Abstract
Two alkaliphilic strains, designated FJAT-45086T and FJAT-45122T, were isolated from alkali soli in Nima County, Tibet, China. Both strains were Gram-positive, rod-shaped and shared low 16S rRNA gene sequence similarity with the members of the genus Bacillus. They contained meso-diaminopimelic acid as the cell-wall diamino acid and MK-7 as the menaquinone. The major fatty acids (>5%) of strain FJAT-45086T were anteiso-C15:0, C16:0, iso-C15:0, C16:1ω11c and anteiso-C17:0, whereas strain FJAT-45122T consisted of iso-C15:0, anteiso-C15:0, iso-C17:1ω10c, iso-C17:0, anteiso-C17:0, C16:0 and C16:1ω11c. The genome G + C content of strains FJAT-45086T and FJAT-45122T was 37.8 and 38.2 mol%, respectively. The polar lipids of strain FJAT-45086T were diphosphatidyl glycerol (DPG), phosphatidyl glycerol (PG), phosphatidyl ethanolamine (PE), and phosphatidyl choline (PC), whereas strain FJAT-45122T consisted of DPG, PG, phosphatidyl methyl ethanolamine (PME) and an unidentified aminophospholipids (UAPL). The average nucleotide identity values of strains FJAT-45086T and FJAT-45122T were below the cut-off level (95–96%) for species delineation. Based on the results, strains FJAT-45086T and FJAT-45122T represent two novel species of the genus Bacillus, for which the names Bacillus alkalisoli sp. nov., and Bacillus solitudinis sp. nov., are proposed. The type strain, FJAT-45122T (=DSM 104631T = CCTCC AB 2016254T), FJAT-45086T (=DSM 104056T = CCTCC AB 2016232T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alkaliphiles generally refer to microorganisms that grow at high pH values (Horikoshi 1999). Recent increased interest in alkaliphilic Bacillus species has led to the discovery of many novel Bacillus species (Nogi et al. 2005; Borsodi et al. 2011; Liu et al. 2019) and at the time of writing, there are more than 35 recognized alkaliphilic Bacillus species (https://www.bacterio.net/bacillus.html). During the course of investigating microbial diversity of saline soil in Nima County, Tibet, China, two alkaliphilic and moderately halophilic Bacillus strains designated FJAT-45086T and FJAT-45122T were isolated. The 16S rRNA gene sequence analysis showed that these strains shared low sequence similarity with the members of the genus Bacillus, and as a result, we sought to establish their taxonomic position based on phenotypic, biochemical, phylogenetic, chemotaxonomic, and comparative genome analysis.
Materials and methods
Sample collection, isolation, and preservation
Strains FJAT-45086T and FJAT-45122T were isolated from saline soil in Nima County, Tibet, China (87°18.378N, 31°57.655E). The sample was serially diluted and an aliquot (100 μL) was spread on Horikoshi-1 medium (DSMZ medium1081). The plate was incubated at 30 ºC for 1 week. The colonies obtained were repeatedly re-streaked on the same medium until pure colonies were obtained and stored as glycerol suspensions (20%, w/v) at − 80 °C and in lyophilized form in skimmed milk at room temperature.
Phenotypic, microscopic, and growth conditions
Gram staining and the KOH lysis test were carried out according to the methods described by Gregersen (1978) and Smibert and Krieg (1994). Colony morphology was observed on Horikoshi-1 medium after 48 h of incubation under optimal growth conditions. The size of the cells was determined by transmission electron microscopy (Hitachi, Japan). Endospores were examined according to Schaeffer–Fulton staining method (Murray et al. 1994).
Growth at various temperatures (10, 15, 20, 25, 30, 37, 45, and 50 °C), NaCl concentrations (0, 1, 3, 5, 7, 10, 15, 20, 25%, w/v) and pH [6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, 10.0, 10.5, 11.0, 12.0, 13.0; with the buffer solutions of KH2PO4/NaOH (pH 6.0–8.0), and NaHCO3/Na2CO3 (pH 8.0–13.0)] were tested in LB medium. The growth was measured by measuring the optical density (OD) values at a wavelength of 600 nm using the UV spectrophotometry (SHIMADZU UV-2550, Japan).
Catalase activity was determined by investigating bubble production with 3% (v/v) H2O2, and oxidase activity was determined using 1% (v/v) tetramethyl-p-phenylenediamine. Cell growth under anaerobic conditions was determined in a CO2 incubator on anaerobically prepared media. Hydrolysis of starch and Tweens (20, 40, 60, and 80) were performed according to the conventional methods described by Smibert and Krieg (1994). Other physiological and biochemical characteristics were confirmed using API 20E, API 50 CHB and API ZYM strips (BioMérieux, France) following the manufacturer’s instructions.
The sensitivity of antibiotics such as clindamycin (2 µg), streptomycin (300 µg), vibramycin (30 µg), erythromycin (15 µg), gentamycin (120 µg), rifampicin (5 µg), vancomycin (30 µg), azithromycin (15 µg), penicillin (10 µg), cefazolin (30 µg), ampicillin (10 µg), chloromycetin (30 µg), tetracycline (30 µg), oxacillin (1 µg), amikacin (30 µg), polymyxin B (300 µg), gentamycin (10 µg), kanamycin (30 µg), and neomycin (30 µg) were also tested.
16S rRNA gene sequence and phylogenetic analysis
The genomic DNA was extracted using DNA extraction kit (Shanghai Generay Biotech Co., Ltd, China) according to the manufacturer’s instructions. The 16S rRNA gene was amplified and sequenced using primers and the conditions described previously (Dong et al. 2019). The obtained 16S rRNA gene sequence was compared with available sequences of cultured species in the EZBioCloud server (https://www.ezbiocloud.net/) (Yoon et al. 2017). Phylogenetic trees were constructed using the neighbor-joining (NJ) (Saitou and Nei 1987), maximum-likelihood (ML) (Felsenstein 1981) and maximum-parsimony (MP) (Fitch 1971) methods implemented with MEGA version 7 (Kumar et al. 2016) after multiple alignments of data by CLUSTAL_X (Thompson et al. 1997) using Kimura 2-parameter model (Kimura 1980). The reliability of each branch was evaluated by bootstrap analysis based on 1000 replications (Felsenstein 1985).
Chemotaxonomy
Chemotaxonomic characters of strains FJAT-45086T and FJAT-45122T and the reference strains were observed using several standard methods under identical conditions. Analysis of the isomer of diaminopimelic acid was performed according to the procedures described by Hasegawa et al. (1983) and Lechevalier and Lechevalier (1970). Menaquinone was analyzed as described by Collins et al. (1977) using reverse-phase HPLC (Kroppenstedt 1982). Extraction and analysis of polar lipids by two-dimensional TLC was performed according to described by Minnikin et al. (1979). For determination of cellular fatty acids, cells were harvested after cultivation on Horikoshi-1 plates at 30 °C for 48 h. The cellular fatty acids in the cell walls were extracted and analyzed according to the standard protocol of the Microbial Identification System (MIDI) tested by Gas Chromatography (model 7890, Agilent) (Sasser 1990).
Genome sequencing and average nucleotide identity (ANI) values
The genome sequence of strains FJAT-45086T and FJAT-45122T was sequenced by the Beijing Novogene Bioinformatics Technology Co., Ltd (China). Reads of each data set were filtered, and high-quality paired-end reads were assembled using SOAPdenovo (version 2.04) (Luo et al. 2012). The tRNAs were predicted using tRNAscan-SE (Lowe and Eddy 1997). Average nucleotide identity (ANI) values were calculated using Jspecies software (Richter et al. 2016).
Results and discussion
Morphological and biochemical characterization
Strains FJAT-45086T and FJAT-45122T were Gram-stain-positive, motile, rod-shaped, endospore-forming (Supplementary Fig. S1) and facultative anaerobes. Colonies on Horikoshi-1 medium were pale yellow, circular, raised, smooth, and non-transparent. Growth of strain FJAT-45086T was observed at 15–35 °C (optimum 25–30 °C) and pH 6.0–12.0 (optimum pH 9.0), while strain FJAT-45122T grew at 10–40 °C (optimum 30 °C) and pH 7.5–12.0 (optimum pH 8.0–9.0). Tolerance to NaCl of strains FJAT-45086T and FJAT-45122T was up to 10% (w/v).
Catalase and oxidase activities of two strains were positive. Nitrate reduction of FJAT-45122T was positive, but negative for FJAT-45086T. The hydrolysis of Tweens (20 and 60) of strain FJAT-45086T was positive, but negative for strain FJAT-45122T. FJAT-45086T was sensitive to clindamycin, vibramycin, erythromycin, gentamycin, rifampicin, vancomycin, azithromycin, penicillin G, cefazolin, treptomycin, ampicillin, chloromycetin, tetracycline, neomycin, gentamycin, oxacillin, amikacin, kanamycin, and polymyxin B, whereas strain FJAT-45122T was sensitive to clindamycin, streptomycin, vibramycin, erythromycin, gentamycin, rifampicin, vancomycin, azithromycin, penicillin G, cefazolin, ampicillin, chloromycetin, tetracycline, neomycin, oxacillin, amikacin, and kanamycin, but not to polymyxin B and gentamycin. Detailed differentiating features of the strains FJAT-45086T, and FJAT-45122T and their closest members in the genus Bacillus are mentioned in Table 1.
16S rRNA gene sequence and phylogenetic analysis
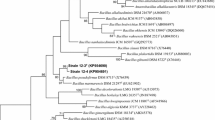
Strain FJAT-45086T had the highest 16S rRNA gene sequence similarity with Bacillus hemicellulosilyticus JCM 9152T (96.5%), while strain FJAT-45122T had the highest 16S rRNA gene sequence similarity with Bacillus halmapalus DSM 8723T (96.9%). The maximum-likelihood tree (Fig. 1) showed that FJAT-45086T and FJAT-45122T clustered with the members of the genus Bacillus. The cluster was further found to be stable when trees were reconstructed using the neighbour-joining and maximum-parsimony algorithms (Supplementary Figs. S2 and S3).
Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences of strains FJAT-45086T, FJAT-45122T and closely related species within the genus Bacillus. Bootstrap values (expressed as percentages of 1000 replications) greater than 50% are shown at branch points. Bar, 0.02 substitutions per nucleotide position. Paenibacillus polymyxa IAM 13419T is used as outgroup
Chemotaxonomy
Strains FJAT-45086T and FJAT-45122T contained meso-2,6-diaminopimelic acid as the cell-wall diamino acid and MK-7 as their menaquinone which was consistent with other Bacillus members (Rao et al. 2019; Dou et al. 2016). The major fatty acids (>5%) of strain FJAT-45086T were anteiso-C15:0, C16:0, iso-C15:0, C16:1ω11c, and anteiso-C17:0, whereas strain FJAT-45122T contained iso-C15:0, anteiso-C15:0, iso-C17:1ω10c, iso-C17:0, anteiso-C17:0, C16:0 and C16:1ω11c. Detailed major fatty acid (>5%) profiles of strains FJAT-45086T and FJAT-45122T and their closely related Bacillus members were mentioned in Table 2. The polar lipids of strain FJAT-45086T were diphosphatidyl glycerol (DPG), phosphatidyl glycerol (PG), phosphatidyl ethanolamine (PE), and phosphatidyl choline (PC), whereas strain FJAT-45122T consisted of DPG, PG, phosphatidyl methyl ethanolamine (PME) and an unidentified aminophospholipids (UAPL) (Supplementary Fig. S4).
Strain FJAT-45086T total genome size was 4,664,253 (bp) with N50 length of 4,601,752, while strain FJAT-45122 T total genome size was 3,892,817 (bp) with an N50 length of 3,829,525 (bp). A total of 104 and 81 tRNAs were predicted for FJAT-45086T and FJAT-45122T, respectively. The genome G + C content of strains FJAT-45086T and FJAT-45122T were 37.8 and 38.2 mol%, respectively. The ANIb and ANIm values between FJAT-45086T, FJAT-45122T and the closest members ranged 67.4–88.2% (Table 1) which were below the cut-off level (95–96%) recommended as the ANI criterion for interspecies identity (Goris et al. 2007; Richter and Rosselló-Móra 2009). Based on the above results, strains FJAT-45086T, and FJAT-45122T represent two novel species of the genus Bacillus, for which the names Bacillus alkalisoli sp. nov. and Bacillus solitudinis sp. nov. are proposed.
Description of Bacillus alkalisoli sp. nov.
Bacillus alkalisoli (al.ka.li.so'li. N.L. n. alkali (from Arabic al-qaliy) alkali; L. neut. n. solum soil; N.L. gen. n. alkalisoli of alkaline soil).
Cells are Gram-staining-positive, motile and facultatively anaerobic. Cell size is 0.5–0.8 × 3.3–5.5 μm. Colonies are pale yellow, circle, smooth, non-transparent, and raised. Endospores are located at the terminal position. The average colony diameter is approximate 1.64 ± 0.29 mm. Grows at 15–35 °C (optimum 25–30 °C), and at pH 6.0–12.0 (optimum pH 9.0). Tolerance to NaCl is up to 10.0% (w/v) (optimum at 4.0%). Catalase and oxidase activities are positive but negative for nitrate reduction. Urease and hydrolysis of Tweens (20, 40, and 60) are positive, but arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, citrate utilization, H2S and indole production and hydrolysis of gelatin, starch, and Tween 80 are negative. In API 50CHB system, positive for d-arabinose, glucose, fructose, mannose, α-methyl-d-glucoside, esculin, maltose, lactose, and 5-keto-d-gluconate. According to API ZYM, produces esterase lipase (C8), naphthol-AS-BI-phosphohydrolase and β-galactosidase, but not esterase (C4), alkaline phosphatase, lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, α-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and β-fucosidase. The cell-wall peptidoglycan contains meso-diaminopimelic acid and MK-7 as the only menaquinone. The polar lipids are diphosphatidyl glycerol (DPG), phosphatidyl glycerol (PG), phosphatidyl ethanolamine (PE), and phosphatidyl choline (PC). The major fatty acids are anteiso-C15:0, C16:0, iso-C15:0, C16:1ω11c, and anteiso-C17:0. The DNA G + C content is 37.8 mol %.
The type strain, FJAT-45086T (=DSM 104056T = CCTCC AB 2016232T), was isolated from alkali soil in Nima County, Tibet, China.
The GenBank accession numbers for the 16S rRNA gene and the genome sequence are KY612311 and NJAX00000000, respectively.
Description of Bacillus solitudinis sp. nov.
Bacillus solitudinis (so.li.tu'di.nis. L. fem. gen. n. solitudinis of a desert).
Cells are Gram-staining-positive, motile, and facultatively anaerobic. Colonies are pale yellow, circle, smooth, non-transparent, and raised. The average colony diameter is approximately 1.15 ± 0.05 mm. Cell size is about 0.2–0.4 × 1.9–2.2 μm. Endospores are located at the terminal position. Grows at 10–40 °C (optimum 30 °C), pH at 7.5–12.0 (optimum pH 8.0–9.0). Tolerance to NaCl is up to 10.0% (w/v) with optimum growth at 1.0–4.0% NaCl (w/v). The catalase, oxidase, and nitrate reduction are positive. The hydrolysis of gelatin, starch, Tween 40 are positive, but negative for H2S, hydrolysis of Tweens (20, 60 and 80) urease, citrate utilization, indole production, arginine dihydrolase, lysine decarboxylase, and ornithine decarboxylase. In API 50 CHB system, positive for glucose, sorbose, rhamnose, sorbitol, arbutin, esculin, lactose, sucrose, trehalose, starch, glycogen, d-turanose, gluconate, and 5-keto-d-gluconate. In API ZYM, positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), chymotrypsin, naphthol-AS-BI-phosphohydrolase, α-glucosidase, but negative for lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, acid phosphatase, α-galactosidase, β-glucuronidase, β-glucosidase, β-galactosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and β-fucosidase. The cell-wall peptidoglycan contains meso-diaminopimelic acid and MK-7 as the menaquinone. The DNA G + C content is 38.2 mol %. The polar lipids are diphosphatidyl glycerol (DPG), phosphatidyl methyl ethanolamine (PME), phosphatidyl glycerol (PG), and unidentified aminophospholipids (UAPL). The major fatty acids are iso-C15:0, anteiso-C15:0, iso-C17:1ω10c, iso-C17:0, anteiso-C17:0, C16:0 and C16:1ω11c.
The type strain, FJAT-45122T (=DSM 104631T = CCTCC AB 2016254T), was isolated from alkali soil in Nima County, Tibet, China.
The GenBank accession numbers for the 16S rRNA gene and the genome sequence are KY612312 and NISO00000000, respectively.
Abbreviations
- ANI:
-
Average nucleotide identity
- DPG:
-
Diphosphatidyl glycerol
- PME:
-
Phosphatidyl methyl ethanolamine
- PG:
-
Phosphatidyl glycerol
- PC:
-
Phosphatidyl choline
- UAPL:
-
Unidentified aminophospholipids
References
Borsodi AK, Pollák B, Kéki Z, Rusznyák A, Kovács AL, Spröer C, Schumann P, Márialigeti K, Tóth EM (2011) Bacillus alkalisediminis sp. nov., an alkaliphilic and moderately halophilic bacterium isolated from sediment of extremely shallow soda ponds. Int J Syst Evol Microbiol 61:1880–1886
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Dong ZY, Narsing Rao MP, Wang HF, Fang BZ, Liu YH, Li L, Xiao M, Li WJ (2019) Transcriptomic analysis of two endophytes involved in enhancing salt stress ability of Arabidopsis thaliana. Sci Total Environ 686:107–117
Dou G, Liu H, He W, Ma Y (2016) Bacillus lindianensis sp. nov., a novel alkaliphilic and moderately halotolerant bacterium isolated from saline and alkaline soils. Antonie Van Leeuwenhoek 109:149–158
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P et al (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Gregersen T (1978) Rapid method for distinction of Gram-negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:123–127
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Horikoshi K (1999) Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev 63:735–750
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded Ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2367
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Liu GH, Rao MPN, Dong ZY, Wang JP, Che JM, Chen QQ, Sengonca C, Liu B, Li WJ (2019) Genome-based reclassification of Bacillus plakortidis Borchert et al. 2007 and Bacillus lehensis Ghosh et al. 2007 as a later heterotypic synonym of Bacillus oshimensis Yumoto et al. 2005; Bacillus rhizosphaerae Madhaiyan et al. 2011 as a later heterotypic synonym of Bacillus clausii Nielsen et al. 1995. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-019-01299-z
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Luo R, Liu B, Xie Y et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. https://doi.org/10.1186/2047-217X-1-18
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Murray RGE, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 21–41
Nogi Y, Takami H, Horikoshi K (2005) Characterization of alkaliphilic Bacillus strains used in industry: proposal of five novel species. Int J Syst Evol Microbiol 55:2309–2315
Rao MPN, Dong ZY, Zhang H, Niu XK, Zhang K, Fang BZ, Xiao M, Kang YQ, Li WJ (2019) Bacillus antri sp. nov., isolated from cave soil. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.003473
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC News 20:16
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC, pp 607–654
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
This work was financially supported by the External cooperative project of Fujian Academy of Agricultural Sciences (Grant no.: DEC201821209), The Science and Technology Innovation Team Program of Fujian Academy of Agricultural Sciences (Grant no.: STIT2017-1-11), the Natural Science Foundation of Fujian Proince, China (Grant no.: 2017J01048), and the Agrobiogenome Program of Fujian Academy of Agricultural Sciences (Grant no.: A2015-3, A2017-4).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that they had no conflict of interest.
Additional information
Communicated by A. Oren.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/ENA/DDBJ accession numbers for 16S rRNA genesequence of strains FJAT-45086T and FJAT-45122T are KY612311 and KY612312, respectively. The Whole Genome Shotgun project of strains FJAT-45086T and FJAT-45122T have been deposited at DDBJ/ENA/GenBank under the accession NJAX00000000 and NISO00000000, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, GH., Narsing Rao, M.P., Dong, ZY. et al. Two novel alkaliphiles, Bacillus alkalisoli sp. nov., and Bacillus solitudinis sp. nov., isolated from saline-alkali soil. Extremophiles 23, 759–764 (2019). https://doi.org/10.1007/s00792-019-01127-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-019-01127-2