Abstract
Triple negative breast cancer (TNBC) is immunohistochemically characterised by the lack of expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor type 2 (HER2). TNBC is known for its poor prognosis and high recurrence probability. There is no effective targeted treatment for TNBC, but only adjuvant chemotherapies. There are two TNBC subtypes, basal-like and non-basal-like, which are defined based on positive cytokeratin (CK) 5/6 and/or epidermal growth factor receptor (EGFR) expression. In particular, CK5/6 expression is reported to correlate with TNBC recurrence. TNBC lacks ER-α expression, but some TNBCs are known to express the androgen receptor (AR). Moreover, although p53 accumulation is detected in various malignant tumors, its influence on adjuvant chemotherapy for patients with TNBC remains unclear. The aim of this study was to assess the combined immunohistochemical expression of CK 5/6, AR, and p53 as a potential prognostic marker of adjuvant chemotherapy for patients with TNBC. The expression of CK5/6, AR, and p53 in formalin-fixed and paraffin-embedded (FFPE) surgical sections from 52 patients with TNBC was analysed by immunohistochemistry (IHC) and the co-expression patterns in individual cells were investigated by immunofluorescent (IF) staining. Low AR expression was correlated with high clinical stage (P < 0.05) and low nuclear grade (P < 0.05). The expression of CK5/6 and p53 did not correlate with clinicopathological features. Patients who needed adjuvant chemotherapy presented the worst prognosis. In particular, when the IHC expression pattern was CK5/6 (−), AR (−), and p53 (+), the disease free survival (DFS) and overall survival (OS) were the worst. On the other hand, patients with AR (+) and p53 (−) TNBC presented a good prognosis. The analysis of the co-expression status of these three markers showed that no cells presented both AR and CK5/6 expression. Furthermore, TP53 mRNA expression was higher in patients with AR-negative TNBC (P < 0.05) and in patients with the worst prognosis (P < 0.05) than in the other patients. These results suggested that, in patients with CK5/6-negative TNBC, AR expression correlated with good prognosis, but p53 accumulation correlated with poor prognosis. The present IHC markers allowed us to predict the post-surgery prognosis of patients with TNBC. In conclusion, TNBCs are heterogeneous. Patients with the CK5/6 (−), AR (−), and p53 (+) TNBC subtype, evaluated by IHC, presented the worst prognosis. These IHC markers will be helpful to follow patients with TNBC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is a heterogeneous disease with different forms that vary in morphology, biology, behaviour, and response to therapy. In 2000, Charles et al. identified several breast cancer subtypes by DNA microarray analysis; the prognosis for each of these subtypes differed significantly [1]. Sorlie et al. categorised breast tumors into five groups, luminal A, luminal B, human epidermal growth factor receptor type 2 (HER2), normal breast-like, and basal-like [2]. These intrinsic subtypes are defined as follows: the luminal A subtype is positive for estrogen receptor α (ER-α) and/or progesterone receptor (PR) and negative for HER2, the luminal B subtype is positive for ER-α and/or PR and positive for HER2 and/or the Ki67 score is over 14 %, the HER2 subtype is positive for HER2 but negative for ER and PR. Within the ER/PR/HER2 triple-negative phenotype, basal-like breast cancers are positive for basal cytokeratin (CK) 5/6, CK14, and CK17 and/or epidermal growth factor receptor (EGFR) and others are subtyped as normal breast-like breast cancers [3]. In clinical studies, CK5 was a more sensitive and specific biomarker than EGFR for basal-like breast cancer [4]. Moreover, positive CK5/6 correlated with worst recurrence in conjunction with c-Met expression and PTEN loss [5] and worst tumor grade [6]. Normal breast-like and basal-like subtypes are known as triple-negative breast cancers (TNBCs) that account for approximately 15 % of all types of breast cancer [7, 8]. This subtyping is important to select effective therapeutic strategies for breast cancer. For instance, anti-estrogens such as tamoxifen are selected to treat patients with the luminal subtypes, which express ER-α and/or PR. Additionally, molecular targeted therapies such as Herceptin® (trastuzumab) are selected to treat patients with HER2 overexpression or HER2 gene amplification. Patients with the TNBC subtype cannot be treated by using the current targeted therapies and usually present a worse prognosis than ER-α-positive breast cancer patients [2, 4, 5]. However, TNBCs still present some heterogeneity in term of biology, prognosis, and treatment sensitivity. TNBC cell lines have been classified by gene expression profiling and the therapeutic response of each subtype cell line was reported to be different [6, 9].
While TNBCs are negative for ER-α and PR expression, androgen receptor (AR) is reported to be expressed in 10–43 % of TNBCs [10, 11]. Sex steroids are thought to play important roles in the development of hormone-dependent human breast carcinoma [12]. Estrogens are the major sex steroids and contribute to tumor growth by binding with ER in breast carcinoma [12]. Invasive ductal breast carcinoma is ER-α negative, but AR positive, which activates the same transcription factors as ER-α [13]. AR expression in breast cancer is associated with an increase in cell proliferation through AR signalling [14]. However, the correlation between AR expression and prognosis remains unclear, because of conflicting reports regarding the influence of AR on tumor progression [7, 11, 15]. Henley et al. suggested that the lack of AR expression may play a critical role in the transformation of high-grade ductal carcinoma in situ (HG-DCIS) to high-grade invasive ductal carcinoma (HG-IDC) [16]. Moreover, androgen exerts a predominant inhibitory effect on the growth of breast cancer cells, both in vitro and in vivo, potentially through the induction of apoptosis [17]. Conversely, loss of AR was associated with poor prognosis in lymph node-positive ER/PR/Her2-negative breast cancers [18]. Additionally, in ER-α–negative breast cancer, the androgenic action may be mediated via direct interaction of dihydrotestosterone (DHT) metabolites with AR, regulating growth inhibition [19]. DHT concentrations have been correlated with TP53 and TP21 mRNA levels in a prostate cancer cell line [20]. In particular, the TP53 gene, which is a tumor suppressor gene located at 17p13, has been reported to be present in more than half of all cancers. It is related to poor prognosis and chemoresistance [21]. TP53 gene mutations are also found in 20–35 % breast carcinomas and are strongly associated with the ‘basal-like’ and ‘HER2+’ subtypes [22]. Although the TP53 gene mutation status in almost all TNBC cell lines has been reported [6, 9], a correlation between TP53 status and prognosis has not been found.
The aim of this study was to investigate the correlation between CK5/6, AR, and p53 expression and cancer prognosis. In particular, we investigated the potential of these markers to predict the prognosis of patients with TNBC after surgery. We assessed the expression of CK5/6, AR, and p53 by immunohistochemistry (IHC) and their co-expression status was observed by morphological immunofluorescence double staining. Moreover, TP53 mRNA levels in AR-positive TNBC or in AR-negative TNBC were also investigated by molecular analysis using laser microdissection (LMD) and quantitative real time reverse transcription-polymerase chain reaction (qRT-PCR) techniques, in order to determine the influence of AR expression on p53.
Materials and methods
Patients
Information from a total of 383 patients who were diagnosed with breast cancer and underwent modified radical mastectomy from 2000 to 2007 at the University Itabashi Hospital was obtained regarding ER, PR, and HER2 status. Of these cases, 52 patients (21.6 %) who were pathologically diagnosed with primary triple-negative invasive breast cancer were recruited in this study. The study was approved by the institutional review board of Nihon University Itabashi Hospital. All specimens were fixed with 20 % formalin and embedded in paraffin. The patients did not receive irradiation, chemotherapy, or hormonal therapy before surgery. Patient characteristics are presented in Table 1.
Immunohistochemistry
Immunohistochemistry (IHC) was carried out on 10 % formalin fixed paraffin embedded (FFPE) tissue sections using SimpleStain MaxPO Multi polymer system (Nichirei Biosciences Inc., Tokyo, Japan) and an autostainer (Histostainer, Nichirei Biosciences Inc.). For antigen retrieval, dewaxed, 4-μm tissue sections were autoclaved in citrate buffer (pH 6.0) for 15 min at 121 °C and cooled at room temperature. After washing several times in PBS pH 7.2, sections were processed to quench the endogenous peroxidase activity with 0.3 % hydrogen peroxide and to block non-specific binding with 1 % goat serum. Sections were incubated with each primary antibody, anti-mouse monoclonal CK5/6 antibody (Clone D5/16B4, 1:100, Dako, Glostrup, Denmark), anti-mouse monoclonal AR antibody (Clone AR441, 1:100, Dako), and anti-mouse monoclonal p53 (Clone DO-7, 1:100, Dako), for 30 min at room temperature. Lung squamous cell carcinoma, normal testis, and brain tumor were used as positive controls for CK5/6, AR, and p53, respectively. The epitope of anti-p53 antibody DO-7 is localised in the amino terminal domain, where rare TP53 gene mutations occur. However, this anti-p53 antibody detects the accumulation of mutated p53 proteins, because wild-type p53 protein is only present in small amounts in normal cells and cannot be detected by IHC with this antibody [23, 24]. Negative controls were not incubated with the primary antibody, but with dilution buffer. The tissue-bound HRP activity was visualized by immersing the sections in 0.005 % 3,3′-diaminobenzidine tetrahydrochloride (DAB) in PBS containing hydrogen peroxide (10 μL/150 mL DAB solution). Each section was counterstained with haematoxylin. In this study, the membranous expression of CK5/6 in more than 10 % of the tumor cells, the nucleic expression of AR in more than 5 % of the tumor cells, and nucleic expression of p53 in more than 1 % of the tumor cells were considered positive [7, 25].
Immunofluorescence
Sections (4-µm thick) were cut and dewaxed with xylene and ethanol. Formalin was then eliminated by treatment with 5 % ammonia and 95 % ethanol for 30 min at room temperature (RT). For antigen retrieval, tissue sections were autoclaved in citrate buffer (pH 6.0) for 15 min at 121 °C and cooled down at RT. After washing, sections were incubated with the three set mixed primary antibody including (A) anti-mouse monoclonal CK5/6 antibody (Clone D5/16B4, 1:100, Dako) and anti-rabbit monoclonal p53 antibody (Clone EPR17343, 1:500, Abcam, Cambridge, UK), (B) anti-mouse monoclonal CK5/6 antibody (Clone D5/16B4, 1:100, Dako) and anti-rabbit monoclonal AR antibody (Clone D6F11, 1:400, Cell Signaling Technologies, Danvers, MA, USA), and (C) anti-mouse monoclonal p53 antibody (Clone DO-7, 1:50, Dako) and anti-rabbit monoclonal AR antibody (Clone D6F11, 1:400, Cell Signaling Technologies) for 30 min at RT. After washing with PBS, sections were incubated with Alexa Fluor® 488 labelled anti-rabbit IgG in goat serum (1:500) and Alexa Fluor® 594 labelled anti-mouse IgG with goat serum (1:500) for 30 min at RT. After washing with PBS, sections were mounted with ProLong® Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific Inc., Waltham, MA, USA). Images were acquired using an Olympus IX71 fluorescence microscope (Olympus, Tokyo, Japan) and color images were generated using the Lumina Vision software (Mitani co., Tokyo, Japan) as follows. The signal of Alexa Fluor® 594 was visualized as red (CK5/6) and Alexa Fluor® 488 as pink (p53) in mixed primary antibody set A, the signal of Alexa Fluor® 594 was visualized as red (CK5/6) and Alexa Fluor® 488 as green (AR) in set B, and the signal of Alexa Fluor® 594 was visualized as pink (p53) and Alexa Fluor® 488 as green (AR) in set C.
Total RNA extraction from microdissected tumor tissue
Eight -μm thick FFPE sections were mounted on membrane film-coated glass slides. After dewaxing with xylene and ethanol, the sections were lightly stained with toluidine blue. The target tumor areas were then microdissected using a laser assisted microdissection system (PALM MBIII-N, Zeiss, Oberkochen, Germany). The microdissected target tumor cells were retrieved precisely into an Eppendorf lid with mineral oil. The laser assisted microdissection procedures have been previously described [26]. Additionally, benign mammary epithelial cells were also microdissected from 2 TNBC tissue sections.
The target tumor cell sample was mixed with 200 μL of denaturing buffer containing 2 % SDS, 0.1 mM EDTA, 10 mM Tris–HCl. The samples were then incubated at 55 °C with proteinase K until sections were completely dissolved. Total RNA was purified with 20 μL 2 M sodium acetate (pH 4.0), 220 μL citrate saturated phenol (pH 4.3), and 60 μL chloroform-isoamyl alcohol, centrifuged for 15 min at 15,000 rpm, and the upper aqueous layer was transferred into new tubes. Two hundred microliter of isopropanol and 2 μL of glycogen were added as a carrier and the samples were stored at −80 °C for more than 30 min, centrifuged at 14,000 rpm, washed with 70 % ethanol, and air dried on ice. They were then dissolved with 5–10 μL of RNase free water and quantified at the 260 nm optical density using Nanodrop 1000 (Thermo Fisher Scientific Inc.). Total RNA samples were stored at −80 °C until use. Both genomic DNA elimination and cDNA synthesis were performed by using QuantiTect Reverse Transcription Kit (QIAGEN, GmbH, Hilden, Germany) according to the manufacturer’s instructions.
Measurement of TP53 mRNA expression by quantitative real-time RT-PCR
Quantities of mRNA for TP53 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control were measured by quantitative real-time RT-PCR (qRT-PCR). qRT-PCR was performed with SYBR Green PCR Master Mix (Thermo Fisher Scientific Inc.). The forward and reverse primers were as follows: TP53 that was coding the proline rich rare mutated domain near the amino terminus of TP53 5′-gaagacccaggtccagatga-3′ and 5′-ttctgggaagggacagaaga-3′, and GAPDH 5′-ggaaggtgaaggtcggagtca-3′ and 5′-gtcattgatggcaacaatatccact-3′. qRT-PCR amplification and data analysis were performed by using ABI Prism 7000 Sequence Detection System (Thermo Fisher Scientific Inc.), with a 20 μL final reaction mixture containing 900 nmol/L each primer, 1× SYBR Green PCR Master Mix (Thermo Fisher Scientific Inc.). The reaction mixture was preheated at 95 °C for 10 min, followed by 45 cycles at 95 °C for 15 s and 60 °C for 1 min. Each TP53 mRNA relative value against the quantity of GAPDH mRNA was measured by using the ΔΔCt method [27].
Statistical analysis
Statistical analyses were performed using SPSS® Statistics version 20.0 (IBM Japan, Tokyo, Japan). The significance of the difference between the immunohistochemical expression of each target protein and patients’ clinicopathological status was evaluated by Chi squared test and Chi squared test with Yates’ correction. Correlation between TP53 mRNA expression levels and patient prognosis was determined by Mann–Whitney’s U test. While all patients, for whom complete clinical information was obtained, were investigated and followed up for 6–12 years, the number of patients who had events like recurrence and/or death was not enough to be analysed by Kaplan–Meier log-rank test. In this study, survival analyses were performed by using the data of disease free survival rate (DFS, %) and overall survival rate (OS, %).
Results
Detection of CK5/6, AR, and p53 by immunohistochemistry
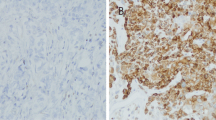
In specimens from patients with TNBC, CK5/6 was expressed in the cell membrane (Fig. 1a) in 28 cases (53.8 %, Table 1), but did not correlate with clinicopathological features. AR was expressed in the cytoplasm and nucleus of tumor cells (Fig. 1b) in 24 cases (46.2 %, Table 1). The number of patients with positive AR expression was significantly lower in the high clinical stage group (P < 0.05), and in the worst nuclear grade group (P < 0.05). AR tended to be higher in CK5/6-negative cases (58.3 %, Table 1), but no significant correlation was observed. Although p53 was expressed in the tumor cell nucleus (Fig. 1c) in 37 cases (71.2 %, Table 1), no correlation with clinicopathological features was detected. On the other hand, in benign mammary epithelium cells, strong CK5/6 expression and lower AR and p53 expression were detected (Fig. 1d–f, respectively).
Immunohistochemical expression of CK5/6, AR, and p53 (×40). Positive expression, measured by IHC, in TNBC cases is shown in a, b, and c. CK5/6 is expressed in the tumor cytoplasm and membrane (a). AR is expressed in the tumor cell nucleus and slightly expressed in the cytoplasm (b). p53 is expressed in the tumor cell nucleus (c). Benign regions are shown in d (strong membranous expression of CK5/6), e (low AR expression), and f (low p53 expression). Negative controls are shown in g (CK5/6), h (AR), and i (p53). The bar in each figure indicates 50 μm
CK5/6, AR, and p53 co-expression status
CK5/6, AR, and p53 co-expression status was assessed by immunofluorescence double staining as shown in Fig. 2. In the present study, the three mixed primary antibody set were used and the images were visualized using three colors as red (CK5/6), green (AR), and pink (p53). These color images were merged. Each typical type of TNBC with AR-only nucleic staining (Fig. 2a), p53-only nucleic staining (Fig. 2b), and CK5/6-only membranous and/or cytoplasmic staining (Fig. 2c) are shown. Double-positive TNBC types expressed both AR and p53 in the tumor cell nucleus (Fig. 2e, arrows) and CK5/6 and p53 in the cytoplasm and nucleus, respectively (Fig. 2f, arrows). Although there were few cases in which both CK5/6 and AR were expressed (Fig. 2g), membranous CK5/6 expression and nuclear AR expression were observed in different cells. Moreover, because cells with CK5/6, AR, and p53 triple expression were not found, the triple positive type corresponded to the heterogeneous combination of these expression types.
Merged color images of immunofluorescence staining were showed the expression pattern of CK5/6, AR, and p53 (×100 oil). a AR (+, green, nuclei), p53 (−), and CK5/6 (−) type. b AR (−), p53 (+, pink, nuclei), and CK5/6 (−) type. c AR (−), p53 (−), and CK5/6 (+, red, tumor cytoplasm and membrane) type, d AR (−), p53 (−), and CK5/6 (−) type, and nuclei were stained with DAPI (blue). e AR (+, green, nuclei), p53 (+, pink, nuclei), and CK5/6 (−) type (arrows). f AR (−), p53 (+, pink, nuclei), and CK5/6 (+, red, tumor cytoplasm and membrane) type (arrows). g AR (+, green, nuclei), p53 (−), CK5/6 (+, red, tumor cytoplasm and membrane) type. However, green and red images were found in individual cells (color figure online)
Immunohistochemical expression and patients’ prognosis
Most patients with TNBC can only be treated by surgery and the indication for adjuvant chemotherapy is determined according to the National Comprehensive Cancer Network Practice Guidelines in Oncology (NCCN guidelines®). For instance, adjuvant chemotherapy after surgery is recommended to patients with tumor larger than 1 cm and with primary tumor classified as pT1 (2 cm or less), pT2 (more than 2 cm, but not more than 5 cm), or pT3 (more than 5 cm). The pathological classification requires the resection and examination of at least the low axillary lymph node. Patients can then be classified as pN0 (no regional lymph node metastasis), pN1mi (micrometastasis larger than 0.2 mm and/or more than 200 cells, but none larger than 2 mm), or with positive lymph node metastasis. We investigated the correlation between AR and p53 expression and the prognosis of patients with TNBC receiving or not adjuvant chemotherapy. Patients who did not receive adjuvant chemotherapy presented a good prognosis (Table 2) regardless of AR and p53 expression. On the other hand, patients who received adjuvant chemotherapy presented a significantly worst DFS (P < 0.01) and OS (P < 0.05). DFS and OS tended to be worst in the patient group with positive p53 expression (P = 0.06 and 0.08, respectively). The correlation between CK5/6, AR, and p53 expression and prognosis of patients with TNBC who received adjuvant chemotherapy is presented in Table 3. The correlation between CK5/6 expression and prognosis was not significant. In CK5/6-positive patients with TNBC, AR-negative patients presented the worst DFS, but no significant correlation was observed. In CK5/6-negative patients, p53-positive patients presented the worst DFS and OS, but no significant correlation was detected. Furthermore, the prognosis of patients with TNBC correlated with the combined expression of AR and p53 in both CK5/6-positive and -negative cases (Table 4). The worst prognostic group corresponded with negative CK5/6 and AR expression, but positive p53 expression (P < 0.05). However, no significance was detected by Chi squared test with Yate’s correction, because of the small sample size. Patients with positive AR and negative p53 expression survived (100 %) instead of CK5/6 expression.
TP53 mRNA expression levels and prognosis of patients with CK5/6-negative TNBC
In this study, TP53 mRNA expression levels were measured in the tumor cells that were microdissected from FFPE sections of CK5/6-negative patients with TNBC. TP53 mRNA expression levels were significantly higher in malignant tumor cells microdissected from tissue sections patients with TNBC with good and poor prognosis than in benign mammary epithelial cells from the tissue sections of 2 patients with TNBC (0.02 ± 0.04 and 0.005 ± 0.004, respectively, P = 0.04, Fig. 3). TP53 mRNA expression levels tended to be higher in patients who had the worst prognosis (surviving with tumor and/or not-surviving) than in patients without tumor (0.07 ± 0.07 and 0.007 ± 0.01, respectively, P = 0.05, Fig. 3).
TP53 mRNA expression levels were significantly higher in malignant tumor cells microdissected from tissue sections of patients with CK5/6-negative TNBC, including patients with well and worse prognosis, than in no-malignant mammary epithelial cells from 2 TNBC tissue sections (P = 0.04). A well prognosis indicates that the patients survived without tumor and a worse prognosis indicates that the patients with cancer recurrence and/or who did not survive. TP53 mRNA expression levels tended to be higher in patients who had the worse prognosis than in patients who had a well prognosis (P = 0.05)
The results of the correlation between TP53 mRNA expression levels, patients’ prognosis, and AR expression are shown in Fig. 4. TP53 mRNA expression levels were higher in patients with cancer recurrence than in patients without recurrence (0.05 ± 0.06 and 0.007 ± 0.02, respectively, P = 0.04, Fig. 4a); the levels were also higher in patients who died than in patients who survived (0.07 ± 0.06 and 0.007 ± 0.01, respectively, P = 0.03, Fig. 4b). TP53 mRNA expression was significantly higher in the AR-negative group than in the AR-positive group (0.09 ± 0.06 and 0.01 ± 0.02, respectively, P = 0.04, Fig. 4a) in patients with cancer recurrence. Moreover, in patients who did not survive, TP53 mRNA expression was significantly higher in the AR-negative group than in the AR-positive group (0.10 ± 0.06 and 0.02 ± 0.00, respectively, P = 0.03, Fig. 4b). In both CK5/6- and AR-negative patients with TNBC, TP53 mRNA expression level was significantly higher in both patients with cancer recurrence and in those who did not survive.
TP53 mRNA expression levels were measured in microdissected tumor cells from CK5/6-negative TNBC cases that needed adjuvant chemotherapy. a In patients with TNBCs with recurrence, TP53 mRNA expression level was higher than in patients without cancer recurrence (P = 0.04). In particular, TP53 mRNA level was higher in AR-negative tumors (P = 0.04) in patients with recurrence. b TP53 mRNA expression level was higher in the non-surviving patients than in those who survive (P = 0.03). In particular, TP53 mRNA expression was higher in AR-negative tumors (P = 0.03) from patients who did not survived
Discussion
Cancer is heterogeneous, and genetic information is important to choose effective therapeutic strategies [4]. In breast cancer, different subtypes have been classified by using data from genomic studies [1]. For instance, hormonal breast cancer therapy is selected for patients who express hormone receptors (ER-α and PR), and molecular targeted therapy is prescribed to patients who present HER2 amplification. On the other hand, most patients with TNBC (without ER, PR, and HER2 expression) are known to present the worst prognosis and no specific and effective therapy is available for such cases. However, it does not mean that all patients with TNBC present a poor prognosis. In fact, there are individual differences in the prognosis after surgery. In this study, we investigated whether or not CK5/6, AR, and p53 correlated with the prognosis of patients with TNBC after surgery. In our presented TNBC cases, low risk patients, who required no adjuvant chemotherapy according to the clinical guidelines, presented a good prognosis. Only some of the high-risk patients, who required adjuvant chemotherapy according to the clinical guidelines, presented the worst prognosis. We attempted to classify the high-risk patients who needed adjuvant chemotherapy based on the immunohistochemical staining for CK5/6, AR, and p53. These immunohistochemical markers have been heavily used in breast cancer. CK5/6 is one of the basal-like subtype markers and its correlation with worst recurrence of patients with TNBC has been reported [28]. Loss of AR is associated with poor prognosis in patients with lymph node-positive ER/PR/Her2-negative breast cancer [18]. While most of the anti p53 antigens detect the amino terminus domain of p53 protein, wild type p53 protein is not detected because wild type p53 exists in small amount in normal cells, but mutant p53 proteins are detectable in malignant cells [23, 24, 29]. In the present study, the patient group with negative CK5/6 and AR expression but positive p53 expression had the worst DFS and OS. Additionally, the co-expression pattern of CK5/6 and AR in a single cell was not found. Within the same tumor, the cells of invasive ductal carcinoma did not co-expressed CK5/6 and AR. Recently, AR expression was shown to correlate with EGFR in patients with TNBC and the antiproliferative effect of anti-androgen therapy with an EGFR inhibitor was reported [30]. While both CK5/6 and EGFR are basal-like breast cancer markers, the correlation with AR expression may be quite different. Moreover, Pintens et al. reported that, within 25 patients with TNBC, 16.0 % of the cases presented EGFR (HER1) gene amplification and these were EGFR positive, but negative for CK5/6 expression [31]. Patients with CK5/6-negative but EGFR-positive TNBC may have to be classified as an independent subtype to consider the indication for EGFR inhibitors. However, the present results suggest that patients with AR-positive and p53-negative TNBC presented a good prognosis, although AR in breast cancer was reported to increase cell proliferation via AR signalling [14].
Recently, TNBC cell lines were classified by using molecular analyses [1, 6, 32]. According to TNBC subtypes, based on genetic analyses, different chemotherapeutic responses were studied in vitro [32]. In the present study, we attempted to classify patients with TNBC using immunohistochemical staining for CK5/6, AR, and p53 as prognostic markers for adjuvant chemotherapy in vivo. A simple IHC panel can be easily used in clinical practice. Although TP53 gene mutations is reported in almost all TNBC cell lines [6, 9], no significant correlation between TP53 mutations and poor prognosis has been found. In the CK5/6-negative TNBC group, TP53 mRNA expression level was higher in the AR-negative group than in the AR-positive group and in patients who died. Androgens are known as the main regulators of prostate growth and differentiation [20]. However, the biphasic effects of androgens and AR activities on prostate cell proliferation have been reported [20, 21, 33]. TP53 and TP21 mRNA levels depend on the concentration of dihydrotestosterone (DHT) in HNTEP cells. In cell lines with wild type TP53 gene treated with DHT, the active AR signalling pathway regulates p53 expression and induces p53-dependent apoptosis and growth inhibition [20, 21]. The lowest concentration of DHT induced a decrease in TP53 and TP21 mRNA expression in HNTEP cells [20]. However, in TP53 mutant cells, the active AR signalling pathway did not regulate p53-dependent apoptosis, but drug sensitivity increased [21]. In any case, TP53 and TP21 expression levels seem to depend on AR signalling activity. On the other hand, AR downregulation through suppression of p300, which is involved in p53 degradation through ubiquitin [34], increased p53-dependent apoptosis [33]. Usually, a decrease in AR expression induces p21 and p53 downregulation and tumor progression. In the present TNBC cases, TP53 mRNA levels were higher in the AR-negative group, but patients presented the worst prognosis. The present qRT-PCR method detected both wild type and mutant TP53 mRNA. However, in benign mammary epithelium, p53 expression, measured by IHC, was quite low, as was TP53 mRNA expression. Thus, the wild type p53 is expressed in small amounts in benign cells. The IHC and qRT-PCR methods used herein allowed the detection of mutant p53 protein accumulation and mutant TP53 mRNA. Thus, in patients with AR-negative TNBC, the tumor progressed despite a higher TP53 mRNA expression, suggesting that the aberrant p53 protein accumulation occurs as a result of mutant TP53 mRNA overexpression. Further studies are required to investigate the TP53 gene mutation status. Our results show that TP53 mRNA expression level was increased in patients with the worst prognosis. These results suggest that the prognosis of high-risk patients after surgery can be predicted by assessing the expression pattern of CK5/6, AR, and p53 by IHC. Our results suggest that the worst prognosis is associated with the following expression pattern: CK5/6 (−), AR (−), and p53 (+). Recent molecular subtyping has become an important tool for the classification of various cancers. However, morphological molecular analyses, in particular IHC, allow the individual evaluation of molecular expression status under the microscope, and can be applied for routine clinical tests since it is a simple, effective, and low cost method. In conclusion, TNBCs are heterogeneous and the expression status of CK5/6, AR, and p53 assessed by IHC influences the effect of adjuvant chemotherapy. The present results indicate the potential of the evaluation of, in particular, AR and p53 expression status in CK5/6-negative TNBC to predict patients’ prognosis after surgery. These results suggest that patients with CK5/6 (−), AR (+) and p53 (−) TNBC presented the best prognosis, but patients with CK5/6 (−), AR (−), and p53 (+) TNBC presented the worst prognosis for adjuvant chemotherapy.
References
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406:747–752
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98:10869–10874
Reis-Filho JS, Tutt AN (2008) Triple negative tumours: a critical review. Histopathology 52:108–118
Cuenca-López MD, Montero JC, Morales JC, Prat A, Pandiella A, Ocana A (2014) Phospho-kinase profile of triple negative breast cancer and androgen receptor signaling. BMC Cancer 14:302 (Epub ahead of print)
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA (2007) Triple-negative breast cancer; clinical features and patterns of recurrence. Clin Cancer Res 13:4429–4434
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121:2750–2767
Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO (2007) Prognostic markers in triple-negative breast cancer. Cancer 109:25–32
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. Cancer J Clin 64:9–29
Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10:515–527
Mrklić I, Pogorelić Z, Capkun V, Tomić S (2013) Expression of androgen receptors in triple negative breast carcinomas. Acta Histochem 115:344–348
McGhan LJ, McCullough AE, Protheroe CA, Dueck AC, Lee JJ, Nunez-Nateras R, Castle EP, Gray RJ, Wasif N, Goetz MP, Hawse JR, Henry TJ, Barrett MT, Cunliffe HE, Pockaj BA (2014) Androgen receptor-positive triple negative breast cancer: a unique breast cancer subtype. Ann Surg Oncol 21:361–367
Suzuki T, Miki Y, Takagi K, Hirakawa H, Moriya T, Ohuchi N, Sasano H (2010) Androgens in human breast carcinoma. Med Mol Morphol 43:75–81
Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS (2011) Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J 30:3019–3027
Cops EJ, Bianco-Miotto T, Moore NL, Clarke CL, Birrell SN, Butler LM, Tilley WD (2008) Antiproliferative actions of the synthetic androgen, mibolerone, in breast cancer cells are mediated by both androgen and progesterone receptors. J Steroid Biochem Mol Biol 110:236–243
Fioretti FM, Sita-Lumsden A, Bevan CL, Brooke GN (2014) Revising the role of the androgen receptor in breast cancer. J Mol Endocrinol 52:R257–R265
Hanley K, Wang J, Bourne P, Yang Q, Gao AC, Lyman G, Tang P (2008) Lack of expression of androgen receptor may play a critical role in transformation from in situ to invasive basal subtype of high-grade ductal carcinoma of the breast. Hum Pathol 39:386–392
Kimura K, Markowski M, Bowen C, Gelmann EP (2001) Androgen blocks apoptosis of hormone-dependent prostate cancer cells. Cancer Res 61:5611–5618
Conzen SD (2008) Minireview: nuclear receptors and breast cancer. Mol Endocrinol 22:2215–2228
Nahleh Z (2008) Androgen receptor as a target for the treatment of hormone receptor-negative breast cancer: an unchartered territory. Future Oncol 4(1):15–21
Pozzobon A, Schneider L, Brum IS (2012) Androgen-modulated p21 and p53 gene expression in human non-transformed epithelial prostatic cells in primary cultures. Int J Mol Med 30:967–973
Serafin AM, Akudugu JM, Bohm L (2002) Drug resistance in prostate cancer cell lines is influenced by androgen dependence and p53 status. Urol Res 30:289–294
Bertheau P, Espié M, Turpin E, Lehmann J, Plassa LF, Varna M, Janin A, de Thé H (2008) TP53 status and response to chemotherapy in breast cancer. Pathobiology 75:132–139
Bennett WP, Hollstein MC, Metcalf RA, Welsh JA, He A, Zhu SM, Kusters I, Resau JH, Trump BF, Lane DP, Harris CC (1992) p53 mutation and protein accumulation during multistage human esophageal carcinogenesis. Cancer Res 52:6092–6097
Kihana T, Tsuda H, Teshima S, Okada S, Matsuura S, Hirohashi S (1992) High incidence of p53 gene mutation in human ovarian cancer and its association with nuclear accumulation of p53 protein and tumor DNA aneuploidy. Jpn J Cancer Res 83:978–984
Soreid JA, Lea OA, Varhaug JE, Skarstein A, Kvinnsland S (1992) Androgen receptors in operable breast cancer: relation to other steroid hormone receptors, correlations to prognostic factors and predictive value for effect of adjuvant tamoxifen treatment. Eur J Surg Oncol 18:112–118
Nakanishi Y, Shimizu T, Tsujino I, Obana Y, Seki T, Fuchinoue F, Ohni S, Oinuma T, Kusumi Y, Yamada T, Takahashi N, Hashimoto S, Nemoto N (2013) Semi-nested real-time reverse transcription polymerase chain reaction methods for the successful quantitation of cytokeratin mRNA expression levels for the subtyping of non-small-cell lung carcinoma using paraffin-embedded and microdissected lung biopsy specimens. Acta Histochem Cytochem 46:85–96
Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DTW, Jordan RCK (2002) Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analysis. Mod Pathol 15:979–987
Inanc M, Ozkan M, Karaca H, Berk V, Bozkurt O, Duran AO, Ozaslan E, Akgun H, Tekelioglu F, Elmali F (2014) Cytokeratin 5/6, c-Met expressions, and PTEN loss prognostic indicators in triple-negative breast cancer. Med Oncol 31:801
Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, IeM Shih, Kurman RJ (2011) Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol 24:1248–1253
Cuenca-Lopez MD, Montero JC, Morales JC, Prat A, Pandiella A, Ocana A (2014) Phospho-kinase profile of triple negative breast cancer and androgen receptor signaling. BMC Cancer 14:302
Pintens S, Neven P, Drijkoningen M, Van Belle V, Moerman P, Christiaens MR, Smeets A, Wildiers H, Vanden Bempt I (2009) Triple negative breast cancer: a study from the point of view of basal CK5/6 and HER-1. J Clin Pathol 62:624–628
Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ (2007) The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol 1:84–96
Tan J, Chen B, He L, Tang Y, Jiang Z, Yin G, Wang J, Jiang X (2012) Anacardic acid (6-pentadecylsalicylic acid) induces apoptosis of prostate cancer cells through inhibition of androgen receptor and activation of p53 signaling. Chin J Cancer Res 24:275–283
Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, Zhang PJ, Li AL, Zhang XM (2009) Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci USA 106:3788–3793
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maeda, T., Nakanishi, Y., Hirotani, Y. et al. Immunohistochemical co-expression status of cytokeratin 5/6, androgen receptor, and p53 as prognostic factors of adjuvant chemotherapy for triple negative breast cancer. Med Mol Morphol 49, 11–21 (2016). https://doi.org/10.1007/s00795-015-0109-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00795-015-0109-0