Abstract
Transfer of DNA has been shown to be involved in genome evolution. In particular with respect to the adaptation of bacterial species to high temperatures, DNA transfer between the domains of bacteria and archaea seems to have played a major role. In addition, DNA exchange between similar species likely plays a role in repair of DNA via homologous recombination, a process that is crucial under DNA damaging conditions such as high temperatures. Several mechanisms for the transfer of DNA have been described in prokaryotes, emphasizing its general importance. However, until recently, not much was known about this process in prokaryotes growing in highly thermophilic environments. This review describes the different mechanisms of DNA transfer in hyperthermophiles, and how this may contribute to the survival and adaptation of hyperthermophilic archaea and bacteria to extreme environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hyperthermophilic (superheat-loving) organisms are extremophiles and thrive at temperatures around 80 °C or higher. The first hyperthermophile discovered in Yellowstone National Park by Thomas Brock was Sulfolobus acidocaldarius (Brock et al. 1972). Since then, over 90 hyperthermophilic species have been discovered (Stetter 2006a, b, 2013), most of them belonging to the domain of archaea, but some hyperthermophilic bacteria have also been characterized. Throughout evolution, hyperthermophilic organisms were able to adapt to changing environments such as up-shifts in temperature. Horizontal gene transfer (HGT) probably played an important role in the adaptation to more extreme environments. Except from a role in evolution, exchange of DNA might have also functioned in other mechanisms like DNA repair. DNA exchange is a widespread phenomenon that occurs in all domains of life. This emphasizes its significance for life on earth.
Three main mechanisms for transferring DNA have been described for archaea and bacteria: natural transformation, conjugation and transduction. Natural transformation is the uptake of DNA from the external environment, mostly emerging from lysed cells. Incoming DNA can be degraded and/or can be incorporated into the chromosomal DNA. The recipient cell is usually in charge of what DNA is and what DNA is not taken up (Chen et al. 2005; Lorenz and Wackernagel 1994; Thomas and Nielsen 2005). Conjugation on the other hand is a more invasive mechanism in which the donor has control over the transferred DNA. It requires direct contact between two cells that are not necessarily closely related. Mostly, small plasmids are transferred. Conjugation is suggested to be the main mechanism responsible for HGT (Halary et al. 2010; Norman et al. 2009; Wozniak and Waldor 2010). Transduction involves viruses that function as vehicles enabling DNA exchange between closely related species (Lang et al. 2012; Thomas and Nielsen 2005). Other mechanisms not belonging to these three include: the transfer of DNA via gene transfer agents (GTAs) which are virus-like elements encoded by the host genome (Lang et al. 2012), membrane vesicles (Gaudin et al. 2012) and nanotubes which are cellular protrusions that can bridge neighboring cells (Dubey and Ben-Yehuda 2011). Finally, DNA exchange has been shown to occur between hyperthermophilic Sulfolobus species (Grogan 1996). This DNA exchange is mediated by UV-inducible pili from S ulfolobus (Ups) (Fröls et al. 2008). The different DNA transfer methods among hyperthermophiles are listed in Table 1. This review will describe the fate and function of the DNA transferred between hyperthermophilic prokaryotes; furthermore, it will explain in what way the above-mentioned mechanisms contributed to DNA transfer between prokaryotes in hot environments.
Horizontal gene transfer and evolution
Bacteria and archaea are not able to reproduce sexually; instead they use binary fission for reproduction where DNA is transferred vertically from the mother to the daughter cells. In theory, this does not lead to genetic diversity. Yet, gene-mutation, -loss and -duplication can occur, introducing genetic variation. Moreover, archaea and bacteria can obtain DNA from their environment or nearby organisms and incorporate it into their own genomic DNA (Dickerson 1980). Early studies showing the spread of antibiotic resistances (Datta and Kontomichalou 1965) already made scientists believe that transfer of DNA occurred among bacteria. However, despite these observations, the importance of HGT in genome evolution was underappreciated for decades (Cohan 1994a, b). Therefore, early models of evolution were only based on vertical transfer of DNA (Levin 1981). Much later studies involving comparative genomics showed the extent to which HGT actually played a role in genome evolution (Jain et al. 2003; Koonin and Wolf 2008; Ochman et al. 2000). Nowadays it has become clear that throughout evolution, archaea and bacteria exchanged genes which allowed them to adapt to the changes in the dynamic environments they live in (Guglielmini et al. 2013).
Horizontally transferred DNA can be incorporated into the chromosome using different methods (Thomas and Nielsen 2005; Zaneveld et al. 2008). Besides homologous recombination (HR) between similar DNA molecules, which is executed by DNA repair proteins (described in the following section), other mechanisms exist. Integration can for instance be performed by site-specific integrases such as those found in the Sulfolobus spindle-shaped virus 1 (SSV1) or conjugative Sulfolobus plasmid pNOB8 (described in the “Transduction and HGT among hyperthermophilic viruses” and “Conjugation” sections, respectively) (She et al. 2004, 2006). Moreover, transposition might facilitate HGT (Zaneveld et al. 2008), transposons or insertion sequences (ISs) carrying genes from a previous host might integrate into the genome of another organism that contains the same transposable elements. HR between the ISs or transposons then leads to incorporation of the DNA. An example of the latter mechanism is a 16-kb DNA fragment flanked by ISs that has been transferred between Pyrococcus furiosus and Thermococcus litoralis (Diruggiero et al. 2000).
Bioinformatic methods designed to detect HGT, identify unusual base composition and codon usage within genomes (Garcia-Vallvé et al. 2000; Lawrence and Ochman 1998; Nakamura et al. 2004), or perform phylogenetic analyses on individual genes (Beiko et al. 2005; Puigbò et al. 2010). Studies using these methods estimated that at least 20 % of the bacterial genes and 40 % of the archaeal genes were transferred during evolution (Popa and Dagan 2011; Thomas and Nielsen 2005).
HGT occurs across species boundaries, even between organisms from different domains of life. The latter contributed tremendously to the adaptation of bacteria to hot environments, which is illustrated by the fact that hyperthermophilic bacteria obtained more genes originating from archaea than generally observed with mesophilic bacteria. For instance, the hyperthermophilic Aquifex aeolicus and Thermotoga maritima acquired 16.2 and 24 % of their genes, respectively, from archaea. In comparison, mesophilic Bacillus subtilis, Sunechocystis species, Borrelia burgdorferi and Escherichia coli obtained only 5 % or less archaeal genes (Aravind et al. 1998; Nesbø et al. 2001). Though it must be noted that HGT might, next to the cause of adaptation, also be the consequence of being hyperthermophily as was hypothesized by Nesbø et al. (2001).
The importance of HGT for adaptation to changing environments is illustrated by a genome comparison between Deinococcus radiodurans and Thermus thermophilus (Omelchenko et al. 2005). The two related species diverged from a common, probably mesophilic ancestor (Weisburg et al. 1989), but have surprisingly different phenotypes that correspond to their distinct natural habitats: D. radiodurans is a mesophile that is extremely resistant to ionizing radiation (Anderson et al. 1956), whereas T. thermophilus is a thermophile that shows a rather average sensitivity to ionizing radiation (Oshima and Imahori 1974). It was shown that in both species, adaptation to the highly dissimilar environments was mediated by the loss, and more importantly gain, of several genes during evolution. D. radiodurans obtained bacterial genes that enhanced its ability to survive different kinds of stresses. T. thermophilus on the other hand, acquired numerous archaeal genes that contributed to its thermophilic adaptation. Of these, about 50 % are encoded on a mega plasmid which has an increased plasticity. This is illustrated by the fact that between related T. thermophilus HB8 and T. thermophilus HB27, gene content and order of the plasmid differ substantially. However, both plasmids are implicated in growth at higher temperatures. They both encode several proteins involved in the putative mobile thermophilic-specific DNA repair system. In addition, a gene encoding a reverse gyrase was found on the mega plasmid of T. thermophilus HB8. On the other hand, in the megaplasmid of T. thermophilus HB27, a pseudogene of the reverse gyrase was found suggesting that this gene was once acquired from a hyperthermophilic organism by a common ancestor of the two strains, but was decayed from T. thermophilus HB27 (Brüggemann and Chen 2006; Omelchenko et al. 2005).
The reverse gyrase is found in many thermophiles and in all hyperthermophiles and is probably the best-documented example of HGT being involved in adaptation to hot environments (Déclais et al. 2000; Forterre 2002). Phylogenetic analyses suggest that an early archaeal reverse gyrase gene was acquired by an ancient bacterium. This was then followed by distribution of the gene within the Thermotogales and Aquificales via HGT (Brochier-Armanet and Forterre 2007; Gribaldo and Brochier-Armanet 2006). Its contribution to life at higher temperatures is illustrated by the fact that a reverse gyrase deletion strain of Thermococcus kodakaraensis KOD1 shows growth defects especially at higher temperatures (Atomi et al. 2004). It might therefore be possible that it is only essential for life above 90 °C. Moreover, moderately thermophilic Nautilia profundicola Am-H lives in an environment near hydrothermal vents with fluctuating temperatures; the reverse gyrase of this strain showed a significant upregulation upon up-shifts in temperature (Campbell et al. 2009). The reverse gyrase was previously thought to increase the DNA stability at high temperature by introducing positive coils into the DNA (Kikuchi and Asai 1984). However, not all hyperthermophiles have positively coiled DNA. More recent studies therefore suggest that the reverse gyrase is rather involved in the protection of DNA against degradation which occurs at high temperatures (Kampmann and Stock 2004; Napoli et al. 2004). Although the exact function of the reverse gyrase in this process is not yet fully resolved, its acquisition was almost certainly an important step in the evolution of many thermophilic bacteria (Heine and Chandra 2009).
Adaptations to environmental changes can also be rather subtle. On Vulcano Island (Italy) for instance, there are several shallow vents which have dissimilar and fluctuating physiological conditions (Capasso et al. 1999; Sedwick and Stuben 1996). The genome sequences of several Pyrococcus isolates from these vents revealed extensive genome rearrangements in specific genomic “hot spots” containing mobile genetic elements. It is therefore thought that a number of HGT events took place, possibly contributing to the adaptation to the rapidly changing environments on Vulcano Island (White et al. 2008).
Rachel Whitaker et al. used Sulfolobus islandicus as a model organism to study evolutionary biology in hyperthermophilic archaea (Zhang et al. 2013). By comparing the genomes of 12 different S. islandicus strains from a single hotspring in Kamchatka (Russia), they investigated the rate and mode of recent evolutionary events. Two coexisting groups of S. islandicus species were found that seem to have exchanged DNA mainly within the groups (Cadillo-Quiroz et al. 2012). This is a clear indication of speciation (Dykhuizen and Green 1991). Given the fact that Sulfolobus species exchange DNA in species-specific aggregates (Ajon et al. 2011), the barrier between the two S. islandicus groups could be the inability to successfully create mating pairs (Cadillo-Quiroz et al. 2012). This species-specific recognition is probably driven by Ups-pili and glycosylation patterns (van Wolferen and Albers, unpublished), as will be described later. Strains from the two different S. islandicus groups might therefore show variations in these two structures. Future studies need to confirm whether or not strains from the two different groups are indeed unable to form mixed aggregates. Moreover, glycosylation patterns and Ups-pili between strains from the two different groups need to be compared.
Taken together, genome evolution has been essential for the adaptation of hyperthermophiles to changing and extreme environments in which HGT seems to have been the main player. Without the ability to exchange DNA between species, hyperthermophilic bacteria probably would not exist.
DNA transfer for repair
Next to the crucial role DNA transfer played in genome evolution, other destinations of transferred DNA have become evident. It has been suggested that prokaryotes use imported similar DNA to repair their own DNA with HR (Bernstein et al. 1981). HR is the genetic recombination between two similar or identical molecules of DNA, and is the only efficient mechanism for accurately repairing double stranded DNA breaks (DSBs). It is dependent on another copy of the damaged DNA. This other copy can be the second chromosome that is naturally present during the G2 phase. However, taking up other DNA evidently increases the chances of having a non-damaged homologous template for DNA repair (Bernstein et al. 2012). Bernstein et al. (2012) even propose that DNA repair is the primary function of DNA transfer. For recent reviews describing the DSB repair via HR in bacteria and archaea, see Ayora et al. (2011) and White (2011).
Many bacteria induce their competence genes upon DNA damage which underlines the role that transferred DNA might play in DNA repair. In Streptococcus pneumoniae, competence is induced upon treatment with DNA damaging agents mitomycin C and fluoroquinolone (Claverys et al. 2006). Competence of Helicobacter pylori is induced by ciprofloxacin which causes DSBs (Dorer et al. 2010). B. subtilis shows DNA uptake upon UV stress (Michod et al. 1988), and Legionella pneumophila strongly induces competence upon the addition of several different DNA damaging agents (Charpentier et al. 2011). The only competence system studied in detail in thermophilic prokaryotes is that of Thermus species; this system, however, was shown to be constitutively active (Hidaka et al. 1994). Hyperthermophilic Sulfolobus species on the other hand, exchange DNA upon DSB induction with UV-light or chemical compounds, like bleomycin (Fröls et al. 2008). The mechanism behind this exchange is still unclear, but possibly it is related to the competence system as will be described later. Charpentier et al. (2011) speculate that DNA transfer might have evolved as a DNA damage response in SOS-deficient bacteria. Among the above-mentioned bacteria, S. pneumoniae, H. pylori and L. pneumophila indeed do not show an SOS response. This hypothesis might also be true for Sulfolobus species, as archaea in general have so far not been shown to induce SOS responses. It remains to be shown if other hyperthermophilic bacteria and archaea also display DNA damage-inducible DNA transfer mechanisms that may or may not be similar to that found in Sulfolobus species.
Further evidence supporting the idea that transferred DNA functions in DNA repair is the fact that DNA uptake is often strongly biased towards DNA from the same or closely related species (Seitz and Blokesch 2012). In the Pasteurellaceae and Neisseriae it was shown that this bias is provoked by recognizing so-called DUS sequences that are present multiple times in their own DNA (Danner et al. 1980; Elkins et al. 1991; Fitzmaurice et al. 1984; Mell et al. 2012; van Passel 2008). In Vibrio cholerae on the other hand, the competence system is strongly induced by the presence of the species-specific auto inducer CAI-1, which also leads to a higher uptake of DNA from the same species (Suckow et al. 2011). Other bacterial species make use of restriction modification systems that degrade foreign DNA (Murray 2002) and increase thereby the chances of incorporating self-DNA. Likewise, CRISPR-Cas systems can target foreign DNA, and thereby hamper HGT, as was shown for Staphylococcus epidermidis (Marraffini and Sontheimer 2008). A recent study even showed that the presence of CRISPR loci seems to be linked to the presence of an active competence system (Jorth and Whiteley 2012). Uptake of self-DNA can additionally be increased by the induction of cell death of neighboring cells from the same species (fratricide), as has been shown for S. pneumoniae (Guiral et al. 2005; Håvarstein et al. 2006) and probably also H. pylori (Dorer et al. 2010). Lysed cells provide free DNA that can be taken up for possible DNA repair. Lastly, in the archaeal Sulfolobus species, DNA exchange occurs within species-specific cell aggregates (Ajon et al. 2011). In that way, only species-specific DNA is exchanged.
A bias towards species-specific DNA also occurs at the level of HR. In general, this process evidently gets less efficient for more dissimilar sequences to integrate into the DNA (Majewski and Cohan 1998, 1999; Wolf et al. 2001; Zawadzki et al. 1995). Therefore, in all prokaryotic species there is a natural bias towards integration of DNA from the same or closely related species.
In (hyper)thermophilic organisms one could hypothesize that DNA repair mechanisms are of particular importance as the rates of spontaneous DNA mutations are elevated at high temperatures (Lindahl 1993). However, the genomic mutation rate in the hyperthermophilic archaeon S. acidocaldarius was shown to be equal to mesophilic organisms (Grogan et al. 2001). Given the fact that DNA stability is more important at higher temperatures, mutations probably have a more drastic effect in hot environments. In (hyper)thermophiles, mutation rates should therefore theoretically be even lower than in mesophiles to give the same effect. By extrapolating the mutation rate of used markers to the whole genome, recent studies indeed showed that the rate of base substitutions in S. acidocaldarius and T. thermophilus is lower compared to mesophilic organisms (Drake 2009). Spontaneous mutations in the DNA of (hyper)thermophiles must consequently be repaired so rapidly that they cannot be measured. Thus, efficient DNA repair systems seem to be present; this may involve efficient HR mechanisms that use transferred DNA as a template. In general, the role of DNA transfer in DNA repair of prokaryotes might be far more important than previously thought.
DNA as a nutrient source
An alternative fate for imported DNA might be the use of nucleotides as a nutrient source (Palchevskiy and Finkel 2009; Redfield 1993a). This idea is supported by the fact that B. subtilis and Haemophilus influenzae induce competence upon nutrient starvation and not upon, for instance DNA damage (Redfield 1993a, b). Moreover, non-homologous or partially degraded DNA molecules might not be suitable for HR and therefore better used as nutrient source (MacFadyen et al. 2001; Redfield et al. 1997; Redfield 1988). Proof for this theory is amongst others the observation that E. coli is able to grow with DNA as sole carbon and energy source, which is dependent on the presence of homologs of competence genes in H. influenzae and Neisseria gonorrhoeae (Finkel and Kolter 2001; Palchevskiy and Finkel 2006).
In hyperthermophilic environments, unprotected DNA is degraded faster than in mesophilic surroundings (Lindahl 1993). One could therefore speculate that bad-quality DNA that is commonly present is taken up by competent hyperthermophiles and used as a nutrient source. However, no evidences in agreement with this hypothesis have been shown so far. Moreover, it seems unlikely that the main function of DNA uptake is to serve as a food source when far less energy consuming processes are capable of taking up nutrients just as well. Hence, it seems more plausible that only DNA that cannot be incorporated into the genome is used as a source of nutrients.
Conjugation
Conjugation is the unidirectional transfer of DNA between cells by a process requiring cell–cell contact (de la Cruz et al. 2010; Gomis-Rüth and Coll 2006). This process has a host range larger than observed for transformation or transduction (Guglielmini et al. 2013) and is thought to be the main mechanism involved in HGT (Halary et al. 2010; Norman et al. 2009; Wozniak and Waldor 2010). During conjugation, DNA transfer is carried out by the donor cell by means of a bridge-like connection between two cells; the recipient cell seems to have little control in this process (Pérez-Mendoza and de la Cruz 2009). Mostly plasmids and ICEs (integrating conjugative elements) are transferred. Conjugation has thereby contributed significantly to the rapid spread of antibiotic resistance, virulence, and social traits among prokaryotes (Gomis-Rüth and Coll 2006; Guglielmini et al. 2013; Schröder and Lanka 2005).
Conjugation systems form the largest subfamily of the Type IV secretion systems (T4SS); the latter also include DNA uptake and release systems and protein translocation systems (Schröder and Lanka 2005). The T4SS is a large protein complex spanning the complete bacterial cell envelope. Proteins essential for T4SSs are: a TraU/VirB4 ATPase energizing both the assembly of the system as well as the substrate transfer, and 12–20 mating-pair formation proteins (MPFs) promoting physical contact with the recipient cell (Schröder and Lanka 2005). Besides the general T4SS proteins, conjugation-specific proteins include a relaxase and a type IV coupling protein (T4CP) (de la Cruz et al. 2010). The relaxase initiates conjugative transfer by binding and nicking the DNA at the origin of transfer (oriT). The T4CP then couples the DNA to the channel-forming T4SS that subsequently transfers the nucleoprotein complex through the membrane of the donor cell and delivers it into the recipient cell (de la Cruz et al. 2010; Vogelmann et al. 2011). A gene cluster encoding a VirB4 ATPase, a T4CP and a relaxase is considered to encode a putative conjugative system; without the relaxase, the system is thought to be a protein-exporting T4SS (Guglielmini et al. 2011).
In all different conjugation systems, assembly of the pilus is essential to draw cells close to each other and allow exchange of macromolecules from cell to cell. The best-characterized T4SS surface structures are the conjugative pili from Gram-negative bacteria. There is a huge diversity in composition and structure of these filaments. Pili encoded by the F plasmid from E. coli are long (2–20 μm) and flexible, with a diameter of 8 nm, while P-pili from IncP plasmid RP4 are very short, rigid and have diameters from 8 to 12 nm (Alvarez-Martinez and Christie 2009; Lawley et al. 2003; Silverman 1997).
Conjugation in hyperthermophilic Bacteria
Thermophilic Thermus species are equipped with a highly efficient natural competence system (described in the following section). Recently, it was speculated that also conjugation occurs among Thermus species (César et al. 2011). Given the low solubility of oxygen in geothermal environments, certain T. thermophilus strains express a nitrate reductase, which makes anaerobic growth possible. This nitrate reductase is encoded in the nir-nar-nor cluster, present as a variable region on mega plasmid pTT27 (Alvarez et al. 2011; Bricio et al. 2011). It was shown that T. thermophilus strains NAR1, HB27 and PRQ25 were able to exchange pTT27 among each other, thereby enabling anaerobic growth (Alvarez et al. 2011; Ramírez-Arcos et al. 1998). In addition, when a hygromycin resistance marker (hyg) was placed on pTT27, this gene could be transferred to non-competent cells with frequencies of up to 10−2 (César et al. 2011). These findings support the presence of an active conjugation system. However, no conjugation-like homologous have been found in the above-mentioned T. thermophilus strains. Therefore, another unrelated conjugation system must be present in these species (César et al. 2011). A recently sequenced megaplasmid pTHTHE1601 from T. thermophilus SG0.5JP17-16 revealed the presence of a putative VirB operon, possibly involved in conjugation. Proteins encoded by this operon include TrbC/VirB2 (Ctr1), VirB4 (Ctr3) and VirD4 (Ctr7) ATPases. Future experiments need to confirm the participation of this T4SS in conjugation (César et al. 2011).
Conjugation in hyperthermophilic Archaea
Based on bioinformatics methods (hidden Markov models) scoring for co-localization of genes encoding a virB4/traU ATPase, a T4CP, a relaxase and MPF-specific proteins, only two archaeal conjugative elements could be found: one ICE encoded in the chromosome of the thermoacidophilic Aciduliprofundum boonei, and one conjugative plasmid in Haloarcula marismortui (pNG500). However, many VirB4 homologues were found encoded on archaeal chromosomes or plasmids, often associated with T4CP-like proteins. Because conjugative plasmids were found in Sulfolobus species (described below) it might be possible that unknown relaxases exist in archaea (Guglielmini et al. 2011).
Around 3 % of all isolated Sulfolobus strains contain self-transmissible conjugative plasmids (Prangishvili et al. 1998). The best-characterized plasmids are pNOB8 from Japanese Sulfolobus strain NOB8H2 (Schleper et al. 1995; She et al. 1998) and pING1 isolated from S. islandicus strain HEN2P2 (Prangishvili et al. 1998; Stedman et al. 2000). The first archaeal plasmid shown to be horizontally transferred is a relatively large plasmid (45 kb) pNOB8. It can propagate easily in liquid cultures of its original host in mating mixtures with Sulfolobus solfataricus and S. islandicus via conjugative-like transfer mechanism. pNOB8 has very high copy numbers of between 20 and 40 plasmids per chromosome. When pNOB8 is transformed to Sulfolobus cells; the cells become a donor and are able to transfer the plasmid to other cells. The same was observed for recipient cells containing the transmitted plasmid (Schleper et al. 1995). Interestingly, upon mixing a donor and recipient strain, cellular aggregates could be observed. Moreover, electron microscopy revealed intercellular cytoplasmic bridges that connect two or more cells (Fig. 1a) (Schleper et al. 1995), which resemble those found in halophilic Haloferax volcanii. The cytoplasmic bridges of H. volcanii mediate the bidirectional transfer of chromosomal DNA representing the first known mating system in archaea (Rosenshine et al. 1989). pNOB8 was also shown to be able to integrate site-specifically into the host genome within a tRNA gene (in the so-called attP site) using a self-encoded integrase (She et al. 2004).
a Scanning electron micrographs of donor strain S. solfataricus NOB8H2 (upper left) and 1:1 mating mixtures with S. solfataricus NOB8H23, showing cell aggregates and intercellular bridges. Reprinted from Schleper et al. (1995). b Transmission electron micrographs from cross sections of nanotubes from Thermococcus sp., scale bar 200 nm (a) and 100 nm (b) (a and b). Nanotube associated with a MV, scale bar, 100 nm (c). Scanning electron micrograph of a network of cells linked by nanotubes, scale bar 500 nm. Inset partially disrupted nanotube showing an artificially twisted internal core extruding from an outer envelope; scale bar, 100 nm) (d). Reprinted from Marguet et al. (2013)
Several other archaeal conjugation plasmids have been characterized during the last two decades. All were found in Sulfolobus species and they are grouped into pKEF or pARN plasmids (Alvarez-Martinez and Christie 2009; Greve et al. 2004). In all analyzed conjugative plasmids only a few genes encode proteins homologous to bacterial conjugation proteins, including TraG/VirD4, TrbE/VirB4 and partitioning proteins ParA and ParB. Therefore, despite the fact that conjugational exchange of plasmids has been observed between Sulfolobus species, the exact mechanism is still unclear.
Importantly, Sulfolobus species were also shown to bidirectionally exchange chromosomal DNA, possibly via a conjugation-like mechanism (Aagaard et al. 1995; Ajon et al. 2011; Ghane and Grogan 1998; Schmidt et al. 1999). This mechanism is mediated by UV-inducible pili and will be discussed separately in one of the following sections.
Overall, conjugational exchange of plasmid DNA has been observed both in hyperthermophilic bacteria and archaea; however, only very few of the classical conjugation proteins are found in these species. It seems therefore that conjugation at high temperatures involves different specialized machineries that sofar have not been identified and characterized.
Natural transformation
Natural transformation refers to the uptake of exogenous DNA by naturally competent species (Dubnau 1999). It is a widespread phenomenon in bacteria that has been described for at least 70 bacterial species from all major taxonomic groups. In contrast to conjugation, competence does not require mobile genetic elements (Johnsborg et al. 2007). As previously discussed, imported DNA can be used for different cellular processes, such as DNA repair, nutrition and the introduction of genetic diversity (Seitz and Blokesch 2012). The process of natural transformation starts with the induction of competence, followed by the binding of environmental DNA, which is subsequently fragmented whereupon it enters the cells. If the entered DNA shows homology to certain genomic DNA regions it can be integrated into the chromosome (Averhoff 2009).
Despite the fact that competence systems are common in bacteria, initiation and regulation of the systems differ between species. Some organisms have highly regulated competence that is triggered by certain signals like pheromones, nutrient limitation or high cell density. Competence in Gram-positive B. subtilis and S. pneumonia has been studied in detail and seems to be a general response to stress (Johnsborg et al. 2007; Lorenz and Wackernagel 1994). In Gram-negative bacteria, regulation of competence is less well understood. However, it is known that H. influenza induces competence upon starvation (Herriott et al. 1970) and Vibrio cholera upon growth on chitin (Meibom et al. 2005). Moreover, as described previously, many bacteria show induction of competence upon DNA damage (Charpentier et al. 2011; Dorer et al. 2010; Michod et al. 1988), and thus competence may serve for DNA repair via HR. The only well-studied thermophilic bacteria harboring natural competence systems belong to the genus Thermus. These competence systems seem to be constitutively active (Hidaka et al. 1994), as was also shown for mesophilic N. gonorrhoeae (Dubnau 1999). Bacterial competence is in general subject to regulation by complex signal transduction pathways [for reviews see Claverys et al. (2006), Seitz and Blokesch (2012)].
Due to their vastly different cell envelopes, DNA uptake mechanisms in Gram-positive and -negative bacteria structural differ, in particular, the presence of a secretin ring is unique for the outer membrane of Gram-negative bacteria. However, most of the other proteins are shared (reviewed in Krüger and Stingl 2011) indicating a large similarity in DNA uptake mechanisms. Most bacterial competence systems are composed of a DNA-translocation complex that is coupled to a type IV pilus (T4P) or an evolutionary related type II secretion system (T2SS), proteins in both systems include a pre-pilin peptidase, a secretion ATPase, a polytopic transmembrane protein and pilin- or pseudopilin-subunits. The DNA uptake systems of H. pylori and Campylobacter jejuni form an exception as they are related to the dissimilar T4SSs (described in the “Conjugation” section) (Hofreuter et al. 2000; Stingl et al. 2009).
The exact role of the pili or pseudopili in competence is still not well understood, but their presence is essential for successful DNA uptake (reviewed in Krüger and Stingl 2011). It has been suggested that DNA is brought close to the cell surface by binding to the (pseudo)pili that subsequently retract. Until recently, this hypothesis was questioned as no binding of DNA to (pseudo)pili had ever been observed (Assalkhou et al. 2007; Provvedi and Dubnau 1999). However, a very recent paper described the binding of minor pilin ComP of competent Neisseria meningitidis to DUS sequences suggesting that pili are indeed bringing DNA to the cell surface (Cehovin et al. 2013). For detailed reviews about bacterial competence systems, see Averhoff and Friedrich (2003), Chen and Dubnau (2004), Claverys et al. (2009), Johnsborg et al. (2007) and Krüger and Stingl (2011).
Naturally competent hyperthermophilic Bacteria
Of the at least 70 naturally competent bacteria that are found so far (Johnsborg et al. 2007), six are (hyper)thermophilic. One of these is the thermophilic cyanobacterium Thermosynecoccus elongates BP-1 (Onai et al. 2004), all others belong to the Thermus genus, these are: T. aquaticus, T. caldophilus, T. flavus, T. thermophilus (Koyama et al. 1986) and T. scotoductus (Gounder et al. 2011). With transformation frequencies of up to 10−2, hyperthermophilic T. thermophilus was shown to have the most efficient DNA uptake system of all studied naturally competent bacteria (Koyama et al. 1986). This was confirmed by the fact that DNA uptake had a speed of around 40 kb/s per cell, which is extremely fast (Schwarzenlander and Averhoff 2006). In comparison, other bacteria like B. subtilis and H. influenza show rates of 4 and 16 kb/s per cell, respectively (Deich and Smith 1980; Dubnau 1991). T. thermophilus has for that reason become a model organism for studying natural transformation in bacteria.
Additional properties that make the T. thermophilus DNA uptake system of great interest is that it is constitutively active (Hidaka et al. 1994) and that it is equipped with a broad substrate specificity. T. thermophilus HB27 does not show any bias towards certain types of DNA; genetic material from all domains of life can be taken up with equal efficiencies (Schwarzenlander et al. 2009; Schwarzenlander and Averhoff 2006). From this point of view, one could speculate that the Thermus competence system is involved in HGT rather than in DNA repair. This hypothesis is strengthened by the fact that throughout evolution Thermus species have indeed undergone several HGT events; these events are suggested to be involved in the gain of thermophilicity (Averhoff and Müller 2010; Omelchenko et al. 2005). It is therefore hypothesized that Thermus species developed a highly efficient competence system to acquire all sorts of DNA, which allowed them to adapt through the incorporation of foreign, probably primarily archaeal, genes.
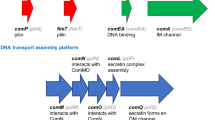
Through whole-genome comparisons and mutational analyses, 16 competence genes were identified in T. thermophilus (Friedrich et al. 2001, 2002, 2003). Genome comparisons further revealed that these genes are conserved among all sequenced Thermus species (César et al. 2011). The proteins encoded by those genes are divided into three groups: type IV pili-related proteins (PilA1–4, PilD, F, C, Q), DNA translocator proteins (ComEA, ComEC, DprA) and novel Thermus-specific proteins (ComZ, PilM, N, O, W) (Averhoff 2009) (Fig. 2a). Among the type IV pili-related proteins there is a conserved pre-pilin signal peptidase PilD which is, as was shown in many other bacteria and archaea, responsible for the maturation of the pilin subunits (Nunn and Lory 1991). Four predicted pilin subunits are present: PilA1, PilA2, PilA3 and PilA4, which are essential for pili formation (Friedrich et al. 2002). So far, no ATPase energizing the assembly of the type IV pili has been identified in T. thermophilus. However, a deletion mutant of a second traffic-ATPase, PilF, resulted in piliated but non-competent cells (Friedrich et al. 2001), PilF is therefore thought to be a retraction ATPase functioning similar to PilT in N. gonorrhoeae (Maier et al. 2002; Wolfgang et al. 1998). Other type IV pili-related proteins are: the inner membrane protein PilC, anchoring the pili into the inner membrane, and the outer membrane protein PilQ whose structure was recently elucidated (Burkhardt et al. 2011). The latter is similar to secretins in other Gram-negative bacteria and was shown to bind DNA. PilQ is thought to form a membrane channel that guides the DNA through the outer membrane to the DNA translocator which is present in the inner membrane (Burkhardt et al. 2012; Rumszauer et al. 2006; Schwarzenlander et al. 2009).
Proposed DNA transfer systems in (hyper)thermophilic Thermus and Sulfolobus species. a The natural competence system of T. thermophilus: Pilin subunits PilA1–4 are processed by signal peptidase PilD and subsequently build into a pilus. PilC anchors the structure into the inner membrane. DNA enters the cell envelope via outer membrane protein PilQ. Upon pili retraction energized by the ATPase PilF, DNA enters the periplasmic space and it is handed over by ComEA to channel-forming ComEC transporting the DNA further into the cytoplasm. Before entering the cell, one strand of the DNA is degraded by a yet unknown nuclease while the other strand of incoming DNA is guided by DprA to RecA. Accessory proteins PilM, PilN, PilO, PilW and ComZ, are either involved in the assembly of the transporter and/or assist transport of DNA across the inner membrane. b The UV-inducible pili system in of Sulfolobales (Ups-system): Pilin subunits UpsA and UpsB are processed by signal peptidase PibD and subsequently build into a pilus, energized by ATPase UpsE. UpsF anchors the structure into the membrane. OM outer membrane, PG peptidoglycan, IM inner membrane, M membrane
The three DNA translocator proteins found in T. thermophilus are all homologous to conserved competence proteins of other bacteria: ComEA is a protein anchored to the inner membrane that binds DNA and subsequently guides it to ComEC. ComEC forms a channel in the inner membrane and transports the DNA into the cytosol. During this process, incoming DNA is probably linearized by an unknown nuclease that degrades one of the two strands. Single stranded DNA is subsequently thought to be handed over to RecA by DprA as shown for S. pneumoniae (Mortier-Barrière et al. 2007). As RecA is essential for transformation (Martin et al. 1995), transferred DNA might then be integrated into the genomic DNA via a RecA-dependent process.
Not much is known about the Thermus-specific competence proteins; PilM, N, O and ComZ are all present in the cytoplasmic membrane (Friedrich et al. 2003; Rumszauer et al. 2006) and are suggested to be involved in the assembly of the DNA-translocation system (Averhoff and Müller 2010). PilW is thought to be responsible for proper localization of PilQ in the outer membrane (Rumszauer et al. 2006). As T. thermophilus lives at extremely high temperatures, the unique competence proteins might be an evolutionary adaptation to these special environmental conditions (Averhoff and Müller 2010). For detailed reviews about the competence system in T. thermophilus see, Averhoff (2009), Averhoff and Müller (2010) and César et al. (2011).
Naturally competent hyperthermophilic Archaea
Only four archaeal species have so far been found to be naturally competent (Bertani and Baresi 1987; Sato et al. 2003; Waege et al. 2010; Worrell et al. 1988), including two hyperthermophilic organisms. The first naturally transformable hyperthermophilic archaeon was Thermococcus kodakarensis (Sato et al. 2003), a sulphur reducing euryarchaeon that lives in marine hydrothermal vents and has an optimal growth temperature of 85 °C. It can be transformed naturally with linear and circular DNA, which is subsequently integrated into the genomic DNA. The observed transformation frequencies of 10−7 are rather low (Sato et al. 2003, 2005). Another naturally competent member of the Thermococcales is P. furiosus which grows at temperatures around 100 °C (Waege et al. 2010). Also this species can be transformed with linear and circular DNA and shows transformation frequencies of up to 10−3 approaching those of highly competent bacteria (Lipscomb et al. 2011). Among the crenarchaeota, so far no naturally competent organisms have been found. Nevertheless, Grogan and Stengel (2008) were able to show that the hyperthermophilic S. acidocaldarius is able to constitutively take up short single stranded oligonucleotides. Though, as earlier experiments revealed that no DNA uptake could be observed when cells were mixed with purified marker DNA, culture supernatant or lysed cells (Grogan 1996), one could question if we can speak here about true natural competence for S. acidocaldarius.
The mechanisms of natural transformation in (hyperthermophilic) archaea have not been studied, and the systems involved have not been identified as homologs of bacterial competence systems have sofar not been found (Averhoff 2009; Claverys et al. 2009). However, the complete DNA sequence of hyperthermophilic crenarchaeon Thermoproteus tenax, revealed the presence of a few homologs of bacterial genes involved in competence (dprA and comF). Since these have not been detected in any other archaea, they were probably acquired from bacteria via HGT (Siebers et al. 2011). However, the existence of a functional bacterial-like competence system in T. tenax needs to be proven. It is likely that naturally transformable archaea that did not acquire competence genes from bacteria have other unrelated DNA uptake mechanisms, which will be a challenge for future studies.
Transduction and HGT among hyperthermophilic viruses
Transduction refers to the transfer of DNA from one cell to another by means of viruses and was first described for Salmonella typhimurium (Zinder and Lederberg 1952). In this process, viruses accidentally package host DNA along with their own DNA and subsequently infect other prokaryotes. In that way, surviving host cells might acquire genetic material from previous hosts (Canchaya et al. 2003). Transduction therefore probably contributed significantly to HGT. Whole-genome analyses indeed suggest that phages played an important role in the acquisition of new genes thereby promoted genetic diversity (Bordenstein and Reznikoff 2005; Brüssow et al. 2004; Ochman et al. 2000; Pallen and Wren 2007). Moreover, when two different viruses infect the same host simultaneously, HGT might not only occur between previous and current hosts, but also among the viruses (Krupovic et al. 2011), leading to viral genetic diversity.
Similar to viruses, gene transfer agents (GTAs), which are virus-like elements encoded by the host-genome, contribute to HGT. It has been demonstrated that GTAs can horizontally transfer a kanamycin resistance gene very efficiently between bacterial species (McDaniel et al. 2010), which implies a general importance of GTAs in HGT. However, so far no GTAs have been found in hyperthermophilic organisms.
Since 1959, about 6,300 prokaryotic viruses have been described morphologically (Ackermann 1996, 2001, 2007; Ackermann and Prangishvili 2012; Eisenstark 1967). Most of these are bacterial viruses (98.5 %), however, over the past few years increasing numbers of archaeal viruses have been described, especially those infecting hyperthermophilic crenarchaea. All studied hyperthermophilic viruses were shown to contain DNA and not RNA, but a recent study revealed the presence of viral RNA in acidic hot springs in Yellowstone National Park, USA, probably originating from viruses infecting hyperthermophilic archaea. These unidentified RNA viruses might form a novel group of phages (Bolduc et al. 2012). For a detailed review about bacterial and archaeal viruses, see Krupovic et al. (2011).
Viruses infecting hyperthermophilic Bacteria
Within the (hyper)thermophilic bacteria, so far only a relatively small number of phages has been found. In total 27 hyperthermophilic bacteriophages have been morphologically described, all infecting Thermus species (Ackermann and Prangishvili 2012). These include: Inoviridae, Myoviridae, Siphoviridae, and Tectiviridae. The genomes of myovirus φYS40 (Naryshkina et al. 2006; Sakaki and Oshima 1975), siphoviruses P23-45, P74-26 (Minakhin et al. 2008; Yu et al. 2006), IN93 and icosahedral virus P23-77 (Jaatinen et al. 2008; Jalasvuori et al. 2009) were sequenced which contributed greatly to the understanding of regulation of transcription and translation as well as the evolution of Thermus phages. The sequence of P23-77 showed an evolutionary link to another Thermus phage, IN93. Interestingly, it also revealed evolutionary relationships to haloarchaeal plasmid pHH205, an integrated Haloarcula provirus, and the Haloarcula virus SH1. These similarities include homologies between two major capsid proteins and a putative packaging ATPase (Jalasvuori et al. 2009). A possible explanation for their relatedness could be that the haloarchaeal and Thermus viruses evolutionarily diverged from each other along with their hosts. Subsequently, the viruses may have evolved into plasmids or proviruses. Another hypothesis is that viruses were able to cross the domain barrier and could thereby mediate HGT between the two domains (Jalasvuori et al. 2009). In the course of this process, other host genes might also have been transferred.
The DNA sequence from another bacteriophage φIN93 revealed the presence of an IS that was shown to originate from its host T. thermophilus (Matsushita and Yanase 2009). Upon a future infection, the virus might again transfer this IS to another organism, leading to HGT.
Viruses infecting hyperthermophilic Archaea
Among the studied hyperthermophilic viruses, most were found to infect members of the crenarchaeota, including: Acidianus, Aeropyrum, Pyrobaculum, Stygiolobus, Sulfolobus and Thermoproteus species. These viruses were shown to be highly diverse both on morphological and genomic level and have been classified into eight viral families: Ampullaviridae, Bacaudaviridae, Clavaviridae, Fuselloviridae, Globuloviridae, Guttaviridae, Lipothrixviridae and Rudiviridae (Ackermann and Prangishvili 2012).
The spindle-shaped fuselloviruses that infect Sulfolobus and Acidianus species are among the best-studied crenarchaeal viruses (Lipps 2006). The genome of Sulfolobus spindle-shaped virus 1 (SSV1), isolated from Sulfolobus shibatae was the very first archaeal viral genome being sequenced (Palm et al. 1991). Since then, 13 more fusellovirus genome sequences have been published (Held and Whitaker 2009; Redder et al. 2009). As was shown for conjugative plasmid pNOB8, SSVs can stably integrate in a Sulfolobus chromosome via integration within a tRNA gene (in the so-called attP site), thereby forming a provirus (Held and Whitaker 2009; Muskhelishvili et al. 1993). This site-specific integration is thought to be an important mechanism for HGT and genome evolution. The SSV1-type integrase that is essential for this integration, is next to the pNOB8-type integrase (described in the “Conjugation” section) of particular interest in studies on the integration of crenarchaeal mobile genetic elements (She et al. 2006; She et al. 2004). SSVs might in addition be involved in the HGT of plasmids; upon an SSV-infection, non-conjugative pRN-like plasmids could be encapsulated into virus-like particles that are released from the cell. Subsequently, they can spread with the help of the viruses and integrate into a following host genome using a self-encoded SSV1-type integrase (Arnold et al. 1999; Wang et al. 2007). Although they cannot infect S. acidocaldarius, SSVs are not host species dependent, it was shown that they can infect different Sulfolobales (Ceballos et al. 2012): Consequently, virus-mediated HGT might have occurred between the different hosts of SSVs (Ceballos et al. 2012).
Next to the Fuselloviridae, members from the Rudi- and Lipothrixviridae also infect Sulfolobus and Acidianus species; these are helical viruses with a linear dsDNA genome (Prangishvili et al. 2006a). Comparative genomics revealed that members from the two virus families horizontally exchanged genes. Interestingly, it was shown that both viruses also obtained genes from their hosts (Peng et al. 2001). The latter include genes encoding a dUTPase: a thymidylate synthase (ThyX) and a Holliday junction resolvase (Peng et al. 2001; Prangishvili et al. 2006b). This acquisition of host genes could have possibly led to HGT between different hosts. Comparable to the fusseloviruses, a lipothrixvirus also shows a relationship with an integrative plasmid. Acidianus filamentous virus 1 (AFV1) mediates the horizontal spread of pAH1 between different hosts. The mechanism involved is so far not known (Basta et al. 2009).
Other crenarchaeal viruses involved in HGT are the Sulfolobus turreted icosahedral viruses (STIVs). A sequence comparison between STIV and related STIV2, revealed a similar genome organization as well as the loss and gain of several genes (Happonen et al. 2010). STIV2 shares genes with other archaeal viruses, including the Fuselloviridae, and it also contains a conserved crenarchaeal gene encoding a DNA-binding protein. Moreover, Thermococcus kodakarensis virus 4 (TKV4) and Methanococcus voltae virus (MVV) were shown to encode capsid proteins and genome packaging ATPases that were similar to those from STIV-like viruses (Krupovic and Bamford 2008). It is therefore clear that HGT has occurred between these archaeal viruses (Koonin 2009; Koonin and Wolf 2008). This transfer of DNA might have been mediated by viruses structurally related to STIV able to infect both eury- and crenarchaeota (Krupovic et al. 2011).
Only two hyperthermophilic euryarchaeal viruses have been described, both infecting members from the order of thermococcales: Pyrococcus abyssi virus 1 (PAV1) (Geslin et al. 2003, 2007) and Thermococcus prieurii virus 1 (TPV1) (Gorlas et al. 2012). They are morphologically similar to the Fuselloviridae and infect cells without causing lysis. Interestingly, the PAV1 genome shows homology to several different plasmids and integrating elements from other archaeal species; these genes are all encoded on approximately the same half of the viral genome. Genes encoding capsid proteins are located on the other half of the genome. This genetic organization could be due to a fusion of a plasmid and a virus, or otherwise, PAV1 might have given rise to plasmids and integrating elements. As was described for SSV1 and AFV1 viruses, PAV1-like viruses might have been involved in the horizontal transfer of plasmids (Krupovic et al. 2010).
In general, it seems clear that virus infections contributed significantly to HGT between viruses as well as their hosts. In the course of evolution this has led to a highly diverse virus population as well as dynamic prokaryotic genomes. Future studies might give more detailed insights in the impact that viruses have on the evolution of their hosts especially in hyperthermophilic environments.
Membrane vesicles, nanopods and nanotubes
The release of membrane vesicles (MVs) is an important physiological process for organisms from all domains of life. MVs mediate the intercellular transfer of several different biological compounds including DNA. They might therefore have contributed to HGT. Both bacterial and archaeal MVs are produced from the cell surface (Deatherage and Cookson 2012). Not much is known about how this is achieved in Gram-positive bacteria, but in Gram-negative bacteria the release of vesicles is thought to be promoted by temporal disruptions of the interaction between the outer membrane and the peptidoglycan (Deatherage and Cookson 2012). In archaea on the other hand, in Sulfolobales the release of MVs appears to occur by membrane scission events that are facilitated by ESCRT-III proteins (Ellen et al. 2009). In other archaeal species that do not contain ESCRT-III homologues, such as Thermococcus, the mode of vesicle formation is unknown. For recent reviews about MV formation in bacteria and archaea, see Deatherage and Cookson (2012), Kulp and Kuehn (2010) and Mashburn-Warren and Whiteley (2006).
Several bacteria were shown to produce DNA-containing vesicles (Dorward et al. 1989; Kahn et al. 1982; Kolling and Matthews 1999; Pérez-Cruz et al. 2013; Renelli et al. 2004; Rumbo et al. 2011; Yaron et al. 2000). However, no MV release has been observed for hyperthermophilic bacteria. Hyperthermophilic archaea on the other hand do generate MVs. Sulfolobus species were shown to produce MVs harboring toxins killing other Sulfolobus strains (sulfolobicins) (Ellen et al. 2009; Prangishvili et al. 2000). Moreover, Ignicoccus species produce many periplasmic MVs (Rachel et al. 2002). Euryarchaeota from the order of Thermococcales commonly release MVs that were previously thought to be viruses. Some of these MVs were shown to be associated with DNA that is highly resistant to DNAse treatment and high temperatures (Soler et al. 2008). MVs from Thermococcus nautilus contain the endogenous plasmid pTN1 (Soler et al. 2011). If a derivative from this plasmid, shuttle vector pLC70 (Santangelo et al. 2008), is transformed to T. kodakarensis, the cells start to release MVs containing the same plasmid. These MVs can be subsequently used to again transfer pLC70 into plasmid-free cells. As the transfer of DNA was insensitive to DNAse treatment, DNA must have been present inside the MVs. MVs might therefore function in both the protection as well as the transfer of DNA (Gaudin et al. 2012). Comparative genomics showed that HGT has taken place between Thermococcus and Thermotoga. Moreover, it is thought that certain plasmids have been horizontally transferred between the Thermococcales and Methanococcales (Krupovic et al. 2013). Future studies need to determine whether or not MVs can also be transferred efficiently between different species. This would imply an important role of MVs in the above-described HGT events (Marguet et al. 2013).
An interesting structure, first described for the Gram-negative soil bacterium Delftia sp. Cs1, is the so-called nanopod (Shetty et al. 2011). Nanopods are prokaryotic organelles, used for projecting MVs several micrometers away from the cell. Very recently, both T. gammatolerans and T. kodakarensis were shown to have tubular structures with a row of internal vesicles resembling bacterial nanopods (Marguet et al. 2013). Similar to the nanopods from Delftia sp. Cs1, the nanopods from Thermococcus species might also project MVs and could thereby increase the distance and perhaps also efficiency of MV transfer between cells (Marguet et al. 2013). Next to nanopods, also nanotubes can be found in Thermococcales (Marguet et al. 2013). Nanotubes are intra- or inter-species tubular cytoplasmic bridges between neighboring cells and were first described for B. subtilis, S. aureus and E. coli. As the structures are between 30 and 100 nm wide, all sorts of molecules including DNA can be transferred between connected cells (Dubey and Ben-Yehuda 2011). Very recently, nanotubes with a diameter of 60–80 nm have been observed in the hyperthermophilic Thermococcus sp. 5-4 (Marguet et al. 2013) (Fig. 1b). Given their similar appearance, these archaeal structures might also be involved in exchange of biomolecules, as is observed for bacterial nanotubes. In that way they might facilitate DNA transfer between cells from similar or different species. As nanotubes now have been observed in a range of bacteria and archaea, they appear to be more widespread than previously thought. Other tubular structures observed in hyperthermophilic archaea are the cannulae found between Pyrodictium cells (König et al. 1988). Cryo-electron tomography, however, revealed that the cannulae enter the periplasmic space, but not the cytoplasm (Nickell et al. 2003). It is therefore unlikely that they transfer DNA.
UV-inducible pili of Sulfolobales (Ups)
A further DNA transfer mechanism that involves type IV pili is a UV-induced system in hyperthermophilic Sulfolobales (Schmidt et al. 1999). The type IV pili that are involved in this DNA exchange are the so-called Ups-pili (UV-inducible pili of Sulfolobales) (Fröls et al. 2008). These pili were shown to mediate UV-induced cellular aggregation during which chromosomal marker exchange could be observed. With marker recombination frequencies of up to 10−2 the efficiency of DNA transfer is very high (Ajon et al. 2011). Next to UV stress, the DNA damaging agent bleomycin could also induce cellular aggregation. Thus, the trigger for pili formation and subsequent cellular aggregation is therefore probably the formation of DNA DSBs (Fröls et al. 2008). Since Ups-knockout strains showed decreased survival rates upon UV treatment (Ajon et al. 2011), the Ups-pili are proposed to be part of a unique HR-based “community” DNA repair mechanism (Fröls et al. 2009). Correspondingly, the formation of cellular aggregates only takes place between cells from the same Sulfolobus species ensuring species-specific DNA exchange essential for DNA repair (Ajon et al. 2011). Recent data suggest that the intraspecies recognition of Sulfolobus species is determined by S-layer glycosylation patterns and the Ups-pilin subunits (van Wolferen and Albers, unpublished). Additional studies on the molecular basis of this self-recognition will give more insights in how intraspecies communication is achieved and what are the barriers that drive speciation (Cadillo-Quiroz et al. 2012).
The Ups-gene cluster is conserved among all Sulfolobales and encodes five proteins: UpsX, a hypothetical protein; UpsE, a secretion ATPase; UpsF, an integral membrane protein; and UpsA/B, two putative pilin subunits containing class III pre-pilin signal peptides (Fig. 2b). Directly downstream to the ups gene cluster, three genes are present encoding predicted DNA processing proteins. These are: an endonuclease III homologue, a ParB-like protein and an ATP-dependent helicase. Deletion of any of these genes in S. acidocaldarius results in lower survival rates upon UV treatment (van Wolferen and Albers, unpublished), we therefore speculate that they are involved in either in the DNA uptake or the HR-mediated DNA repair pathway that is linked to the Ups-system. A role in HR seems more likely as the helicase ortholog from S. solfataricus (Sso0112) was shown to catalyze the processing of Holliday junctions (Valenti et al. 2012).
Because type IV pili are often involved in the competence systems, one could speculate that uptake of DNA from lysed cells takes place rather than direct DNA exchange. However, the exchange of DNA was shown to be insensitive to DNAse treatment. Moreover, efforts to obtain recombinants by mixing cells with supernatant, lysed cells or marker DNA failed (Grogan 1996). This demonstrates that exchange of DNA indeed occurs directly from one living cell to another. Nevertheless, the mode of DNA transfer and the role of the Ups-pili in this process are still unclear. One could imagine that the Ups-pili are only involved in recognizing other cells from the same species and the initiation of a physical interaction. DNA transfer could subsequently occur via a yet unknown mechanism. Conjugation-like DNA transfer could for instance occur or cell fusion events might take place such as those observed during pNOB8 transfer (Schleper et al. 1995). However, when plating Ups-gene inactivation mutants directly on top of each other, which theoretically would also bring cells in a close proximity, no recombinants can be obtained (Ajon et al. 2011). It can therefore be speculated that the Ups-pili fulfill an active role in DNA exchange. The pili might function in a similar manner as type IV pili involved in natural transformation. In competent bacteria, however, retraction of the pili is almost always essential and in Sulfolobus species so far no retraction ATPase could be found. Furthermore, an unsolved puzzle is the directionality of the DNA transfer in Sulfolobus species. All cells appear to be serving as donor and as recipient (Grogan 1996) which makes sense as all cells are genotypically similar. Following the DNA damage hypothesis, it would be most logical for a cell with damaged DNA to acquire DNA from another cell, possibly functioning as a template for HR. As it seems that there is little to no specificity in the chromosomal DNA that can be transferred between cells, it is hard to use chromosomal markers to study directionality in more detail.
Besides a role in cellular aggregation, Ups-pili were also shown to be involved in biofilm formation (Koerdt et al. 2010). Moreover, Ups-pili may play a role in HGT. Even though Sulfolobus species appear to aggregate only species specifically, in nature rare intra-species DNA exchange might have occurred, thereby promoting HGT. Overall, we can say that Ups-mediated DNA transfer has proven to be a very interesting and unique mechanism of the Sulfolobales. So far, evidences in favor of a role in DNA repair are strong. Future studies might improve our understanding about intraspecies recognition, the mode of DNA transfer and confirm the function of DNA repair.
Summary and concluding remarks
Transfer of DNA has been shown to occur in all domains of life although this process can take place by means of different mechanisms. The widespread occurrence stresses the universal significance of DNA transfer for life on earth. Here, we focused on why and how DNA is transferred among hyperthermophilic prokaryotes. DNA transfer at high temperatures may serve an important role in evolution. HGT has proven to be a powerful driving force for prokaryotic adaptation to changing environments such as rising temperatures. Moreover, as DNA degradation is increased at higher temperatures, specialized DNA repair mechanisms involving DNA uptake may play an important role in cell survival. A third but more controversial role of DNA uptake is the use of nucleotides as a nutrient source.
Numerous DNA transfer mechanisms have been described for hyperthermophilic microorganisms. Some are based on the direct contact of neighboring cells, such as conjugation and the production of nanotubes or UV-induced pili, others involve the direct uptake of DNA from the environment (natural transformation). Lastly, vehicles such as viruses or vesicles might function in DNA transfer. One could speculate that especially at higher temperatures, DNA transfer methods that involve direct cellular contact or DNA-protecting vehicles are favored. In that way, unprotected DNA is not exposed directly to the DNA damaging surroundings. Nevertheless, uptake of free DNA has been described for several (hyper)thermophilic microorganisms, including the Thermus thermophilus competence system. The latter mechanism is the most efficient competence system found thus far in nature, which might relate to its proposed role in the adaptation to the high temperatures. In this respect, a high uptake rate also ensures limited exposure of free DNA to the surroundings.
Future research will undoubtedly expand the array of DNA transfer mechanisms among hyperthermophiles. Though more importantly, observed transfer processes need to be unravelled mechanistically, as their exact functioning is often still far from well understood. Interesting examples include conjugation and natural transformation among hyperthermophilic archaea which are both thought to differ extremely from their bacterial counterparts.
As genetic manipulation of hyperthermophilic bacteria and archaea has shown to be very challenging, further studies on their DNA transfer methods may advance the development of such tools. For example, the natural competence of T. thermophilus (Koyama et al. 1986), T. kodakarensis (Santangelo et al. 2008, 2010; Sato et al. 2003, 2005) and P. furiosis (Basen et al. 2012; Lipscomb et al. 2011) enabled the development of genetic tools. Moreover, the SSV1-based shuttle vector pMJ05 enabled reporter gene studies and the expression of proteins in S. solfataricus (Jonuscheit et al. 2003). Future studies will therefore not only increase our knowledge about the extremely diverse DNA transfer mechanisms functioning at high temperatures, but will also promote the study of other aspects in hyperthermophiles using these genetic systems.
References
Aagaard C, Dalgaard JZ, Garrett RA (1995) Intercellular mobility and homing of an archaeal rDNA intron confers a selective advantage over intron- cells of Sulfolobus acidocaldarius. Proc Natl Acad Sci USA 92:12285–12289
Ackermann HW (1996) Frequency of morphological phage descriptions in 1995. Arch Virol 141:209–218
Ackermann HW (2001) Frequency of morphological phage descriptions in the year 2000. Brief review. Arch Virol 146:843–857
Ackermann HW (2007) 5500 Phages examined in the electron microscope. Arch Virol 152:227–243
Ackermann HW, Prangishvili D (2012) Prokaryote viruses studied by electron microscopy. Arch Virol 157:1843–1849
Ajon M, Fröls S, van Wolferen M et al (2011) UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol Microbiol 82:807–817
Alvarez L, Bricio C, Gómez MJ, Berenguer J (2011) Lateral transfer of the denitrification pathway genes among Thermus thermophilus strains. Appl Environ Microbiol 77:1352–1358
Alvarez-Martinez CE, Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808
Anderson AW, Nordon HC, Cain RF et al (1956) Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol 10:575–578
Aravind L, Tatusov RL, Wolf YI et al (1998) Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet 14:442–444
Arnold HP, She Q, Phan H et al (1999) The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol Microbiol 34:217–226
Assalkhou R, Balasingham S, Collins RF et al (2007) The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 153:1593–1603
Atomi H, Matsumi R, Imanaka T (2004) Reverse gyrase is not a prerequisite for hyperthermophilic life. J Bacteriol 186:4829–4833
Averhoff B (2009) Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol Rev 33:611–626
Averhoff B, Friedrich A (2003) Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch Microbiol 180:385–393
Averhoff B, Müller V (2010) Exploring research frontiers in microbiology: recent advances in halophilic and thermophilic extremophiles. Res Microbiol 161:506–514
Ayora S, Carrasco B, Cárdenas PP et al (2011) Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol Rev 35:1055–1081
Basen M, Sun J, Adams MWW (2012) Engineering a hyperthermophilic archaeon for temperature-dependent product formation. mBio 3:e00053–12
Basta T, Smyth J, Forterre P et al (2009) Novel archaeal plasmid pAH1 and its interactions with the lipothrixvirus AFV1. Mol Microbiol 71:23–34
Beiko RG, Harlow TJ, Ragan MA (2005) Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA 102:14332–14337
Bernstein H, Byers G, Michod R (1981) Evolution of sexual reproduction: importance of DNA repair, complementation, and variation. Am Nat 117:537–549
Bernstein H, Bernstein C, Michod RE (2012) DNA repair as the primary adaptive function of sex in bacteria and eukaryotes. In: Kimura S et al (eds) DNA repair: new research, Nova Science Publishers Inc, pp 1–49. ISBN: 978-1-62100-756-2
Bertani G, Baresi L (1987) Genetic transformation in the methanogen Methanococcus voltae PS. J Bacteriol 169:2730–2738
Bolduc B, Shaughnessy DP, Wolf YI et al (2012) Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol 86:5562–5573
Bordenstein SR, Reznikoff WS (2005) Mobile DNA in obligate intracellular bacteria. Nat Rev Microbiol 3:688–699
Bricio C, Alvarez L, Gómez MJ, Berenguer J (2011) Partial and complete denitrification in Thermus thermophilus: lessons from genome drafts. Biochem Soc Trans 39:249–253
Brochier-Armanet C, Forterre P (2007) Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea 2:83–93
Brock TD, Brock KM, Belly RT, Weiss RL (1972) Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol 84:54–68
Brüggemann H, Chen C (2006) Comparative genomics of Thermus thermophilus: plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J Bacteriol 124:654–661. doi:10.1016/j.jbiotec.2006.03.043
Brüssow H, Canchaya C, Hardt W-D (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602
Burkhardt J, Vonck J, Averhoff B (2011) Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J Biol Chem 286:9977–9984
Burkhardt J, Vonck J, Langer JD et al (2012) Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation. J Biol Chem 287:8484–8494
Cadillo-Quiroz H, Didelot X, Held NL et al (2012) Patterns of gene flow define species of thermophilic Archaea. PLoS Biol 10:e1001265
Campbell BJ, Smith JL, Hanson TE et al (2009) Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet 5:e1000362
Canchaya C, Fournous G, Chibani-Chennoufi S et al (2003) Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424
Capasso G, Favara R, Francofonte S, Inguaggiato S (1999) Chemical and isotopic variations in fumarolic discharge and thermal waters at Vulcano Island (Aeolian Islands, Italy) during 1996: evidence of resumed volcanic activity. J Volcanol Geoth Res 88:167–175
Ceballos RM, Marceau CD, Marceau JO et al (2012) Differential virus host-ranges of the Fuselloviridae of hyperthermophilic Archaea: implications for evolution in extreme environments. Front Microbiol 3:295
Cehovin A, Simpson PJ, McDowell MA et al (2013) Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci USA 110:3065–3070
César CE, Álvarez L, Bricio C et al (2011) Unconventional lateral gene transfer in extreme thermophilic bacteria. Int Microbiol 14:187–199
Charpentier X, Kay E, Schneider D, Shuman HA (2011) Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol 193:1114–1121
Chen I, Dubnau D (2004) DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249
Chen L, Brugger K, Skovgaard M et al (2005) The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol 187:4992–4999
Claverys J-P, Prudhomme M, Martin B (2006) Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60:451–475
Claverys J-P, Martin B, Polard P (2009) The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33:643–656
Cohan FM (1994a) Genetic exchange and evolutionary divergence in prokaryotes. Trends Ecol Evol 9:175–180
Cohan FM (1994b) The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am Nat 143:965–986
Danner DB, Deich RA, Sisco KL, Smith HO (1980) An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311–318
Datta N, Kontomichalou P (1965) Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239–241
de la Cruz F, Frost LS, Meyer RJ, Zechner EL (2010) Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40
Deatherage BL, Cookson BT (2012) Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun 80:1948–1957
Déclais AC, Marsault J, Confalonieri F et al (2000) Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J Biol Chem 275:19498–19504
Deich RA, Smith HO (1980) Mechanism of homospecific DNA uptake in Haemophilus influenzae transformation. Mol Gen Genet 177:369–374
Dickerson RE (1980) Evolution and gene transfer in purple photosynthetic bacteria. Nature 283:210–212
Diruggiero J, Dunn D, Maeder DL et al (2000) Evidence of recent lateral gene transfer among hyperthermophilic archaea. Mol Microbiol 38:684–693
Dorer MS, Fero J, Salama NR (2010) DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog 6:e1001026
Dorward DW, Garon CF, Judd RC (1989) Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol 171:2499–2505
Drake JW (2009) Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet 5:e1000520
Dubey GP, Ben-Yehuda S (2011) Intercellular nanotubes mediate bacterial communication. Cell 144:590–600
Dubnau D (1991) Genetic competence in Bacillus subtilis. Microbiol Rev 55:395–424
Dubnau D (1999) DNA uptake in bacteria. Annu Rev Microbiol 53:217–244
Dykhuizen DE, Green L (1991) Recombination in Escherichia coli and the definition of biological species. J Bacteriol 173:7257–7268
Eisenstark A (1967) Bacteriophage techniques. In: Maramorosch K, Koprowski H (eds) Methods in virology, vol 1. Academic Press, NY, pp 449–525
Elkins C, Thomas CE, Seifert HS, Sparling PF (1991) Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol 173:3911–3913
Ellen AF, Albers S-V, Huibers W et al (2009) Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 13:67–79
Finkel SE, Kolter R (2001) DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol 183:6288–6293
Fitzmaurice WP, Benjamin RC, Huang PC, Scocca JJ (1984) Characterization of recognition sites on bacteriophage HP1c1 DNA which interact with the DNA uptake system of Haemophilus influenzae Rd. Gene 31:187–196
Forterre P (2002) A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet 18:236–723
Friedrich A, Hartsch T, Averhoff B (2001) Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl Environ Microbiol 67:3140–3148
Friedrich A, Prust C, Hartsch T et al (2002) Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl Environ Microbiol 68:745–755
Friedrich A, Rumszauer J, Henne A, Averhoff B (2003) Pilin-like proteins in the extremely thermophilic bacterium Thermus thermophilus HB27: implication in competence for natural transformation and links to type IV pilus biogenesis. Appl Environ Microbiol 69:3695–3700
Fröls S, Ajon M, Wagner M et al (2008) UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol 70:938–952
Fröls S, White MF, Schleper C (2009) Reactions to UV damage in the model archaeon Sulfolobus solfataricus. Biochem Soc Trans 37:36–41
Garcia-Vallvé S, Romeu A, Palau J (2000) Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res 10:1719–1725
Gaudin M, Gauliard E, Schouten S et al (2012) Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ Microbiol Rep 5:109–116
Geslin C, Le Romancer M, Erauso G et al (2003) PAV1, the first virus-like particle isolated from a hyperthermophilic Euryarchaeote, “Pyrococcus abyssi”. J Bacteriol 185:3888–3894
Geslin C, Gaillard M, Flament D et al (2007) Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi. J Bacteriol 189:4510–4519
Ghane F, Grogan DW (1998) Chromosomal marker exchange in the thermophilic archaeon Sulfolobus acidocaldarius: physiological and cellular aspects. Microbiology 144:1649–1657
Gomis-Rüth FX, Coll M (2006) Cut and move: protein machinery for DNA processing in bacterial conjugation. Curr Opin Struct Biol 16:744–772
Gorlas A, Koonin EV, Bienvenu N et al (2012) TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol 14:503–516
Gounder K, Brzuszkiewicz E, Liesegang H et al (2011) Sequence of the hyperplastic genome of the naturally competent Thermus scotoductus SA-01. BMC Genomics 12:577
Greve B, Jensen S, Brügger K et al (2004) Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea 1:231–239
Gribaldo S, Brochier-Armanet C (2006) The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci 361:1007–1022
Grogan DW (1996) Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J Bacteriol 178:3207–3211
Grogan DW, Stengel KR (2008) Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/lambdaRed and Sulfolobus acidocaldarius recombination systems. Mol Microbiol 69:1255–1265
Grogan DW, Carver GT, Drake JW (2001) Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA 98:7928–7933
Guglielmini J, Quintais L, Garcillán-Barcia MP et al (2011) The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7:e1002222
Guglielmini J, de la Cruz F, Rocha EPC (2013) Evolution of conjugation and Type IV secretion systems. Mol Biol Evol 30:315–331
Guiral S, Mitchell TJ, Martin B, Claverys J-P (2005) Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci USA 102:8710–8715
Halary S, Leigh JW, Cheaib B et al (2010) Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci USA 107:127–132
Happonen LJ, Redder P, Peng X et al (2010) Familial relationships in hyperthermo- and acidophilic archaeal viruses. J Virol 84:4747–4754
Håvarstein LS, Martin B, Johnsborg O et al (2006) New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol Microbiol 59:1297–1307
Heine M, Chandra SBC (2009) The linkage between reverse gyrase and hyperthermophiles: a review of their invariable association. J Microbiol 47:229–234
Held NL, Whitaker RJ (2009) Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol 11:457–466
Herriott RM, Meyer EM, Vogt M (1970) Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101:517–524
Hidaka Y, Hasegawa M, Nakahara T, Hoshino T (1994) The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci Biotechnol Biochem 58:1338–1339
Hofreuter D, Odenbreit S, Püls J et al (2000) Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res Microbiol 151:487–491
Jaatinen ST, Happonen LJ, Laurinmäki P et al (2008) Biochemical and structural characterisation of membrane-containing icosahedral dsDNA bacteriophages infecting thermophilic Thermus thermophilus. Virology 379:10–19
Jain R, Rivera MC, Moore JE, Lake JA (2003) Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol 20:1598–1602
Jalasvuori M, Jaatinen ST, Laurinavicius S et al (2009) The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J Virol 83:9388–9397
Johnsborg O, Eldholm V, Håvarstein LS (2007) Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol 158:767–778
Jonuscheit M, Martusewitsch E, Stedman KM, Schleper C (2003) A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol Microbiol 48:1241–1252
Jorth P, Whiteley M (2012) An evolutionary link between natural transformation and CRISPR adaptive immunity. mBio 3:
Kahn ME, Maul G, Goodgal SH (1982) Possible mechanism for donor DNA binding and transport in Haemophilus. Proc Natl Acad Sci USA 79:6370–6374
Kampmann M, Stock D (2004) Reverse gyrase has heat-protective DNA chaperone activity independent of supercoiling. Nucleic Acids Res 32:3537–3545
Kikuchi A, Asai K (1984) Reverse gyrase—a topoisomerase which introduces positive superhelical turns into DNA. Nature 309:677–681
Koerdt A, Gödeke J, Berger J et al (2010) Crenarchaeal biofilm formation under extreme conditions. PLoS ONE 5:e14104
Kolling GL, Matthews KR (1999) Export of virulence genes and shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol 65:1843–1848
König H, Messner P, Stetter KO (1988) The fine structure of the fibers of Pyrodictium occultum. FEMS Microbiol Lett 49:207–212
Koonin EV (2009) On the origin of cells and viruses: primordial virus world scenario. Ann NY Acad Sci 1178:47–64
Koonin EV, Wolf YI (2008) Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719
Koyama Y, Hoshino T, Tomizuka N, Furukawa K (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol 166:338–340
Krüger N-J, Stingl K (2011) Two steps away from novelty—principles of bacterial DNA uptake. Mol Microbiol 80:860–867
Krupovic M, Bamford DH (2008) Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology 375:292–300
Krupovic M, Forterre P, Bamford DH (2010) Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol 397:144–160
Krupovic M, Prangishvili D, Hendrix RW, Bamford DH (2011) Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev 75:610–635
Krupovic M, Gonnet M, Hania WB et al (2013) Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS ONE 8:e49044
Kulp A, Kuehn MJ (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184
Lang AS, Zhaxybayeva O, Beatty JT (2012) Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 10:472–482
Lawley T, Klimke W, Gubbins M, Frost L (2003) F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15
Lawrence JG, Ochman H (1998) Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA 95:9413–9417
Levin BR (1981) Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99:1–23
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362:709–715
Lipps G (2006) Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus. Extremophiles 10:17–28
Lipscomb GL, Stirrett K, Schut GJ et al (2011) Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol 77:2232–2238
Lorenz MG, Wackernagel W (1994) Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev 58:563–602
MacFadyen LP, Chen D, Vo HC et al (2001) Competence development by Haemophilus influenzae is regulated by the availability of nucleic acid precursors. Mol Microbiol 40:700–707
Maier B, Potter L, So M et al (2002) Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci USA 99:16012–16017
Majewski J, Cohan FM (1998) The effect of mismatch repair and heteroduplex formation on sexual isolation in Bacillus. Genetics 148:13–18
Majewski J, Cohan FM (1999) Adapt globally, act locally: the effect of selective sweeps on bacterial sequence diversity. Genetics 152:1459–1474
Marguet E, Gaudin M, Gauliard E et al (2013) Membrane vesicles, nanopods and/or nanotubes produced by hyperthermophilic archaea of the genus Thermococcus. Biochem Soc Trans 41:436–442
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845
Martin B, García P, Castanié MP, Claverys JP (1995) The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol Microbiol 15:367–379
Mashburn-Warren LM, Whiteley M (2006) Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol 61:839–846
Matsushita I, Yanase H (2009) A novel insertion sequence transposed to thermophilic bacteriophage {phi}IN93. J Biochem 146:797–803
McDaniel LD, Young E, Delaney J et al (2010) High frequency of horizontal gene transfer in the oceans. Science 330:50
Meibom KL, Blokesch M, Dolganov NA et al (2005) Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827
Mell JC, Hall IM, Redfield RJ (2012) Defining the DNA uptake specificity of naturally competent Haemophilus influenzae. Nucleic Acids Res 40:8536–8549
Michod RE, Wojciechowski MF, Hoelzer MA (1988) DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics 118:31–39
Minakhin L, Goel M, Berdygulova Z et al (2008) Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23-45 and P74-26: siphoviruses with triplex-forming sequences and the longest known tails. J Mol Biol 378:468–480
Mortier-Barrière I, Velten M, Dupaigne P et al (2007) A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130:824–836
Murray NE (2002) Immigration control of DNA in bacteria: self versus non-self. Microbiology 148:3–20
Muskhelishvili G, Palm P, Zillig W (1993) SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol Gen Genet 237:334–342
Nakamura Y, Itoh T, Matsuda H, Gojobori T (2004) Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet 36:760–766
Napoli A, Valenti A, Salerno V et al (2004) Reverse gyrase recruitment to DNA after UV light irradiation in Sulfolobus solfataricus. J Biol Chem 279:33192–33198
Naryshkina T, Liu J, Florens L et al (2006) Thermus thermophilus bacteriophage phiYS40 genome and proteomic characterization of virions. J Mol Biol 364:667–677
Nesbø CL, L’Haridon S, Stetter KO, Doolittle WF (2001) Phylogenetic analyses of two “Archaeal” genes in Thermotoga maritima reveal multiple transfers between archaea and bacteria. Mol Biol Evol 18:362–375
Nickell S, Hegerl R, Baumeister W, Rachel R (2003) Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography. J Struct Biol 141:34–42
Norman A, Hansen LH, Sørensen SJ (2009) Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci 364:2275–2289
Nunn DN, Lory S (1991) Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA 88:3281–3285
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Omelchenko MV, Wolf YI, Gaidamakova EK et al (2005) Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol Biol 5:57
Onai K, Morishita M, Kaneko T et al (2004) Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: a simple and efficient method for gene transfer. Mol Gen Genet 271:50–59
Oshima T, Imahori K (1974) Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol 24:102–112
Palchevskiy V, Finkel SE (2006) Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188:3902–3910
Palchevskiy V, Finkel SE (2009) A role for single-stranded exonucleases in the use of DNA as a nutrient. J Bacteriol 191:3712–3716
Pallen MJ, Wren BW (2007) Bacterial pathogenomics. Nature 449:835–842
Palm P, Schleper C, Grampp B et al (1991) Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250
Peng X, Blum H, She Q et al (2001) Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226–234
Pérez-Cruz C, Carrión O, Delgado L et al (2013) A new type of outer membrane vesicles produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl Environ Microbiol AEM. doi: 10.1128/AEM.03657-12
Pérez-Mendoza D, de la Cruz F (2009) Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genomics 10:71
Popa O, Dagan T (2011) Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol 14:615–623
Prangishvili D, Albers SV, Holz I et al (1998) Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid 40:190–202
Prangishvili D, Holz I, Stieger E et al (2000) Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J Bacteriol 182:2985–2988
Prangishvili D, Forterre P, Garrett RA (2006a) Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4:837–848
Prangishvili D, Garrett RA, Koonin EV (2006b) Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res 117:52–67
Provvedi R, Dubnau D (1999) ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol Microbiol 31:271–280
Puigbò P, Wolf YI, Koonin EV (2010) The tree and net components of prokaryote evolution. Genome Biol Evol 2:745–756
Rachel R, Wyschkony I, Riehl S, Huber H (2002) The ultrastructure of Ignicoccus: evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea 1:9–18
Ramírez-Arcos S, Fernández-Herrero LA, Marín I, Berenguer J (1998) Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J Bacteriol 180:3137–3143
Redder P, Peng X, Brügger K et al (2009) Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol 11:2849–2862
Redfield RJ (1988) Evolution of bacterial transformation: is sex with dead cells ever better than no sex at all? Genetics 119:213–221
Redfield RJ (1993a) Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered 84:400–404
Redfield RJ (1993b) Evolution of natural transformation: testing the DNA repair hypothesis in Bacillus subtilis and Haemophilus influenzae. Genetics 133:755–761
Redfield RJ, Schrag MR, Dean AM (1997) The evolution of bacterial transformation: sex with poor relations. Genetics 146:27–38
Renelli M, Matias V, Lo RY, Beveridge TJ (2004) DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150:2161–2169
Rosenshine I, Tchelet R, Mevarech M (1989) The mechanism of DNA transfer in the mating system of an archaebacterium. Science 245:1387–1389
Rumbo C, Fernández-Moreira E, Merino M et al (2011) Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Ag Chemother 55:3084–3090
Rumszauer J, Schwarzenlander C, Averhoff B (2006) Identification, subcellular localization and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J 273:3261–3272
Sakaki Y, Oshima T (1975) Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus HB8. J Virol 15:1449–1453
Santangelo TJ, Cubonová L, Reeve JN (2008) Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl Environ Microbiol 74:3099–3104
Santangelo TJ, Cubonová L, Reeve JN (2010) Thermococcus kodakarensis genetics: TK1827-encoded beta-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl Environ Microbiol 76:1044–1052
Sato T, Fukui T, Atomi H, Imanaka T (2003) Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:210–220
Sato T, Fukui T, Atomi H, Imanaka T (2005) Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol 71:3889–3899
Schleper C, Holz I, Janekovic D et al (1995) A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J Bacteriol 177:4417–4426
Schmidt KJ, Beck KE, Grogan DW (1999) UV stimulation of chromosomal marker exchange in Sulfolobus acidocaldarius: implications for DNA repair, conjugation and homologous recombination at extremely high temperatures. Genetics 152:1407–1415
Schröder G, Lanka E (2005) The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1–25
Schwarzenlander C, Averhoff B (2006) Characterization of DNA transport in the thermophilic bacterium Thermus thermophilus HB27. FEBS J 273:4210–4218
Schwarzenlander C, Haase W, Averhoff B (2009) The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ Microbiol 11:801–808
Sedwick P, Stuben D (1996) Chemistry of shallow submarine warm springs in an arc-volcanic setting: Volcano Island, Aeolian Archipelago, Italy. Mar Chem 53:15
Seitz P, Blokesch M (2012) Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363
She Q, Phan H, Garrett RA et al (1998) Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2:417–425
She Q, Shen B, Chen L (2004) Archaeal integrases and mechanisms of gene capture. Biochem Soc Trans 32:222–226
She Q, Zhu H, Xiang X (2006) Integration mechanisms: possible role in genome evolution. In: Garrett RA, Klenk H-P (eds) Archaea. Blackwell Publishing Ltd, Malden, pp 113–124
Shetty A, Chen S, Tocheva EI et al (2011) Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS ONE 6:e20725
Siebers B, Zaparty M, Raddatz G et al (2011) The complete genome sequence of Thermoproteus tenax: a physiologically versatile member of the Crenarchaeota. PLoS ONE 6:e24222
Silverman PM (1997) Towards a structural biology of bacterial conjugation. Mol Microbiol 23:423–429
Soler N, Marguet E, Verbavatz J-M, Forterre P (2008) Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res Microbiol 159:390–399
Soler N, Gaudin M, Marguet E, Forterre P (2011) Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem Soc Trans 39:36–44
Stedman KM, She Q, Phan H et al (2000) pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J Bacteriol 182:7014–7020
Stetter KO (2006a) Hyperthermophiles in the history of life. Philos Trans R Soc Lond B Biol Sci 361:1837–1843
Stetter KO (2006b) History of discovery of the first hyperthermophiles. Extremophiles 10:357–362
Stetter KO (2013) A brief history of the discovery of hyperthermophilic life. Biochem Soc Trans 41:416–420
Stingl K, Muller S, Scheidgen-Kleyboldt G et al (2009) Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA 107:1184–1189
Suckow G, Seitz P, Blokesch M (2011) Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol 193:4914–4924
Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721
Valenti A, De Felice M, Perugino G et al (2012) Synergic and opposing activities of thermophilic RecQ-like helicase and topoisomerase 3 proteins in Holliday junction processing and replication fork stabilization. J Biol Chem 287:30282–30295
van Passel MWJ (2008) An intragenic distribution bias of DNA uptake sequences in Pasteurellaceae and Neisseriae. Biol Direct 3:12
Vogelmann J, Ammelburg M, Finger C et al (2011) Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J 30:2246–2254
Waege I, Schmid G, Thumann S et al (2010) Shuttle vector-based transformation system for Pyrococcus furiosus. Appl Environ Microbiol 76:3308–3313
Wang Y, Duan Z, Zhu H et al (2007) A novel Sulfolobus non-conjugative extrachromosomal genetic element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology 363:124–133
Weisburg WG, Giovannoni SJ, Woese CR (1989) The Deinococcus-Thermus phylum and the effect of rRNA composition on phylogenetic tree construction. Syst Appl Microbiol 11:128–134
White MF (2011) Homologous recombination in the archaea: the means justify the ends. Biochem Soc Trans 39:15–19
White JR, Escobar-Paramo P, Mongodin EF et al (2008) Extensive genome rearrangements and multiple horizontal gene transfers in a population of Pyrococcus isolates from Vulcano Island, Italy. Appl Environ Microbiol 74:6447–6451
Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV (2001) Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res 11:356–372
Wolfgang M, Lauer P, Park HS et al (1998) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29:321–330
Worrell VE, Nagle DP, McCarthy D, Eisenbraun A (1988) Genetic transformation system in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J Bacteriol 170:653–656
Wozniak RAF, Waldor MK (2010) Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563
Yaron S, Kolling GL, Simon L, Matthews KR (2000) Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol 66:4414–4420
Yu MX, Slater MR, Ackermann H-W (2006) Isolation and characterization of Thermus bacteriophages. Arch Virol 151:663–679
Zaneveld JR, Nemergut DR, Knight R (2008) Are all horizontal gene transfers created equal? Prospects for mechanism-based studies of HGT patterns. Microbiology 154:1–15
Zawadzki P, Roberts MS, Cohan FM (1995) The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics 140:917–932
Zhang C, Krause DJ, Whitaker RJ (2013) Sulfolobus islandicus: a model system for evolutionary genomics. Biochem Soc Trans 41:458–462
Zinder ND, Lederberg J (1952) Genetic exchange in Salmonella. J Bacteriol 64:679–699
Acknowledgments
M.vW was supported by a grant from the German Science Foundation (DFG, AL1206/3-1). M.A was supported by an ALW grant from the Dutch Science Organization (NWO). S.V.A. received support from intramural funds of the Max Planck Society.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Rights and permissions
About this article
Cite this article
van Wolferen, M., Ajon, M., Driessen, A.J.M. et al. How hyperthermophiles adapt to change their lives: DNA exchange in extreme conditions. Extremophiles 17, 545–563 (2013). https://doi.org/10.1007/s00792-013-0552-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-013-0552-6