Abstract
The genes of ectoine biosynthesis pathway were identified in six species of aerobic, slightly halophilic bacteria utilizing methane, methanol or methylamine. Two types of ectoine gene cluster organization were revealed in the methylotrophs. The gene cluster ectABC coding for diaminobutyric acid (DABA) acetyltransferase (EctA), DABA aminotransferase (EctB) and ectoine synthase (EctC) was found in methanotrophs Methylobacter marinus 7C and Methylomicrobium kenyense AMO1T. In methanotroph Methylomicrobium alcaliphilum ML1, methanol-utilizers Methylophaga thalassica 33146T , Methylophaga alcalica M8 and methylamine-utilizer Methylarcula marina h1T, the genes forming the ectABC–ask operon are preceded by ectR, encoding a putative transcriptional regulatory protein EctR. Phylogenetic relationships of the Ect proteins do not correlate with phylogenetic affiliation of the strains, thus implying that the ability of methylotrophs to produce ectoine is most likely the result of a horizontal transfer event.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As known, microorganisms living in fluctuating salinity environments have developed several strategies to survive during osmotic up- and downshifts. The majority of moderately halophilic and halotolerant bacteria respond to an external salinity increase by accumulation of low-molecular weight organic compounds, so-called compatible solutes. Even if accumulated in exceedingly high concentrations (up to several moles per liter), compatible solutes do not interfere with cellular metabolism. Different classes of compatible solutes, such as amino acids, ectoines, betaines, sugars, and polyols, have been described. The cyclic imino acid ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidine carboxylic acid) serves as a compatible solute in a variety of moderately halophilic and halotolerant bacteria (Galinski et al. 1985; Severin et al. 1992; Galinski 1995; Ventosa et al. 1998; Kempf and Bremer 1998; Grant 2004; Oren 2002, 2008). Ectoine is widely used in biotechnology, cosmetics and medicine as a multifunctional bioprotectant (Lippert and Galinski 1992; Buenger 1999; Göller and Galinski 1999; Graf et al. 2008; Knap et al. 1999). Detailed study on organization/regulation of the ectoine biosynthetic pathway in various bacterial producers is an active research area. Ectoine biosynthesis represents a branch in the pathway for synthesis of the aspartate family amino acids and includes three additional special enzymes: diaminobutyric acid (DABA) acetyltransferase (EctA, EC 2.3.1.178), DABA aminotransferase (EctB, EC 2.6.1.76) and ectoine synthase (EctC, EC 4.2.1.108) (Peters et al. 1990; Ono et al. 1999). The genes coding for these enzymes are usually located in the gene clusters ectABC or ectABC–ask (Louis and Galinski 1997; Kuhlmann and Bremer 2002; Vargas et al. 2008; Schwibbert et al. 2010; Lo et al. 2009).
Aerobic methylotrophic bacteria (methylotrophs, also known as C1-utilizers) are able to use single carbon compounds such as methane, methanol or methylamine as a sole source of carbon and energy (Anthony 1982; Hanson and Hanson 1996). Methylotrophic bacteria were found by us in a variety of aquatic and terrestrial habitats, including saline ecosystems (Lidstrom 2006; Trotsenko and Khmelenina 2002; Kalyuzhnaya et al. 2008). Recently, we have shown that the ability to synthesize ectoine plays a key role in osmoadaptation of aerobic methylotrophic bacteria (Doronina et al. 1998, 2003a, b; Khmelenina et al. 1999, 2010). However, the biochemistry and genetics of ectoine biosynthesis were characterized only in halotolerant obligate methanotroph Methylomicrobium alcaliphilum 20Z (synonym Methylobacter alcaliphilus) (Reshetnikov et al. 2005, 2006; Mustakhimov et al. 2010). In this study, the nucleotide sequences of the ectoine biosynthetic genes were determined in six species of methylotrophic bacteria isolated from various saline habitats. We found that organization of the ect genes in either the ectABC or ectR–ectABC–ask gene cluster well correlated with the salt tolerance of the strains. Our results indicate that in methylotrophic species, the ability to synthesize ectoine was most likely acquired via lateral gene transfer.
Materials and methods
Bacterial strains and culture conditions
Type cultures Methylomicrobium kenyense AMO1 (NCIMB 13566T = VKM B-2464T), Methylophaga thalassica (ATCC 33146T = NCIMB 2163T = VKM B-2057T) and Methylarcula marina h1T (=VKM B-2159) were obtained from the All-Russian Collection of Microorganisms (VKM, Russia). Two aerobic methanotrophs Methylomicrobium alcaliphilum ML1 and Methylobacter marinus 7C were isolated from an alkaline lake Mono Lake (USA) and a hypersaline lake Burlinskoe (Kulunda Steppe of Russia), respectively, and taxonomically described recently (Khmelenina et al. 2010). The methanol-utilizer Methylophaga alcalica M8 isolated from Buryatyan soda lake (Russia) shared high homology (99.9% of the 16S rRNA sequence identity and 87% DNA–DNA hybridization with the type strain of this species M. alcalica M39 (ATCC BAA-297T) (Doronina N.V., personal communication). The methanotrophic bacteria (strains ML1, 7C and AMO1) were grown under methane/air atmosphere (1:1) in a liquid nitrate mineral salt medium at 30°C as earlier described (Khmelenina et al. 1999, 2010; Sorokin et al. 2000). The methanol-utilizers M. thalassica ATCC 33146 and M. alcalica M8 were grown in nitrate mineral salt medium containing 0.5% (v/v) methanol and 10 μg l−1 vitamin B12 as described by Doronina et al. (2003a). M. marina h1T was grown in mineral salt medium supplemented with 0.5% (w/v) methylamine (Doronina et al. 2000). For growth of the alkaliphiles M. alcaliphilum ML1, M. kenyense AMO1 and M. alcalica M8, the pH of the medium was adjusted to 9.0 with NaHCO3/Na2CO3 buffer (final concentration of 0.05 M). All media were supplemented with appropriate NaCl concentrations. E. coli TOP10 (Invitrogen, USA) was grown at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml).
Analysis of intracellular ectoine content
The extraction of intracellular solutes from freeze-dried cells with methanol was done as described by Khmelenina et al. (1999). Ectoine content in the cell extracts was measured by high-performance liquid chromatography (HPLC) (Eshinimaev et al. 2007).
Identification of ectoine biosynthesis genes
Genomic DNAs from methylotrophs were prepared as earlier described (Kalyuzhnaya et al. 1999). Previously described degenerate primer sets Tra1/Tra4 and Tra3/CR were used for amplification of ectB and ectC, respectively, from chromosomal DNA of M. alcalica M8 (Reshetnikov et al. 2006). Assuming that in M. alcalica M8, the ectoine biosynthesis genes are organized in a cluster as ectABC–ask, the new degenerate primer AspV for the region 792–813 bp from the ask start codon was designed on the basis of conserved regions of the ect and ask genes from M. alcaliphilum 20Z and Vibrio cholerae. The ectA–ectB region of the gene cluster was amplified by using an inverse PCR (IPCR). Samples of M. alcalica DNA were digested by Acc65I or Mph1103I and self-ligated to generate circular DNA molecules. The latter were used in IPCRs with M8Y/M8X and M8askR/M8askF primer sets and the resulting fragments were sequenced.
Degenerate primers Tra3 and CR were used to amplify fragments containing the 3′ end of ectB and 5′ end of ectC genes from DNA samples of five other strains of methylotrophic bacteria. Based on the resultant sequences of the PCR products, the following homologous primers were designed: MTZ for M. thalassica ATCC 33146, RTect for M. alcaliphilum ML1, AMOR for M. kenyense AMO1 and 7CF/7CR for Mb. marinus 7C (Table 1). In addition, a new degenerate primer ATZ was designed based on EctA sequences of M. alcalica M8 and M. alcaliphilum 20Z.
The ATZ and MTZ primer set was used to obtain a PCR fragment containing the 3′ end of ectA and 5′ end of ectB from M. thalassica ATCC 33146. For further characterization of the ect genes, M. thalassica DNA was digested by Acc65I and the restriction fragments were ligated. Using IPCR with MTF or MTRev primers (Table 1), a fragment of 1400 bp was obtained and sequenced. For deciphering of the locus upstream ectA, DNA was digested by VspI and the restriction fragments were ligated. Circular DNA molecules were used in IPCRs with primer sets MTr1/MTr2 and the resulting fragments were sequenced. To decipher the 3′ end of ectC and 5′ end of ask, the primers MTX and AspV were used for PCR. The newly generated sequence information was used to design primers MT and MTF (Table 1) for IPCR with the self-ligated Mph1103I-fragment. Additional IPCR with MTXR/MTXF primers and self-ligated ClaI fragments was performed, thus resulting in the identification of the 3′ end of the ask gene.
Primers ATZ and AMOR were used for amplification of the ectA–ectB fragment from M. kenyense AMO1 (Fig. S1, E). An amplicon of ~1280 bp containing the 3′-region of ectA and 5′-region of ectB was sequenced. Two IPCR rounds were employed: (1) circular DNA molecules obtained after digestion of DNA with MunI, and IPCR with homologous primers AMOZ (corresponding to ectC) and AMOF (corresponding to ectB) allowed sequencing the 3′ end of ectC and its flanking sequence, (2) IPCR with homologous primers AMOA and AMOF or by using template circular DNA obtained after digestion by AclI. DNA fragments of, respectively, 1200 bp (3′-end of ectC) and 800 bp (5′-end of ectA) were sequenced.
Vectorette PCR was used for identification of nucleotide sequences of ect genes as described by Ko et al. (2003). Samples of M. marinus 7C genomic DNA were digested by EcoRI. After annealing and ligation of a double strand vectorette unit (vect 57TTAA which complemented vect 53) to DNA digested by EcoRI, a vectorette PCR amplification with primer 7CF was performed (Table 1). For the second vectorette PCR, genomic DNA was digested by Acl I. After annealing and ligation of vectorette unit (vect 57CG, which also complemented vect 53), 7CR was used in a second round of PCR amplification. The generated fragments were sequenced and assembled as described below.
An ectA–ectB fragment from M. alcaliphilum ML1 DNA was amplified using the ATZ/RTect primers set. To obtain a complete sequence of the ectA gene, IPCR was performed with circular DNA fragments generated by Mph11031 digestion and self-ligation, and the primer pair AtfX and BC (corresponding to ectB). To obtain sequence information for the 3′ end of the ectC and 5′ end of the ask genes, PCR with BC and AspV was carried out. In addition, a vectorette PCR with AtfX primer and DNA fragments generated by HindIII digestion was performed and the resulting fragments were sequenced (Fig. S1).
For identification of ect genes in M. marina h1, two PCRs with degenerate and complementary primers (h1-A/h1R, and h1V/h1F) were carried out. Two vectorette PCRs with EcoRI-digested DNA samples and the specific primers hl-B and hl-ask were performed (Table 1). The generated fragments were sequenced and assembled.
DNA manipulation and sequence analysis
Plasmid DNA isolation, restriction, digestion, DNA ligation, PCR amplification, agarose gel electrophoresis and other routine DNA manipulations were carried out according to Sambrook and Russell (2001). Restriction endonucleases were purchased from Fermentas (Lithuania). The PCR products were purified by using “Wizard SV Gel and PCR Clean-Up System” columns (Promega, USA) and cloned into the EcoRV site of pZErO2.0 (Invitrogen, USA). DNA sequencing of the cloned regions was performed by using an ABI Prism 310 DNA sequencer (Perkin Elmer, USA) and following the supplier’s instruction. DNA and protein sequences were assembled and analyzed with the Lasergene software package (DNASTAR). Searches for homologies were performed at NCBI (http://www.ncbi.nlm.nih.gov/) using the PSI-BLAST program. Protein sequences were aligned with the CLUSTAL X software program (Thompson et al. 1997). Minor corrections in alignment of sequences were also made manually. All positions containing gaps and missing data were eliminated using Complete Deletion algorithm. The resulting sequences included 628 positions per concatenated EctABC sequences.
Phylogenetic analyses were conducted in MEGA v.4.0 (Tamura et al. 2007) using the maximum-parsimony method (Close-Neighbor-Interchange algorithm). The branch lengths were calculated using the average pathway method and represented in the number of changes over the whole sequence.
Accession numbers
The nucleotide sequences of M. alcaliphilum ML1, M. kenyense AMO1T, M. marinus 7C, M. alcalica M8, M. thalassica ATCC 33146T and M. marina h1T were deposited in the GenBank under the accession numbers EU523735, DQ238213, EU523734, EU315063, EU315062 and GU249592, respectively.
Results
Salt tolerance and intracellular ectoine content in methylotrophic bacteria
All the methylotrophic bacteria used herewith were isolated from saline ecosystems. Methanol-utilizers M. alcalica M8, M. thalassica ATCC 33146 and methylamine-utilizer M. marina h1 showed optimal growth at approximately 0.5 and 1 M salt (Doronina et al. 1998, 2003a). Methanotroph M. alcaliphilum ML1 grew well in the range 0.05 and 1.7 M NaCl and optimally at 0.5 M salt. These cultures could be described as slightly halophilic bacteria in accordance with classification by Imhoff (1993). M. marinus 7C and M. kenyense AMO1 were also salt dependent, since these methanotrophs could not grow at all in mineral medium unless it contained 0.2 M NaCl (Khmelenina et al. 2010; Sorokin et al. 2000).
For some cultures, the ability to produce ectoine has been previously demonstrated (Doronina et al. 1998, 2003a). As shown, M. alcalica M8 and M. marina h1 compensated for the salinity of growth medium by accumulation of ectoine up to 235 mg per g of dried cell weight (DCW) at 9% NaCl. We also tested the ability of the other strains of methylotrophic bacteria to produce ectoine in response to increased external osmolarity. They were grown in defined mineral medium at different salinities. When cultures reached OD600 = 1, cell samples were collected and intracellular ectoine content was measured by HPLC. In all cultures tested, the intracellular pool of ectoine increased with elevated salinity. In methanotroph M. alcaliphilum ML1 and methanol-utilizer M. thalassica ATCC 33146 grown at 9% NaCl, the intracellular concentrations of ectoine reached 230 ± 20 and 200 ± 15 mg per g of DCW, respectively. M. marinus 7C and M. kenyense AMO1 grown at the upper limit of salinity (4 and 5% NaCl) accumulated ectoine to 60 and 70 mg per g of DCW. Remarkably, hydroxyectoine was not detected in all species studied.
Organization of the ectoine biosynthesis genes in methylotrophic bacteria
The 3′-end of ectB and 5′-end of ectC gene fragments were successfully amplified from DNA samples of the methylotrophic cultures. The newly obtained sequences were then used to design the homologous primers for IPCR or vectorette PCR as described in “Materials and methods”. The resultant products were sequenced and assembled. DNA fragments from M. alcaliphilum ML1 (4.3 kb), M. alcalica M8 (4.8 kb), M. thalassica ATCC 33146 (6.3 kb) and M. marina h1 (5.1 kb) contained four open reading frames (ORFs). Three of the ORFs encoded proteins with high amino acid sequence identities to the ectoine biosynthesis enzymes, DABA acetyltransferase (EctA), DABA aminotransferase (EctB) and ectoine synthase (EctC) (Fig. S1). The product of the fourth ORF located downstream of ectC showed sequence identity to an aspartokinase gene from V. cholerae (43–48% AA sequence identity), and the ORF was designated as ask. The lengths and molecular masses of these predicted polypeptides were similar among the methylotrophic strains studied. The intergenic regions between ectA stop codon and ectB start codon comprised 42, 54, 60 and 55 bp in M. alcaliphilum ML1, M. alcalica M8, M. thalassica ATCC 33146 and M. marina h1, respectively. A similar 40–131 bp gap was observed between ectB and ectC in three former species, whereas in M. marina h1, ectB, ectC and ask genes were tightly linked, with only 7 and 2 bp between the genes, respectively. The overall organization of the ect genes in M. alcaliphilum ML1, M. alcalica M8, M. thalassica ATCC 33146 and M. marina h1 implied that the four genes might form an ectABC–ask operon.
As shown, the ectR gene, encoding a transcriptional regulatory protein of the MarR family, is located upstream of the ect operon in M. alcalica M8 (Mustakhimov et al. 2009). The EctR-like gene oriented in the opposite direction was also identified upstream of the ectABC–ask cluster in M. alcaliphilum ML1, M. thalassica ATCC 33146 and M. marina h1. The EctRs showed a significant homology to the EctR1 protein from M. alcaliphilum 20Z (99, 60 and 54% AA identity for M. alcaliphilum ML1, M. thalassica ATCC 33146 and M. marina h1, respectively).
A genomic fragment (3.5 kb) of M. kenyense AMO1 contained only three ORFs corresponding to ectA, ectB and ectC. The intergenic region between ectA and ectB consisted of 47 bp and that between ectB and ectC was 170 bp. No additional ORF was found within 600 bp downstream of the ectC gene and ~300 bp upstream of ectA gene.
The sequenced 4.25-kb DNA fragment from M. marinus 7C included tightly linked ectABC genes without intergenic regions (Fig. S1). An additional ORF4 (1371 bp) was detected in the DNA locus 81 bp downstream of ectC. The product of this Orf4 (49 kDa) was similar to the transport proteins belonging to Na+/solute symporter (SSS) and PutP families (Jung 2001) of Mariprofundus ferrooxydans PV-1 ZP_01451115 (56% identity of amino acid sequences), Nitrosococcus oceani ATCC 19707 YP_343605 (46%) and Hyphomonas neptunium ATCC 15444 YP_760353 (43%). No coding sequence in the DNA locus ~300 bp upstream of ectA gene in M. kenyense AMO1 and M. marinus 7C was found.
Hence, two main types of organization of genes for ectoine biosynthesis (ectABC or ectR–ectABC–ask cluster) were found in the methylotrophs studied. Interestingly, differences in organization of the ect genes clearly correlated with salt tolerance of the methylotrophs. A three-gene cluster, ectABC, was identified in two methanotrophs, the neutrophilic M. marinus 7C and alkaliphilic M. kenyense AMO1, and both were capable of growth at salinity no more than 5% NaCl. Remarkably, methylotrophic species that were able to grow at higher salinity (up to 10% NaCl) possess a gene cluster ectR–ectABC–ask.
Phylogenetic analysis of ectoine biosynthesis genes
The amino acid sequences of EctA, EctB and EctC of the methylotrophs studied were individually compared to each other (Table S1). The highest homologies were found between Ect proteins from methanotrophs M. alcaliphilum ML1, M. kenyense AMO1 and methanol-utilizers, M. alcalica M8 and M. thalassica ATCC 33146. However, only low identities of EctABC of M. marinus 7C and M. marina h1 with those of the above-mentioned bacterial species were found, with EctA being the most divergent protein (36–71% identity, Table S1).
The NCBI database searches using the BLAST network service revealed ectABC gene clusters in complete genome sequences of a wide variety of halophilic and halotolerant bacteria. From them, 30 species of gram-negative bacteria additionally contained ask immediately downstream of ectABC, while all sequenced gram-positive halotolerant/halophilic bacteria possessed only the three-gene cluster.
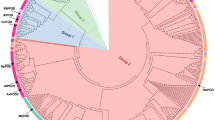
The phylogenetic analysis of concatenated polypeptides representing EctABC sequences from taxonomically diverse groups of halophilic/tolerant bacteria is presented in Fig. 1. Similar topologies were observed for the individual EctA-, EctB- and EctC-based phylogenetic trees (data not shown). The Ect proteins of M. alcaliphilum ML1, M. alcalica M8, M. thalassica ATCC 33146 and M. kenyense AMO1 branched together with the other representatives of Gammaproteobacteria. The nearest neighbors for “methylotrophic” Ect proteins were those of Marinomonas sp. MWYL1 and Teredinibacter turnerae. However, Ect proteins from the gammaproteobacterial methanotroph M. marinus 7C were most closely related to the proteins of zetaproteobacterial Marinoprofundus ferrooxidans (55.3, 80.7 and 78.5% identities of translated amino acid sequences of EctA, EctB and EctC, respectively). In contrast to M. marinus 7C, which contained the cluster ectABC, the ect genes in M. ferrooxidans most likely comprised the four-gene cluster ectABC–ask. Ect proteins from alphaproteobacterial M. marina h1 clustered together with other members of Alphaproteobacteria and shared the highest identity with the enzymes from Ruegeria sp. TM 1040 (~52%) and Roseovarius nubinhibens (~51%) (Fig. 1).
Maximum-parsimony phylogenetic tree based on deduced amino acid sequences from ectABC genes of methylotrophs (underlined) and those from other bacteria possessing ect genes. The bootstrap consensus tree is based on 1,000 replicates (Felsenstein 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The accession numbers of the respective peptides are given in brackets
Figure 2 shows the phylogenetic relationships among homologs of aspartokinase that are co-located with the ect genes on the chromosome and presumably involved in ectoine biosynthesis. Sequences of the ask genes that are part of ect operons in 30 halophilic bacteria were also included in this analysis. Aspartokinases from M. alcaliphilum 20Z, M. alcaliphilum ML1, M. alcalica M8 and M. thalassica ATCC 33146 comprise the separate branch on the phylogenetic tree, being most closely related to the protein of M. ferrooxidans (94.8, 76.7, 66.9, 57.9% AA identities, respectively).
The homologs of the novel negative transcriptional regulator EctR were identified in 20 genomes of sequenced halophilic bacterial species, including methylotroph Methylophaga thiooxydans DMS010. In these cultures, the ectR-like gene located immediately upstream of the ectoine gene cluster. Figure 3 shows the phylogenetic relationships among the newly identified EctR homologs and described regulatory proteins of the MarR family. Putative EctR proteins from M. alcaliphilum ML1, M. alcalica M8, M. thalassica ATCC 33146 and M. marina h1 showed high sequence identities (54–99%) to EctR1 from M. alcaliphilum 20Z (Mustakhimov et al. 2010). EctRs from other halophilic bacteria shared 35.5–55.1% identity with that of strain 20Z. However, all EctR-like proteins shared relatively low identity (≤20%) with other Mar-like transcriptional regulators.
Discussion
In this study, we first identified and compared genetic elements involved in ectoine biosynthesis by slightly halophilic gamma- or alphaproteobacterial methylotrophs. These methano- and methylotrophs were isolated from distantly located saline ecosystems: M. marinus 7C (hypersaline lake in Kulunda Steppe, Russia), M. kenyense AMO1 (soda lake, Kenya), M. alcaliphilum ML1 (Mono Lake, USA), M. alcalica M8 (soda lake, Buryatia), M. thalassica ATCC 33146 and M. marina h1 (Black Sea) (Khmelenina et al. 2010; Sorokin et al. 2000; Doronina et al. 1998, 2000, 2003a). The cultures differed in their osmotolerance and accumulated ectoine as a major osmoprotectant (Doronina et al. 1998, 2000, 2003a; Khmelenina et al. 2010). In these bacteria, ectoine biosynthesis involves DABA acetyltransferase, DABA aminotransferase and ectoine synthase encoded by ectABC genes. The salt-dependent methanotrophs, alkaliphilic M. kenyense AMO1 and neutrophilic M. marinus 7C possess the triad ectABC, but in the remainder methylotrophs ectoine biosynthesis genes clustered with a putative aspartokinase and negative transcriptional regulator EctR (ectR–ectABC–ask). The presence of the ask gene in ectone gene cluster correlated with the enhanced halotolerance of the methylotrophs. The aspartokinase converting aspartate into β-aspartyl phosphate represents the starting point of the pathway for biosynthesis of both aspartate family amino acids (threonine, methionine, lysine) and ectoine. The presence of the ‘osmotically controlled’ aspartokinase would allow ectoine biosynthesis to be independent from the complex regulatory mechanisms that normally control pathways for aspartate family amino acid biosynthesis and might provide an advantage at higher osmolarity.
So far, the correlation between the upper limit of salinity and the presence of the ask gene in the ect operon was observed only for methylotrophic bacteria described in this work. Database searches of genomes of gram-positive halophiles sequenced so far did not reveal that the ask gene co-located with ect operon. Besides ectoine, other organic compatible solutes belonging to the glutamate family of amino acids, mostly proline and glutamine, contributed to osmotic balance at high external salinities in gram-positive bacteria (Kuhlmann and Bremer 2002; Roberts 2005; Saum and Müller 2008; Rajan et al. 2008). For example, the chloride-dependent Halobacillus halophilus produced proline at high salinity of growth media (Saum and Müller 2008). It appears that osmoresponse of true halophiles synthesizing ectoine along with other nitrogen-containing osmoprotectants must include separate regulation of different branching pathways for amino acid biosyntheses to avoid the need for co-expression of ask and ect genes. Photobacterium profundum and Vibrio cholerae genomes contain two or three ask genes in addition to the ect-associated ask. Alternatively, in the chromosomes of three marine species, Oceanobacter sp. RED65, Sphingomonas sp. SKA58 and Lentisphaera araneosa, no ask gene outside the ect gene cluster was found. Since amplitude of the seasonal variation in salinity in the ocean is relatively small, we may speculate that constitutive expression of the ect- and the linked ask genes could provide sufficient activity of Ask enzyme for both osmoprotection and protein biosynthesis.
Hydroxyectoine is known to play an important role in adaptation to high salinity and temperature in a variety of salt-resistant species (Vargas et al. 2008). The enhanced salt tolerance of some gram-negative halophilic bacteria correlates with the occurrence of the ectD gene co-located with the ectoine gene cluster (Schwibbert et al. 2010). For example, Halomonas elongata DSM 2584 having ectABCD operon was capable of growth in the defined glucose–mineral medium at very high salinity as ectoine and hydroxyectoine counterbalance external osmotic pressure (Wohlfarth et al. 1990; Cánovas et al. 1998). Despite that M. alcaliphilum 20Z possessed an ectR–ectABC–ask–ectD gene cluster (Reshetnikov et al. 2006), transcriptomic studies demonstrated that ectD was not a part of the osmotically controlled ect operon (Mustakhimov et al. 2010). Consequently, no hydroxyectoine was detected in M. alcaliphilum 20Z cells grown at high salinity (Mustakhimov et al. 2010). These data suggested that either the ectD was ‘silent’ in the methanotroph, or the gene was expressed under stress conditions other than high salinity. Also, we failed to identify the ectD in the strain M. alcaliphilum ML1, a close relative of M. alcaliphilum 20Z. Although some methylotrophic strains might possess ectD in chromosome, nevertheless our biochemical data showed that none of them accumulated hydroxyectoine. Thus, hydroxyectoine could not provide the relatively high salt tolerance of the methylotrophs studied.
We believe that transcriptional regulatory mechanisms have an additional impact on bacterial halotolerance. The proteins belonging to a multiple antibiotic resistance regulator (MarR) family of transcriptional regulators are widely distributed across bacterial and archaeal domains. Recently, it has been shown that a MarR homolog (named EctR1) negatively controls expression of the ectoine biosysnthesis enzymes in M. alcalica M8 and M. alcaliphilum 20Z (Mustakhimov et al. 2009, 2010). Also, ectR-like sequences were identified upstream the ect genes in methanotroph M. alcaliphilum ML1 and methanol- and methylamine-utilizers M. thalassica ATCC 33146 and M. marina h1. Moreover, co-linkage of the ectR-like gene and ect operon was also demonstrated in 20 halophilic bacterial genomes from non-redundant databases (Fig. 3). Such a wide distribution and common organization of ect operon imply some functional links between these genes. Perhaps, the EctR homologs control ectoine biosynthesis at the transcriptional level in diverse heterotrophic halotolerant/halophilic bacteria. Anyway, gram-positive bacteria and true halophilic gram-negative ones (such as Halomonas elongata, Halorhodospira halophila, Chromohalobacter salexigens) do not contain an ectR-like gene upstream of the ect operon. Contrarily, all of the species presented on the phylogenetic tree for EctR are slightly or moderately halophilic. Moreover, most of them carry either ectR–ectABC–ask or ectR–ectABCD gene cluster (with the exceptions of Cellvibrio japonicus, Alcanivorax borkumensis and Roseobacter sp. MED193 not possessing either ask or ectD gene). Remarkably, in the slightly halophilic autotroph Nitrobacter sp. Nb-311A and heterotroph Bordetella bronchiseptica, the ectoine biosynthesis genes are organized in ectR–ectABCD cluster and this may additionally imply the EctR as a negative regulator of the ect genes expression.
Our phylogenetic analysis showed high sequence identities of the EctABC proteins from both methane- and methanol-utilizing bacteria belonging to the Methylomicrobium and Methylophaga genera. The proteins comprise a coherent group on the phylogenetic tree, being most closely related with EctABC of other representatives of Gammaproteobacteria. Two strains of the M. alcaliphilum species, ML1 and 20Z, shared very high similarity among Ect proteins (94.8, 98.2 and 98.5% identity for EctA, EctB and EctC, respectively). These strains were isolated from two soda lakes located in geographically remote areas (USA and Russia). Such conservation of ect genes may indicate that efficient osmoregulation is a key metabolic function in environmental niches with seasonally fluctuating salinity. Interestingly, amino acid sequences of the Ect proteins from the methanotroph M. kenyense AMO1 have higher sequence identity with those of methanol-utilizing bacteria than other methanotrophs. The sequences of EctABC from gammaproteobacterial M. marinus 7C also possessed very low homology to the ectoine biosynthesis enzymes from other methylotrophs, being closely related to EctABC from autotrophic zetaproteobacterial bacterium Mariprofundus ferrooxydans. As judged from database searches, homologs of the ectoine biosynthetic genes occurred in the autotrophic ammonia-oxidizing archaeon Nitrosopumilus maritimus. However, ectoine accumulation in this organism has not yet been examined. The EctABC proteins deduced from the genome of the Nitrosopumilus maritimus showed a considerable degree of the amino acid sequence identity (54 and 49% for EctB, 49.3 and 48.5% for EctC, 31.3 and 36.7% for EctA) to the corresponding and functionally characterized EctABC proteins from M. alcaliphilum 20Z (Reshetnikov et al. 2006) and Marinococcus halophilus (Louis and Galinski 1997).
Overall, a rather high homology of ect genes in prokaryotes of various taxonomic positions and physiological properties supported the hypothesis of evolutionary conservation of this biochemical pathway (Kuhlmann and Bremer 2002). Lateral transfers of ect genes can be considered as a possible evolutionary event that extended distribution of methylotrophic bacteria across saline ecosystems.
References
Anthony C (1982) The biochemistry of methylotrophs. Academic Press, London, p 251
Eshinimaev BT, Tsyrenzhapova IS, Khmelenina VN, Trotsenko YA (2007) Measurement of the content of the osmoprotectant ectoine in methylotrophic bacteria by means of normal-phase high performance liquid chromatography. Appl Biochem Microbiol 43:193–196 (Moscow, English translation)
Buenger J (1999) Ectoine added protection and care for the skin. Eurocosmetics 7:22–24
Cánovas D, Vargas C, Csonka LN, Ventosa A, Nieto JJ (1998) Synthesis of glycine betaine from exogenous choline in the moderately halophilic bacterium Halomonas elongata. Appl Environ Microbiol 64:4095–4097
Doronina NV, Sakharovsky VG, Drachuk SV, Trotsenko YA (1998) Organic osmoprotectors of aerobic halophilic methylobacteria. Mikrobiologiya 67:457–462 (English translation)
Doronina NV, Trotsenko YA, Tourova TP (2000) Methylarcula marina gen nov., sp. nov. and Methylarcula terricola sp. nov.: novel aerobic, moderately halophilic, facultatively methylotrophic bacteria from coastal saline environments. Int J Syst Evol Microbiol 50:1849–1859
Doronina NV, TsD Darmaeva, Trotsenko YA (2003a) Methylophaga alcalica sp. nov., a novel alkaliphilic and moderately halophilic, obligately methylotrophic bacterium from the East Mongolian saline soda lake. Int J Syst Evol Microbiol 53:223–229
Doronina NV, TsD Darmaeva, Trotsenko YA (2003b) Methylophaga natronica sp. nov., a new alkaliphilic and moderately halophilic, restricted-facultative methylotrophic bacterium from soda lake of the Southern Transbaikal region. Syst Appl Microbiol 26:382–389
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Galinski EA (1995) Osmoadaptation in bacteria. Adv Microbial Physiol 37:273–328
Galinski EA, Pfeiffer HP, Trüper HG (1985) 1,4,5,6,-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid, a novel cyclic acid from halophilic phototrophic bacteria of genus Ectothiorhodospira. Eur J Biochem 149:135–139
Göller K, Galinski EA (1999) Protection of the model enzyme (lactate dehydrogenase) against heat, urea and freeze–thaw treatment by compatible solute addition. J Mol Biol B Enzym 7:37–45
Graf R, Anzali S, Buenger J, Pfluecker F, Driller H (2008) The multifunctional role of ectoine as a natural cell protectant. Clinics Dermatol 26:326–333
Grant WD (2004) Life at low water activity. Phil Trans R Soc Lond B 359:1249–1267
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
Imhoff JF (1993) Osmotic adaptation in halophilic and halotolerant microorganisms. In: Vreeland RH, Hochstein LJ (eds) The biology of halophilic bacteria. The CRC Press, Boca Raton, pp 211–253
Jung H (2001) Towards the molecular mechanism of Na/solute symport in prokaryotes. Biochim Biophys Acta 1505:131–143
Kalyuzhnaya MG, Khmelenina VN, Kotelnikova S, Holmquist L, Pedersen K, Trotsenko YA (1999) Methylomonas scandinavica sp. nov., a new methanotrophic psychrotrophic bacterium isolated from deep igneous rock ground water of Sweden. Syst Appl Microbiol 22:565–572
Kalyuzhnaya MG, Khmelenina VN, Eshinimaev BT, Sorokin D, Fuse H, Lidstrom M, Trotsenko Y (2008) Reclassification of halo(alkali)philic and halo(alkali)tolerant methanotrophs provisionally assigned to the genera Methylomicrobium and Methylobacter and emended description of the genus Methylomicrobium. Int J Syst Evol Microbiol 58:591–596
Kempf B, Bremer E (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170:319–330
Khmelenina VN, Kalyuzhnaya MG, Sakharovsky VG, Suzina NE, Trotsenko YA, Gottschalk G (1999) Osmoadaptation in halophilic and alkaliphilic methanotrophs. Arch Microbiol 172:321–329
Khmelenina VN, Shchukin VN, Reshetnikov AS, Mustakhimov II, Suzina NE, Eshinimaev BT, Trotsenko YA (2010) Structural and functional features of methanotrophs from hypersaline and alkaline lakes. Mikrobiologiya 79:472–482 (English translation)
Knap S, Ladenstain R, Galinski EA (1999) Thermal stabilization of bovine ribonuclease A by naturally occurring osmolyte beta-hydroxyectoine and betaine. Extremophiles 3:191–198
Ko W-Y, Ryan MD, Akashi H (2003) Molecular phylogeny of the Drosophila melanogaster species subgroup. J Mol Evol 57:562–573
Kuhlmann AU, Bremer E (2002) Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl Environ Microbiol 68:772–783
Lidstrom ME (2006) Aerobic methylotrophic prokaryotes. In: Balows A et al (eds) The Prokaryotes, 3rd edn. Springer Verlag, New York, pp 618–634
Lippert K, Galinski EA (1992) Enzyme stabilization by ectoine-type compatible solutes: protection against heating, freezing, and drying. Appl Microbial Biotechnol 37:61–65
Lo C–C, Bonner AC, Xie G, D’Souza M, Jensen AR (2009) Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways. Microbiol Molec Biol Rev 73:594–651
Louis P, Galinski EA (1997) Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143:1141–1149
Mustakhimov II, Reshetnikov AS, Khmelenina VN, YuA Trotsenko (2009) EctR—a novel transcriptional regulator of ectoine biosynthesis genes in the haloalcaliphilic methylotrophic bacterium Methylophaga alcalica. Doklady Biochem Biophys 429:305–308 (Moscow, English translation)
Mustakhimov II, Reshetnikov AS, Glukhov AS, Khmelenina VN, Kalyuzhnaya MG, Trotsenko YA (2010) Identification and characterization of EctR1, a new transcriptional regulator of the ectoine biosynthesis genes in the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. J Bacteriol 192:410–417
Ono H, Sawada K, Khunajakr N, Toa T, Yamamoto M, Hiramoto M, Shinmyo A, Takano M, Murooka Y (1999) Characterization of biosynthetic enzymes for ectoine as a compatible solute in a moderately halophilic eubacterium, Halomonas elongata. J Bacteriol 181:91–99
Oren A (2002) Halophilic microorganisms and their environments. Kluwer Scientific Publishers, Dordrecht
Oren A (2008) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4:2
Peters R, Galinski EA, Truper HG (1990) The biosynthesis of ectoine. FEMS Microbiol Lett 71:157–162
Rajan AL, Joseph TC, Thampuran N, James R, Ashok Kumar K, Viswanathan C, Bansal KC (2008) Cloning and heterologous expression of ectoine biosynthesis genes from Bacillus halodurans in Escherichia coli. Biotechnol Lett 30:1403–1407
Reshetnikov AS, Mustakhimov II, Khmelenina VN, Trotsenko YA (2005) Cloning, purification, and characterization of diaminobutyrate acetyltransferase from the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Biochemistry 70:878–883 (Moscow, English translation)
Reshetnikov AS, Khmelenina VN, Trotsenko YA (2006) Characterization of the ectoine biosynthesis genes in obligate haloalkalotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Arch Microbiol 184:286–296
Roberts MF (2005) Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst 1:5
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NewYork
Saum SH, Müller V (2008) Regulation of osmoadaptation in the moderate halophile Halobacillus halophilus: chloride, glutamate and switching osmolyte strategies. Saline Syst 4:4
Schwibbert K, Marin-Sanguino A, Bagyan I, Heidrich G, Lentzen G, Seitz H, Rampp M, Schuster SC, Klenk HP, Pfeiffer F, Oesterhelt D, Kunte HJ (2010) A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T. Environ Microbiol. doi:10.1111/j.1462-2920.2010.02336.x
Severin J, Wohlfarth A, Galinski EA (1992) The predominant role of recently discovered tetrahydropyrimidines for the osmoadaptation of halophilic eubacteria. J Gen Microbiol 138:1629–1638
Sorokin DY, Jones BE, Kuenen JG (2000) A novel obligately methylotrophic, methane-oxidizing Methylomicrobium species from a highly alkaline environment. Extremophiles 4:145–155
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmaugin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trotsenko YA, Khmelenina VN (2002) Biology and osmoadaptation of haloalkaliphilic methanotrophs. Mikrobiologiya 71:123–132 (English translation)
Vargas C, Argandona M, Reina-Bueno M, Rodriguez-Moya J, Fernandez-Aunion C, Nieto JJ (2008) Unravelling the adaptation responses to osmotic and temperature stress in Chromohalobacter salexigens, a bacterium with broad salinity tolerance. Saline Syst 4:14. doi:10.1186/1746-1448-4-14
Ventosa A, Nieto JJ, Oren A (1998) Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544
Wohlfarth A, Severin J, Galinski EA (1990) The spectrum of compatible solutes in heterotrophic halophilic eubacteria of the family Halomonadaceae. J Gen Microbiol 136:705–712
Acknowledgments
This work was supported by Grants CRDF Rub1-2946-PU-09, RFBR 11-04-00801a, Russian State Contract 16.740.11.0615 and NSF Grant MCB-0604269. The authors thank Nicole Smalley for helpful comments on the earlier draft.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. da Costa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
792_2011_396_MOESM1_ESM.doc
Fig. S1. Sequencing strategies used for identification of ectoine biosynthetic genes in Methylomicrobium alcaliphilum ML1 (C), Methylophphaga alcalica M8 (A), Methylophaga thalassica ATCC 33146 (B), Methylobacter marinus 7C (D), Methylomicrobium kenyense AMO1 (E) and Methylarcula marina h1 (F). The nucleotide sequences of oligonucleotide primers are given in Table 1. (DOC 94 kb)
Rights and permissions
About this article
Cite this article
Reshetnikov, A.S., Khmelenina, V.N., Mustakhimov, I.I. et al. Diversity and phylogeny of the ectoine biosynthesis genes in aerobic, moderately halophilic methylotrophic bacteria. Extremophiles 15, 653 (2011). https://doi.org/10.1007/s00792-011-0396-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00792-011-0396-x