Abstract
Cuproxidases are a subset of the blue multicopper oxidases that catalyze the oxidation of toxic Cu(I) ions into less harmful Cu(II) in the bacterial periplasm. Cuproxidases from psychrophilic, mesophilic, and thermophilic bacteria display the canonical features of temperature adaptation, such as increases in structural stability and apparent optimal temperature for activity with environmental temperature as well as increases in the binding affinity for catalytic and substrate copper ions. In contrast, the oxidative activities at 25 °C for both the psychrophilic and thermophilic enzymes are similar, suggesting that the nearly temperature-independent electron transfer rate does not require peculiar adjustments. Furthermore, the structural flexibilities of both the psychrophilic and thermophilic enzymes are also similar, indicating that the firm and precise bindings of the four catalytic copper ions are essential for the oxidase function. These results show that the requirements for enzymatic electron transfer, in the absence of the selective pressure of temperature on electron transfer rates, produce a specific adaptive pattern, which is distinct from that observed in enzymes possessing a well-defined active site and relying on conformational changes such as for the induced fit mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, the range of biological temperatures has been expanded with the discovery of new strains inhabiting extreme biotopes. Psychrophiles have been found in the brine veins between polar sea ice crystals at −20 °C (Deming 2002). More recently, the bacterium Planococcus halocryophilus isolated from Arctic permafrost was found to divide at −15 °C and to be metabolically active at −25 °C (Mykytczuk et al. 2013), which possibly represents the lowest temperature for sustained life before dormancy. On the other hand, the archaeon Methanopyrus kandleri has been isolated from a hydrothermal vent at 122 °C and was shown to resist exposure to 130 °C (Takai et al. 2008).
Among the numerous cellular adaptations required to thrive at these extreme temperatures, the enzymatic function has attracted much attention for both fundamental research and biotechnological applications. To compensate for the reduction of chemical reaction rates inherent to low temperature, psychrophiles synthesize highly active enzymes. In most cases, such activity improvements arise from a flexible, heat-labile protein structure and a dynamic active site that loosely bind its substrate to decrease the free energy barrier of activation (Feller and Gerday 2003; Siddiqui and Cavicchioli 2006; Struvay and Feller 2012). In contrast, thermophilic enzymes have to maintain a functional native state at elevated temperatures. Their robust protein structure resists unfolding but, as a consequence of the compact and rigid conformation, thermophilic enzymes are generally inactive at room temperature (Kohen et al. 1999; Kumar and Nussinov 2001; Vieille and Zeikus 2001). These adaptations have been explained in terms of activity–stability–flexibility relationships or tradeoffs (Feller and Gerday 2003; Feller 2010). Interestingly, the same types of weak interactions and structural factors are reduced in number and/or strength to provide the dynamic structure in psychrophilic proteins, or are increased to provide the heat-stable structure in thermophilic proteins.
However, noticeable exceptions to the activity–stability–flexibility tradeoff hypothesis for temperature adaptation have been reported. Phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima is extremely stable but also much more active than its mesophilic counterpart from E. coli. This has been related to the requirement for a very fast turnover of its heat-labile substrate at high temperature (Sterner et al. 1996). The chaperonin GroEL from the Antarctic bacterium Pseudoalteromonas haloplanktis is not cold adapted and displays similar stability and activity than that of its mesophilic homolog from E. coli. This has been tentatively related to the requirement for a heat-stable structure and activity of the psychrophilic chaperonin during heat stress of the Antarctic bacterium (Tosco et al. 2003). These examples suggest that different adaptive strategies can be used, depending on the type of reaction catalyzed or on the cellular context. In this respect, the rate of electron transfer in macromolecules is expected to be almost unaffected in the range of biological temperatures. However, a comparative study of extremophilic enzymes catalyzing electron transfer is currently lacking. This aspect has been investigated here using psychrophilic, mesophilic, and thermophilic blue oxidases as models.
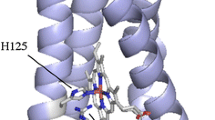
Cuproxidases (cuprous oxidases) are members of the blue multicopper oxidase (MCO) family. In the bacterial periplasm, they catalyze the one-electron oxidation of toxic Cu(I) ions into much less harmful Cu(II) with concomitant four-electron reduction of dioxygen to water (Fig. 1). Cuproxidases display the general fold of MCOs consisting of three repeated cupredoxin domains, with an additional methionine-rich region that confers specificity for Cu(I) (Djoko et al. 2010). Oxidation is mediated via four catalytic copper ions arranged in two centers and designated according to their spectroscopic properties (Solomon et al. 1996). The T1 copper has an absorption peak around 600 nm and gives rise to the blue color typical of MCOs. The T2 and two T3 copper atoms form a trinuclear center (TNC) and give rise to a peak in the region of 330 nm. These catalytic coppers are coordinated by strictly conserved histidine, cysteine, and methionine side chains. According to the reaction mechanism proposed for the cuproxidase CueO (Cu efflux oxidase) from E. coli, Cu(I) ions are gathered via a methionine-rich region, mainly a disordered loop, and an adjacent substrate binding site (sCu). The latter is buried underneath the protein surface and H-bonded to the T1 site, suggesting an electron transfer pathway from sCu to the T1 site, and then to the TNC where oxygen is reduced to water (Solomon et al. 2008; Singh et al. 2011; Cortes et al. 2015). Cuproxidases also display a laccase-like activity (phenol oxidase) which is dependent on a single Cu(II) binding to the sCu site, the latter becoming a mediator of electron transfer between reducing organic substrates and the T1 site (Djoko et al. 2010). This oxidation has been used here for activity measurements.

Figure based on PDB entries 3OD3 and 3NT0 [limited to Cu(I) ions] from Singh et al. 2011
Ribbon diagram of the cuproxidase CueO from E. coli. In magenta, the Met-rich insert binding three Cu(I) ions (orange spheres). According to the proposed mechanism, electrons are transferred from the sCu site (yellow) to the blue T1 copper (blue) and then to the trinuclear center (green) where dioxygen is reduced to water.
To conduct investigations of thermal adaptation in bacterial MCOs, three model enzymes were chosen: CueO from the mesophilic bacterium E. coli (Djoko et al. 2010), because this enzyme has been extensively studied, and its 3D-structure was determined (Roberts et al. 2002; Singh et al. 2011), the thermophilic Tth-MCO, the corresponding MCO from Thermus thermophilus (Miyazaki 2005; Bello et al. 2012) and PhaMOx, a new psychrophilic (cold-adapted) MCO encoded in the genome of the Antarctic bacterium Pseudoalteromonas haloplanktis (Medigue et al. 2005). The three enzymes are homologous periplasmic cuproxidases with laccase-like activity. This study was undertaken to analyze the adaptive strategy of enzymes involving electron transfer in catalysis. We show here that a specific, non-canonical adaptive pattern is observed for these enzymes.
Materials and methods
Expression and purification
Cuproxidases from P. haloplanktis TAC125, E. coli JM109, and T. thermophilus HB27 were produced and purified as described elsewhere. Briefly, the coding sequences were PCR-amplified from genomic DNA without the native Tat signal peptide and cloned into the pET28a expression vector for intracellular production in E. coli BL21(DE3). The psychrophilic PhaMOx was purified by a combination of Q-Sepharose FF, S-Sepharose, and hydroxyapatite column chromatographies. CueO was purified as described (Kim et al. 2001), except that all steps were carried out at 4 °C and Tth-MCO was purified as previously described (Miyazaki 2005). Fully copper-loaded holoenzymes were obtained by addition of a 15-fold excess of Cu(I) from a stock solution of [Cu(I)MeCN)4]+ in acetonitrile, and the excess was removed by gel filtration on PD10 columns eluted with the appropriate buffer. Holoenzymes were used in all experiments, except differential scanning calorimetry (see below).
Differential scanning calorimetry
The PD10 elution buffer for holoenzymes contained 30 mM MOPS and 1 M 3-(1-pyridinio)-1-propanesulfonate (NDSB, a nondetergent sulfobetaine) at pH 7.0. Apoenzymes (Sedlak et al. 2008) were prepared by overnight incubation at 4 °C in the presence of 50 mM ascorbic acid, 50 mM NaCN, and 10 mM EDTA in 30 mM MOPS, 1 M NDSB, pH 7.0. The PD10 elution buffer for apoenzymes contained 30 mM MOPS and 1 M NDSB, pH 7.0. Measurements were performed using a MicroCal VP-DSC instrument at a scan rate of 90 K h−1 and under ~25 psi positive cell pressure with samples at ~1 mg/ml concentration. After cell loading, the protein concentration was determined for the remaining sample by the Bradford method (Pierce). Thermograms were analyzed using the MicroCal Origin software (version 7) to determine the melting point T m, the temperature corresponding to the transition peak, and the calorimetric enthalpy ΔH cal as the area under the transition.
Analytical procedures
The acrylamide-induced quenching of intrinsic fluorescence was measured as described (D’Amico et al. 2003) on an SML-AMINCO Model 8100 spectrofluorimeter (Spectronic Instruments) at an excitation wavelength of 280 nm (1 nm bandpass) and an emission wavelength of 340 nm (2 nm bandpass). Holoenzymes were diluted in 50 mM Tris, pH 7.5 to reach an absorbance at 280 nm of about 0.1. The Cu(II)-dependence of phenol oxidase activity was determined at 25 °C in 50 mM Na-acetate, 10 mM 2,6-dimethoxyphenol, pH 5.0 in the presence of increasing concentrations of CuSO4. The reaction was initiated by substrate addition and monitored by the linear absorbance variation at 477 nm with time. One unit corresponds to 1 µmol of 2,6-dimethoxyphenol oxidized per min, and the K d,app for Cu(II) binding to the sCu site is defined as the concentration of CuSO4 providing half-maximal activity. The temperature dependence of laccase-like activity was recorded in 50 mM Na-acetate, 5 mM 2,2′-azino bis(3-ethylbenzothiazoline-6-sulfonate) (ABTS), 1 mM CuSO4, pH 5.0 in test tubes incubated in a water bath. The reaction was initiated by enzyme addition and absorbance was monitored after 2 min at 415 nm. All activity measurements were performed in triplicates, using a UVIKON XS spectrophotometer (Bio-Tek Instruments).
Results
Primary structure analysis of the homologous cuproxidases
The primary structure of the investigated enzymes displays the typical patterns of cuproxidases, i.e., the strictly conserved ligands of the four catalytic coppers and a Met-rich region involved in Cu(I) binding (Supplementary Fig. S1). When compared with the protein sequence of CueO from E. coli, the psychrophilic PhaMOx shows 53.5 % sequence similarity (24.6 % identity), and the thermophilic Tth-MCO exhibits 59.9 % similarity (30.5 % identity). Tables 1 and 2 show general features of the three MCOs and some peculiarities possibly related to protein adaptation to different thermal environments. The Tat signal sequence gets shorter on going from the psychrophilic to the thermophilic enzyme. Signal peptides of Tat-dependent precursor proteins are optimally adapted to their cognate export apparatus (Blaudeck et al. 2001), and here, the latter has apparently evolved differently in the three bacterial species, possibly as a consequence of their different thermal regimes and membrane compositions. The size of the mature MCOs also decreases on going from the psychrophilic to the thermophilic enzyme. These differences in polypeptide lengths mainly arise from longer and additional loops in the sequence of the psychrophilic PhaMOx and from shorter loops in the 3D structure of the thermophilic Tth-MCO. Thermophilic proteins tend to decrease the size of unstructured regions, as these often destabilize the native state and are preferential sites for unfolding (Kumar and Nussinov 2001). Conversely, long loops in psychrophilic proteins exert a structural constraint on the protein core, possibly enhancing their dynamic conformation (Feller 2013).
The number of proline residues (Table 2) increases from the psychrophilic to the thermophilic enzymes despite the sequence length decreases. This reflects the constraints imposed by this imino acid on the dihedral angles in the polypeptide chain, leading to a better stability of the native state in thermostable proteins. In the case of Tth-MCO, the lower number of Asn and Gln can be related to selection for heat-stable side chains as both residues are prone to thermal deamination (Vieille and Zeikus 2001). Furthermore, its Arg content is higher, reflecting the stabilizing properties of this side chain which is able to perform multiple H-bonds or salt bridges (Mrabet et al. 1992). As a result, the thermophilic enzyme also displays a high pI (even higher than the theoretical value, Table 1) compared with its counterparts that could arise from a high density of positively charged residues at the surface of the protein, possibly contributing to an electrostatic network enhancing thermal stability. As a matter of fact, a large number of salt bridges, composed of 34 ion pairs, have been reported in the 3D structure of Tth-MCO (Bello et al. 2012). Conversely, the more acidic pI of PhaMOx, arising from a high Asp content (Table 2), has been frequently reported in psychrophilic proteins and has been related to free acidic surface side chains destabilizing the outer protein shell via interactions with the solvent (Schiffer and Dotsch 1996). The Glu content of Tth-MCO is high, as also observed in thermophilic genomes (Suhre and Claverie 2003), and is believed to be important in structural stability. Although the prevalence of Glu over Asp in stabilization has been experimentally demonstrated, the molecular origin remains poorly understood (Lee et al. 2004). The Met content decreases with increasing environmental temperature for the three MCOs, and this trend remains identical after removing the number of Met residues involved in Cu(I) binding, (Metcore, Table 2, Supplementary Fig. S1). Methionine provides a less rigid packing of the hydrophobic core (Gassner et al. 1996) and is expected to improve the dynamics of the cold-adapted enzyme, whereas the low Met content in Tth-MCO could contribute to its thermal stability. Finally, the aliphatic index increases from PhaMOx to Tth-MCO, suggesting an increasing involvement of non-polar side chains to stability via improvement of the hydrophobic effect (Kumar and Nussinov 2001).
Structural stability recorded by microcalorimetry
Thermal stability of the cuproxidases in their holo and apo forms was studied by differential scanning calorimetry (DSC). Thermograms obtained for holoenzymes were distorted by a downward drift of the microcalorimetric signal after the transition due to protein aggregation, possibly promoted by the copper ions released during unfolding. Addition of NDSB, a non-detergent sulfobetaine that delays protein aggregation (Collins et al. 2006; D’Amico and Feller 2009), improved the microcalorimetric signals to a certain extent but still only allowed for estimations of the apparent T m (Table 3). In contrast, apoenzymes displayed clear microcalorimetric signals in presence of NDSB (Fig. 2) and allowed determination of the microcalorimetric parameters (Table 3). Denaturation was irreversible in all cases and, therefore, prevented a thermodynamic analysis of unfolding. PhaMOx is the least stable protein followed by CueO and Tth-MCO, as both T m and the calorimetric enthalpy (∆H cal) increase on going from the cold-adapted protein to the heat-stable homolog. The parameter ∆H cal (calculated from the area under the transition) records the total amount of heat required to disrupt all enthalpy-driven interactions during unfolding. Surprisingly, the Tth-MCO apoenzyme displays three well-resolved transitions. These three completely independent stability domains are reminiscent of the three cupredoxin structural domains forming an MCO. This is further supported by the possibility of sequentially unfolding these domains by cooling the sample after each transition, before a new up-scan (Fig. 3). This is in line with previous reports showing a gain in unfolding cooperativity mediated by copper cofactors at the interface of the cupredoxin domains in MCOs (Koroleva et al. 2001; Sedlak et al. 2008). Table 3 also shows the gain in stability (∆T m holo–apo) induced by binding of the copper cofactors. These differences in melting point temperatures basically reflect the copper binding strength (Brandts and Lin 1990), which increases on going from the psychrophilic to the thermophilic enzyme. Such increases in the binding affinity with the environmental temperature are frequently observed in extremophilic proteins (Feller et al. 1999; Feller 2010).
Structural flexibility probed by fluorescence quenching
Fluorescence quenching by acrylamide has been successfully used to probe the molecular permeability or “breathability” of homologous protein structures. As the position of the Trp residues is not strictly conserved in the tertiary structures of the homologous cuproxidases, the variation of quenching with temperature becomes the relevant parameter (Fig. 4). In all extremozymes investigated so far, psychrophilic proteins demonstrated a large quenching effect arising from the deep penetration of the quencher into their flexible structures, whereas thermophilic proteins displayed a weaker quenching effect resulting from their rigid conformation near room temperature (Collins et al. 2003; D’Amico et al. 2003; Georlette et al. 2003; Huston et al. 2008; Xie et al. 2009). However, this pattern is not observed when comparing PhaMOx, CueO, and Tth-MCO (Fig. 4) as both the cold-adapted and the thermophilic cuproxidases demonstrate an identical quenching effect and a higher flexibility as compared with the mesophilic enzyme. In the case of these MCOs, structural stability (Fig. 2) is not correlated with structural flexibility (Fig. 4) using dynamic quenching of fluorescence, although Trp residues are evenly distributed in the sequences, mostly at conserved positions (Supplementary Fig. S1).
Stern–Volmer plots of fluorescence quenching by acrylamide. Fluorescence quenching values at 10 °C (upper panel) and at 35 °C (middle panel) for PhaMOx (circles), CueO (squares), and Tth-MCO (triangles). Lower panel variation of fluorescence quenching between 10 and 35 °C obtained by subtracting the regression lines of the Stern–Volmer plots at individual temperatures
Laccase-like activity of the homologous cuproxidases
Laccase or phenol oxidase activity of cuproxidases is dependent on the presence of a single extra Cu(II) ion which binds in the substrate binding site sCu. The phenol oxidase activity of the three cuproxidases as a function of Cu(II) concentrations was measured in the same conditions, in acetate buffer pH 5.0 at 25 °C with 10 mM 2,6-dimethoxyphenol (2,6-DMP) as substrate (Fig. 5; Table 4). Typically, cold-adapted enzymes are characterized by a high catalytic activity, whereas most thermophilic enzymes are nearly inactive at room temperature. However, in this case, the maximal activity of both the psychrophilic and thermophilic MCOs is similar and lower than that of the mesophilic CueO (Fig. 5). In contrast, the apparent K d for Cu(II) binding to the sCu site (Table 4) follows the generally observed trend for extremophilic proteins: a weak binding for the cold-adapted protein, intermediate for the mesophilic CueO, and a stronger binding for the thermophilic protein. Finally, Cu(II) inhibition at high, non-physiological concentrations (Fig. 5), also indicates different interactions between the cupric ion, the substrate, and the cuproxidases, as shown by the strong inhibition of CueO and the moderate inhibition of PhaMOx and Tth-MCO.
Temperature dependence of activity
The temperature dependence of laccase-like activity with ABTS as substrate (which is less prone to oxidation and precipitation at high temperatures than 2,6-DMP) and in the presence of 1 mM Cu(II) was recorded for the three enzymes (Fig. 6). PhaMOx is active at low to moderate temperatures, CueO displays a maximal activity around 80 °C, whereas the maximum is not yet attained for Tth-MCO below the buffer boiling point. It should be mentioned that the recorded temperature dependence of activity does not reflect the temperature dependence of intrinsic electron transfer in the core enzyme, but rather the effect of temperature on the rate limiting step (electron transfer from the non-natural substrate to the sCu site) and possibly on dioxygen reduction to water. When comparing the temperature dependence of activity (Fig. 6) with the thermal unfolding of holoenzymes (T m in Table 3), it can be seen that CueO loss of activity is concomitant with unfolding (T m is close to maximal activity). In contrast, PhaMOx thermo-inactivation occurs before any detectable unfolding event is recorded by DSC. This can be related to the above-mentioned weaker copper binding in the redox centers (T1 and TNC) and sCu site in the laccase-like reaction (K d,app in Table 4): around 30 °C slight alterations in the copper chelation geometry impair the electron transfer process.
Discussion
Historically, the very weak temperature dependence of electron transfer in biological systems was discovered by the observation that electron transfer between a cytochrome c and a bacteriochlorophyll occurred even at 80 K (Chance and Nishimura 1960). Later, it was proposed that the reaction proceed through quantum mechanical tunneling, which is temperature-independent (DeVault and Chance 1966). According to the Marcus theory (Marcus and Sutin 1985), the rate equation of electron transfer contains an exponential term including T, as for any chemical rate constant, but the various parameters involved render this term negligible. Furthermore, in many bacterial redox centers, the rates actually increase slightly as the temperature is lowered, underlining the complexity of such systems. In the case of the extremophilic blue oxidases, it can be postulated that the rate of electron transfer in the core enzyme is also independent of the environmental temperatures, but it should be kept in mind that this assumption is currently not supported by experimental evidences.
In contrast with many previous studies on extremozymes, we have shown here that the stability of the extremophilic cuproxidases examined is correlated with neither activity, nor flexibility. The primary structures display several insights of adaptations linked to thermal stability (Tables 1, 2). As a matter of fact, the microcalorimetric parameters of stability T m and ∆H cal increase with environmental temperatures (Fig. 2), indicative of an increasing number of stabilizing interactions in the folded structures. Furthermore, the binding affinity of the catalytic coppers also increases from the psychrophilic to the thermophilic MCOs, as inferred from ∆T m (holo–apo) in Table 3. As all copper ligands are strictly conserved in the three cuproxidases (Supplementary Fig. S1), the structures bearing the redox centers should be responsible for these differences in affinity. In the cold-adapted PhaMOx, weaker interactions chelating copper ions in their rigid binding sites would allow lower reorganization energy upon redox changes, still maintaining fast electron transfer reactions at low temperature, whereas strong copper binding by Tth-MCO is required for proper functioning at elevated temperatures. The binding affinity of Cu(II) to the substrate site sCu also increases with environmental temperatures (Table 4): a weak binding for the cold-adapted protein that could be related to improved binding site dynamics as shown in other psychrophilic proteins, and a stronger binding for the thermophilic protein arising from the compact conformation of these proteins. Finally, the temperature dependence of activity (Fig. 6) follows the same trend. The above-mentioned parameters are obviously related to the differences in structural stability between the three cuproxidases and follow the canonical pattern generally reported for extremophilic proteins.
All previous studies using acrylamide-induced fluorescence quenching as a probe for flexibility have invariably correlated this parameter with structural stability (Collins et al. 2003; D’Amico et al. 2003; Georlette et al. 2003; Huston et al. 2008; Xie et al. 2009). In sharp contrast, such correlation is not found in the cuproxidases as flexibility of the cold-adapted PhaMOx and of the thermophilic Tth-MCO is identical, despite large differences in stability. Two explanations can be proposed for this uncommon observation. First, MCOs catalyze oxidation of reducing substrates by an outer-sphere electron transfer mechanism, and hence, large conformational changes linked to substrate binding, and oxidation is not required. Second, MCOs possess an active site containing accurately bound copper cofactors to exert the oxido-reduction function, substrate oxidation by the T1 site and oxygen reduction to water by the trinuclear cluster. The four conserved copper binding motifs are distributed along the amino acid sequence, and the strict maintenance of the copper ions in their specific binding configurations by a rigid conformation of the coordination sphere (and by extension of the protein matrix) allows low reorganization energy. Therefore, increased global flexibility of the cold-adapted enzyme structure to counterbalance the slowdown of molecular motions at low temperatures seems to be both unnecessary and incompatible with electron transfer catalysis. In this respect, whether the low stability of the psychrophilic cuproxidase originates from the lack of selective pressure or is an essential component for its function remains to be clarified. Furthermore, the phenol oxidase activity is also not correlated with stability as both psychrophilic and thermophilic cuproxidases display similar specific activities at 25 °C (Fig. 5; Table 4). The outer-sphere electron transfer process involved in oxidation of reducing substrates is expected to be unaffected at extreme biological temperatures, therefore explaining that activity of MCOs escape the canonical pattern of extremozymes. However, one cannot exclude that activity on the non-natural phenolic substrate is governed by other factors, such as the accessibility of the sCu site.
These results show that the requirements for enzymatic electron transfer produce a specific adaptive pattern, distinct from that observed in enzymes possessing a well-defined active site and relying on conformational changes such as for the induced fit mechanism. This aspect can potentially explain the non-conventional pattern also reported for other proteins involving electron transfer: the uncoupling between stability and flexibility in a psychrophilic ferricytochrome C 552 (Oswald et al. 2014); the higher stability of a psychrophilic cytochrome c 5 as compared with its mesophilic homologue (Takenaka et al. 2010); the exceptional heat resistance of a psychrophilic thioredoxin reductase (Falasca et al. 2012); the similar thermal stability of psychrophilic and mesophilic superoxide dismutases (Merlino et al. 2010). In these examples and in the investigated cuproxidases, the strict maintenance of optimal distances between donors and acceptors (amongst other essential parameters for electron transfer) may have driven evolution toward such activity–stability patterns, in the absence of the selective pressure of temperature on electron transfer rates.
References
Bello M, Valderrama B, Serrano-Posada H, Rudino-Pinera E (2012) Molecular dynamics of a thermostable multicopper oxidase from Thermus thermophilus HB27: structural differences between the apo and holo forms. PLoS One 7:e40700
Blaudeck N, Sprenger GA, Freudl R, Wiegert T (2001) Specificity of signal peptide recognition in Tat-dependent bacterial protein translocation. J Bacteriol 183:604–610
Brandts JF, Lin LN (1990) Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry 29:6927–6940
Chance B, Nishimura M (1960) On the mechanism of chlorophyll-cytochrome interaction: the temperature insensitivity of light-induced cytochrome oxidation in chromatium. Proc Natl Acad Sci USA 46:19–24
Collins T, Meuwis MA, Gerday C, Feller G (2003) Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. J Mol Biol 328:419–428
Collins T, D’Amico S, Georlette D, Marx JC, Huston AL, Feller G (2006) A nondetergent sulfobetaine prevents protein aggregation in microcalorimetric studies. Anal Biochem 352:299–301
Cortes L, Wedd AG, Xiao Z (2015) The functional roles of the three copper sites associated with the methionine-rich insert in the multicopper oxidase CueO from E. coli. Metallomics 7:776–785
D’Amico S, Feller G (2009) A nondetergent sulfobetaine improves protein unfolding reversibility in microcalorimetric studies. Anal Biochem 385:389–391
D’Amico S, Marx JC, Gerday C, Feller G (2003) Activity-stability relationships in extremophilic enzymes. J Biol Chem 278:7891–7896
Deming JW (2002) Psychrophiles and polar regions. Curr Opin Microbiol 5:301–309
DeVault D, Chance B (1966) Studies of photosynthesis using a pulsed laser. I. Temperature dependence of cytochrome oxidation rate in chromatium. Evidence for tunneling. Biophys J 6:825–847
Djoko KY, Chong LX, Wedd AG, Xiao Z (2010) Reaction mechanisms of the multicopper oxidase CueO from Escherichia coli support its functional role as a cuprous oxidase. J Am Chem Soc 132:2005–2015
Falasca P, Evangelista G, Cotugno R, Marco S, Masullo M, De Vendittis E, Raimo G (2012) Properties of the endogenous components of the thioredoxin system in the psychrophilic eubacterium Pseudoalteromonas haloplanktis TAC 125. Extremophiles 16:539–552
Feller G (2010) Protein stability and enzyme activity at extreme biological temperatures. J Phys-Condens Mat 22:323101. doi:10.1088/0953-8984/1022/1032/323101
Feller G (2013) Psychrophilic enzymes: from folding to function and biotechnology. Scientifica 2013:512840. doi:10.1155/2013/512840
Feller G, Gerday C (2003) Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol 1:200–208
Feller G, d’Amico D, Gerday C (1999) Thermodynamic stability of a cold-active α-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38:4613–4619
Gassner NC, Baase WA, Matthews BW (1996) A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc Natl Acad Sci USA 93:12155–12158
Georlette D, Damien B, Blaise V, Depiereux E, Uversky VN, Gerday C, Feller G (2003) Structural and functional adaptations to extreme temperatures in psychrophilic, mesophilic, and thermophilic DNA ligases. J Biol Chem 278:37015–37023
Huston AL, Haeggstrom JZ, Feller G (2008) Cold adaptation of enzymes: structural, kinetic and microcalorimetric characterizations of an aminopeptidase from the Arctic psychrophile Colwellia psychrerythraea and of human leukotriene A(4) hydrolase. Biochim Biophys Acta 1784:1865–1872
Kim C, Lorenz WW, Hoopes JT, Dean JF (2001) Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J Bacteriol 183:4866–4875
Kohen A, Cannio R, Bartolucci S, Klinman JP (1999) Enzyme dynamics and hydrogen tunnelling in a thermophilic alcohol dehydrogenase. Nature 399:496–499
Koroleva OV, Stepanova EV, Binukov VI, Timofeev VP, Pfeil W (2001) Temperature-induced changes in copper centers and protein conformation of two fungal laccases from Coriolus hirsutus and Coriolus zonatus. Biochim Biophys Acta 1547:397–407
Kumar S, Nussinov R (2001) How do thermophilic proteins deal with heat? Cell Mol Life Sci 58:1216–1233
Lee DY, Kim KA, Yu YG, Kim KS (2004) Substitution of aspartic acid with glutamic acid increases the unfolding transition temperature of a protein. Biochem Biophys Res Commun 320:900–906
Marcus RA, Sutin N (1985) Electron transfers in chemistry and biology. Biochim Biophys Acta 811:265–322
Medigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D’Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EP, Rouy Z, Sekowska A, Tutino ML, Vallenet D, von Heijne G, Danchin A (2005) Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res 15:1325–1335
Merlino A, Russo Krauss I, Castellano I, De Vendittis E, Rossi B, Conte M, Vergara A, Sica F (2010) Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis. J Struct Biol 172:343–352
Miyazaki K (2005) A hyperthermophilic laccase from Thermus thermophilus HB27. Extremophiles 9:415–425
Mrabet NT, Van den Broeck A, Van den Brande I, Stanssens P, Laroche Y, Lambeir AM, Matthijssens G, Jenkins J, Chiadmi M, van Tilbeurgh H et al (1992) Arginine residues as stabilizing elements in proteins. Biochemistry 31:2239–2253
Mykytczuk NC, Foote SJ, Omelon CR, Southam G, Greer CW, Whyte LG (2013) Bacterial growth at −15 degrees C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J 7:1211–1226
Oswald VF, Chen W, Harvilla PB, Magyar JS (2014) Overexpression, purification, and enthalpy of unfolding of ferricytochrome c 552 from a psychrophilic microorganism. J Inorg Biochem 131:76–78
Roberts SA, Weichsel A, Grass G, Thakali K, Hazzard JT, Tollin G, Rensing C, Montfort WR (2002) Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci USA 99:2766–2771
Schiffer CA, Dotsch V (1996) The role of protein-solvent interactions in protein unfolding. Curr Opin Biotechnol 7:428–432
Sedlak E, Ziegler L, Kosman DJ, Wittung-Stafshede P (2008) In vitro unfolding of yeast multicopper oxidase Fet3p variants reveals unique role of each metal site. Proc Natl Acad Sci USA 105:19258–19263
Siddiqui KS, Cavicchioli R (2006) Cold-adapted enzymes. Annu Rev Biochem 75:403–433
Singh SK, Roberts SA, McDevitt SF, Weichsel A, Wildner GF, Grass GB, Rensing C, Montfort WR (2011) Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence. J Biol Chem 286:37849–37857
Solomon EI, Sundaram UM, Machonkin TE (1996) Multicopper oxidases and oxygenases. Chem Rev 96:2563–2606
Solomon EI, Augustine AJ, Yoon J (2008) O2 reduction to H2O by the multicopper oxidases. Dalton Trans 30:3921–3932
Sterner R, Kleemann GR, Szadkowski H, Lustig A, Hennig M, Kirschner K (1996) Phosphoribosyl anthranilate isomerase from Thermotoga maritima is an extremely stable and active homodimer. Protein Sci 5:2000–2008
Struvay C, Feller G (2012) Optimization to low temperature activity in psychrophilic enzymes. Int J Mol Sci 13:11643–11665
Suhre K, Claverie JM (2003) Genomic correlates of hyperthermostability, an update. J Biol Chem 278:17198–17202
Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, Hirayama H, Nakagawa S, Nunoura T, Horikoshi K (2008) Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc Natl Acad Sci USA 105:10949–10954
Takenaka S, Wakai S, Tamegai H, Uchiyama S, Sambongi Y (2010) Comparative analysis of highly homologous Shewanella cytochromes c 5 for stability and function. Biosci Biotechnol Biochem 74:1079–1083
Tosco A, Birolo L, Madonna S, Lolli G, Sannia G, Marino G (2003) GroEL from the psychrophilic bacterium Pseudoalteromonas haloplanktis TAC 125: molecular characterization and gene cloning. Extremophiles 7:17–28
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43
Xie BB, Bian F, Chen XL, He HL, Guo J, Gao X, Zeng YX, Chen B, Zhou BC, Zhang YZ (2009) Cold adaptation of zinc metalloproteases in the thermolysin family from deep sea and arctic sea ice bacteria revealed by catalytic and structural properties and molecular dynamics: new insights into relationship between conformational flexibility and hydrogen bonding. J Biol Chem 284:9257–9269
Acknowledgments
This work was supported by the F.R.S-FNRS, Belgium (Fonds de la Recherche Fondamentale et Collective, contract numbers 2.4535.08, 2.4523.11 and U.N009.13 to G.F.) and by the Belgian program of Interuniversity Attraction Poles (iPros P7/44) initiated by the Federal Office for Scientific, Technical and Cultural Affairs. T.C. thanks the FCT and FEDER (POFC-COMPETE) for funding through the Investigador FCT Programme (IF/01635/2014) and project EngXyl (EXPL/BBB-BIO/1772/2013- FCOMP-01-0124-FEDER-041595) as well as strategic funding via UID/BIA/04050/2013. F.R., A.G. and A.C. were FRIA research fellows.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Albers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roulling, F., Godin, A., Cipolla, A. et al. Activity–stability relationships revisited in blue oxidases catalyzing electron transfer at extreme temperatures. Extremophiles 20, 621–629 (2016). https://doi.org/10.1007/s00792-016-0851-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-016-0851-9