction centers of P. minor along with ligand and cofactors in a lipid environment. Here, we are giving the elaborated results of resistant D1/D2 PS-II reaction center analysis; the MD simulation data for the resistant and susceptible PS-II model are tabulated in Table S4a–c.
Simulation in lipid
RMSD analysis
Root mean square deviations of the backbone relative to the minimized and equilibrated structure and relative to the structure before minimization and equilibration (modeled reference) were calculated (Fig. 4). From the plots it is clear that, in the lipid environment, the structure deviates from the starting structure with an RMSD value of approximately ~0.325 nm. This is an acceptable value indicating that the structure is stable during simulation. Differences between the two RMSD plots indicate that the minimized and equilibrated structure at t = 0 ps is slightly different from the starting structure. The RMSD value relative to the energy-minimized structure at t = 0 ps is 0.03 nm, while that relative to the starting structure value is 0.13, and this happens due to imperfect position restraints.
a Root mean square deviation (RMSD) of backbone relative to the minimized and equilibrated structure. b RMSD of backbone relative to PS-II structure before minimization and equilibration. c Radius of gyration plot of PS-II model’s MD simulation in lipid environment. d Potential energy of PS-II model during simulation was almost stable and was of the order of 106 during the entire simulation in lipid environment
Radius of gyration
The radius of gyration (R g) is a measure of protein compactness. In the plot (Fig. 4c) of R g from t = 0 ps up to t = 100 ps, the R g value decreases very rapidly from ~2.683 to 2.635. Thereafter it varies in the range of 2.66 nm–2.63 nm, meaning that the R g values are almost static in this range, indicating a stable protein.
Potential energy
The potential energy plot (Fig. 4d) follows the hypothesis of native structure, meaning thereby that potential energy is negative and of the order of 106, which is an acceptable energy range for such a complex system.
Ramachandran score
All results, i.e., RMSD values, R g and potential energy analysis, show values in the acceptable range, indicating that MD simulation of a membrane protein in a lipid environment is the favorable environment, therefore the stability of the structure is maintained. Ramachandran score of structure in core, allowed and generously allowed regions are 82.9, 15.8 and 1.3, respectively (Fig. S2).
Deviation from template bacterial PS-II
RMSD deviation from template to the constructed D1(R)/D2 reaction center was 1.799 Å and 2.199 Å, for D1(R) protein and D2 protein. The ligands and cofactors have RMSD values slightly less 1.1564 Å. The whole reaction center deviates from its equivalent bacterial reaction center (template) by 1.7175 Å RMSD (Fig. 5). The simulated D1(S)/D2 reaction centers have slightly less deviation from the bacterial reaction center, with RMSD values of 1.025 Å. D1(S) and D2 proteins have RMSD values of 1.858 Å and 1.105 Å, respectively, while ligands and cofactors have an RMSD value of 0.112 Å.
a 3D structure of modeled PS-II reaction center of P. minor along with cofactors. D1 protein is shown in purple and D2 protein is shown in brown. Cofactors are shown in stick notation. b Modeled PS-II reaction center superimposed over the template bacterial reaction center. Template is shown in green and modeled is shown in pink. Ribbon drawing indicates helices, and cofactors are in stick form. Deviation between template and modeled reaction center is 1.7175 Å
The topology of both D1 and D2 revealed seven transmembrane alpha helices. The transmembrane alpha helices of D1 are denoted as 1,2,3,4,5,6, and 7. There are also several other non-transmembrane helices, which are based on bacterial template because the sequence of D1 is well conserved (Fig. 6).
Topology of both D1 and D2 generated using TopPred, a tool at Expasy server [http://www.cbib.u-bordeaux2.fr/pise/toppred.html]. D1 has seven transmembrane helixes spanning residues 35–55, 77–97, 109–129, 144–164, 168–188 and 196–216)
Docking study
The CASTp program was used for cavity area and volume simulation of the binding site in both susceptible and resistant biotype PS-II reaction centers [17]. The binding site has a small cavity that possesses mutated residues S62 and A67.
Docking was performed with AutoDock 3.0 to explore the difference in binding and docking energies of isoproturon and TDI with the S and R models of PS-II reaction center. Docking and binding energies of three best conformations were considered (Table S5a). The difference in docking energies seen in Table S5b and Table S6 explain why TDI is more effective. The best docked complex of the TDI and D1(R)/D2 reaction center was used for pharmacophore- and receptor-based drug design.
Pharmacophore study based on TDI
Ligbuilder was used for the de novo design of a new herbicide using TDI as lead molecule. A pharmacophore simulated by the pocket in resistant D1(R)/D2 is shown in Fig. 7b. A hydrophobic map of the TDI complex with the reaction center is shown in Fig. 7a. The distances between key interaction sites within the binding pocket were also derived and are listed in Table S7.
Proposed molecule for further synthesis
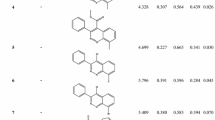
1H-1,2,4-triazole, propan-2-ylbenzene and (1R)-2-amino-1-(dimethylamino)-1-(methylamino)ethanol as fragments of TDI (lead molecule) were taken as a seed for de novo receptor-based ligand designing. These three seeds were grown in the pocket and processed, which finally yielded 200, 200 and 7 molecules. All these molecules were redocked to the binding site and the best five were taken from the set (Tables 1, 2, 3).
Concluding remarks
In our previous work addressing the resistance of P. minor to isoproturon and some other herbicides based on D1(R) protein, some derivatives of isoproturon were synthesized and proved effective in both biotypes. However, the most potent of these, viz. a triazole derivative of isoproturon (TDI) causes growth retardation of wheat for a short time [8]. In order to achieve further improvement of an earlier model, the N and C termini in D1(S/R) protein were both truncated and concatenated and thus a D1/D2 PS-II reaction center was constructed with the aid of phylogenetic analysis to construct a complete D1(S)/(R) protein. Sequence of the D2 protein of P. minor is not available therefore a reference sequence for the same family has been considered as a construct representing the D1/D2 reaction center along with ligands and cofactors to simulation the native environment for herbicide binding. This is the first time that the D1/D2 reaction center of PS-II of P. minor has been modeled and simulated for the design and development of herbicides.
Both susceptible and resistant D1(S)/(R) proteins were modeled after adding the N and C terminal in the amino acid sequence. The N and C termini were taken from wheat’s D1 protein reference sequence obtained from phylogenetic analysis. The structure of PS-II is a complex assembly of multiple peptides, ligands and cofactors. Therefore ligands and cofactors that are very close to the reaction center were considered to maintain their structural integrity during simulation. Finally, two models of the PS-II D1/D2 reaction center were constructed, one model contains susceptible D1(S) along with D2 and cofactors, the second model resistant D1(R) along with D2 and cofactors. Both models were energy minimized and simulated in GROMACS using the ffgmx force field. Since itp files of cofactors were generated by the dundee server, which uses the ffgmx force field, to maintain the compatibility with ligands and cofactors the ffgmx force field was used in GROMACS. As the D1/D2 reaction center is a complex system of membrane proteins, MD simulation was performed in a lipid (POPG) biphasic environment Table S4a–c. The susceptible D1(S)/D2 and resistant D1(R)/D2 reaction center deviate from its equivalent bactreial reaction center by 1.025 Å and 1.7175 Å RMSD, respectively (Fig. 7). The susceptible D1(S) protein containing the PS-II model deviates slightly less from the bacterial reaction center. The final modeled and simulated D1/D2 reaction center contains one β-carotene, one bicarbonate ion, five chlorophyll-a, two phenophytin-a, two plastoquinone and cofactors: iron and OEC (Fig. 7).
From the toplogy of the membrane protein, it is clear that the binding site lies in the stromal region of the first loop ranging from residues 55 to 77, which harbors both mutated residues S62 and A67 (Fig. 7). Isoproturon and TDI are both small hydrophobic molecules that could cross cell membranes due to ideal logP values and low molecular weight. This should favor diffusion of this hydrophobic molecule through the membrane.
From docking studies of isoproturon with susceptible and resistant D1/D2 PS-II reaction centers, it is clear that isoproturon binds more favorably to the susceptible D1(S) protein than to the resistant D1(R) protein (Table S5a). Docking studies have shown that TDI binds effectively to the resistant D1(R) protein with large differences in docking score (Table S5b). However, the docked conformation of isoproturon in the susceptible reaction center is more highly stabilized by hydrogen bonding than in the resistance reaction center. The best docked complex of TDI was used for pharmacophore generation followed by receptor-based drug design.
The key site of interaction and receptor based pharmacophore was simulated (Table S7; Fig. 7), clearly showing the complete loss of a hydrogen bond donor site at the binding site in resistant D1(R) protein, thus giving an explicit idea of the type of modification in the lead molecule. Based on the pharmacophore of the receptor, new molecules were simulated in very stringent conditions. These simulated molecules were further redocked to rank them using molegro virtual molecular docker (Tables 1–3). Structures and SMILE notation of the best five redocked molecules of each seed are given in Table S8. The binding affinity (Pkd) and log P of the simulated molecules are in the range of 6.88–7.90 and of 4.43–5.49, respectively (Tables 1, 2). These molecules also have higher moldock scores. Molecules simulated from 1H-1,2,4-triazole derivatives gave higher moldock scores. Therefore, considering feasible synthesis routes for 1H-1,2,4-triazole based derivatives, more effective and selective molecules could be synthesized and tested. Off target analysis (Table S7a–c) was also performed on the modeled wheat D1 protein (Fig. S3). These in vitro designed molecules showed less affinity with wheat D1 protein. Therefore, designed molecules could be more selective and more potent than the parent triazole derivative of isoproturon. Moreover knowledge from this theoretical PS-II reaction center model could be better utilized for the design and synthesis of new analogs of isoproturon, which are likely to substitute presently used herbicides for overcoming resistance.
Abbreviations
- MD:
-
Molecular dynamics simulation
- PS-II:
-
Photosystem-II
- D1/D2:
-
Photosystem second reaction center
- TDI:
-
Trizole derivative of isoproturon
- D1(R):
-
Resistant D1 protein
- D1(S):
-
Susceptible D1 protein
- POPG:
-
1-Palmitoyl-2-oleoylglycero-3-phosphoglycerol
- Tp:
-
Isotropic pressure coupling
- OEC:
-
Oxygen evolving center
- QB:
-
Plastoquinone B
- CDS:
-
Coding sequence
References
Bhan VM, Chaudhary DBS (1976) Germination, growth and reproducing behavior of Phalaris minor Retz as affected by the date of planting. Indian J Weed Sci 8:126–130
Malik RK, Singh S (1995) Little seed canary grass (Phalaris minor) resistance to isoproturon in India. Weed Technol 9:419–425
Walia US, Brar LS, Dhaliwal BK (1997) Resistant to isoproturon in Phalaris minor, Retz on Punjab. Plant Report 12:138–140
Yadav A, Malik RK, Balyan RS (1997) Studies on alternate herbicides to control isoproturon resistant littleseed canary grass. Pestology 21:26–28
Chhokar RS, Sharma RK (2008) Multiple herbicide resistant in little seed canary grass (Phalaris minor): a threat production in India Weed Bio Management 8:112–123
Singh DV, Gaur AK, Mishra DP (2004) Biochemical and molecular mechanism of resistance against isoproturon in Phalaris minor biotypes: variation in protein and RAPD profiles of isoproturon resistant and susceptible biotypes. Indian J Weed Sci 36:256–259
Tripathi MK, Yadav MK, Gaur AK, Mishra DP (2005) Resistant to isoproturon in Phalaris minor Retz on Punjab. Physiol Mol Biol Plants 11:161–163
Singh DV, Adeppa K, Misra K (2011) Mechanism of isoproturon resistance in Phalaris minor: in silico design, synthesis and testing of some novel herbicides for regaining sensitivity. J Mol Model. doi:10.1007/s00894-011-1169-2
Wu CH, Huang H, Nikolskaya A, Hu Z, Barker WC (2004) The iProClass integrated database for protein functional analysis. Comput Biol Chem 28:87–96
Loll B, Kern J, Zouni WA, Biesiadka J (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438:1040–1044
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Svensson B, Etchebest C, Tuffery P, van Kan P, Smith J, Styring S (1996) A model for the photosystem-II reaction center core including the structure of the primary donor P680. Biochemistry 35:14486–14502
Schlegel B, Sippl W, Holtje HD (2005) Molecular dynamics simulation of bovine rhodopsin: influence of protonation states and different membrane-mimicking environment. J Mol Model 12:49–64
Tieleman DP, Berendsen HJ (1996) Molecular dynamics simulation of fully hydrated DPPC with fully hydrated macroscopic boundary conditions and parameters. J Chem Phys 105:4871–4880
Astuti AD, Refianti R, Mutiara AB (2011) Molecular dynamics simulation on protein using GROMACS. Int J Comput Sci Inf Security 9:16–20
Ibragimova GT, Wade RC (1998) Stability of the beta-sheet of the WW domain: a molecular dynamics simulation study. Biophys J 74:2906–2911
Dundas J, Ouyang J, Tseng A, Binkowski Y, Turpaz LJ (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl Acids Res 34:116–118
Morris GM, Goodsell DS, Huey R, Olson AJ (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2 4. J Comput Aided Mol Design 10:293–304
Wang R, Gao Y, Lai L (2000) LigBuilder: a multiple-purpose program for structure-based drug design. J Mol Model 6:498–516
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1119 kb)
Rights and permissions
About this article
Cite this article
Singh, D.V., Agarwal, S., Kesharwani, R.K. et al. Molecular modeling and computational simulation of the photosystem-II reaction center to address isoproturon resistance in Phalaris minor . J Mol Model 18, 3903–3913 (2012). https://doi.org/10.1007/s00894-012-1386-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1386-3