Abstract
Phenotypic and genotypic analysis was carried out on four iron- and sulfur-oxidizing acidophilic bacteria (the “NO-37 group”) isolated from different parts of the world. 16S rRNA phylogeny showed that they are highly related to each other, but are less related to the type strain of Acidithiobacillus ferrooxidans. The NO-37 group isolates are obligate chemolithoautotrophs, facultative anaerobes, diazotrophic, and psychrotolerant. They are less tolerant of extremely low pH, and in contrast to At. ferrooxidans T, all of the NO-37 group isolates are motile. The GC contents of genomic DNA of the NO-37 group isolates were around 56 mol% and the DNA–DNA hybridization value between genomic DNA of isolate NO-37 and At. ferrooxidans T was 37%. It also appears that the bacteria of the NO-37 group have a different biochemical mechanism for oxidizing ferrous iron than At. ferrooxidans T; the gene coding for the archetypal rusticyanin (RusA) was not detected in any of the NO-37 group isolates, rather a gene coding for a homologous protein (RusB) was amplified from three of the four novel isolates. Isolates of the NO-37 group clearly belong to a species that is different to those already recognized in the genus Acidithiobacillus, for which the name Acidithiobacillus ferrivorans is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Acidithiobacillus currently comprises three recognized bacterial species, At. thiooxidans, At. caldus, and At. ferrooxidans, and one (At. albertensis) whose status is uncertain (Kelly and Wood 2000). They are extremely acidophilic Gammaproteobacteria, and have in common the ability to grow autotrophically using elemental and various reduced forms of sulfur as sole energy source. At. ferrooxidans is unique among acidithiobacilli in also being able to use ferrous iron as an electron donor, though this particular trait is not that uncommon among acidophilic prokaryotes as a whole (Johnson and Hallberg 2008). At. ferrooxidans is, by far, the most widely studied of all acidophilic bacteria as a consequence of its perceived importance in biotechnology (mostly bioprocessing of metal sulfide ores and concentrates; Rawlings and Johnson 2007) and environmental pollution (the generation of acid mine drainage; Nordstrom 2000). At. ferrooxidans is a strict chemoautotroph and has been shown to use hydrogen (Drobner et al. 1990) and formic acid as electron donors in addition to iron and sulfur (Pronk et al. 1991). It is also a facultative anaerobe, being able to grow through ferric iron respiration in anoxic media (Pronk et al. 1992).
It has been known for sometime that iron- and sulfur-oxidizing acidophiles identified as “At. ferrooxidans” include isolates that appear to be distinct species. Pioneering work by Arthur Harrison, in which DNA–DNA hybridization and chromosomal DNA base contents were determined, identified six sub-groups of “At. ferrooxidans,” though two of these had only single strain representatives (Harrison 1982). Later work based on 16S rRNA gene sequence analysis supported the view that different strains of “At. ferrooxidans” exhibit sufficient phylogenetic heterogeneity to warrant reclassification as novel species (e.g., Karavaiko et al. 2003). More recently, Ni et al. (2008) found a relationship between some phenotypic characteristics (tolerance to extremely low pH, specific rates of iron and sulfur oxidation and tolerance to some transition metals) and different phylotypes of “At. ferrooxidans.”
Johnson et al. (2001) isolated several strains of acidophilic iron- and sulfur-oxidizing Gram-negative bacteria from a long-abandoned copper mine in Norway. Sequence analysis of their 16S rRNA genes showed that, while they were closely related to each other, they had only ~98% gene sequence similarity to the type strain of At. ferrooxidans, and so were described as “Acidithiobacillus spp.” rather than strains of At. ferrooxidans. Subsequent sequencing of the 16S rRNA genes of some other putative “At. ferrooxidans” strains held within the Acidophile Culture Collection maintained at Bangor University revealed that several of these were closely related to the Norwegian isolates rather than to At. ferrooxidans T. In addition, the partial 16S rRNA gene sequence of a cold-tolerant “At. ferrooxidans” isolated from Norilsk, Siberia (Kupka et al. 2007), again indicated close affiliation of this strain with the Norwegian isolates.
Here, we delineate a representative strain of the Norwegian isolates and some closely related acidophiles isolated from different geographical locations as strains of a novel species, Acidithiobacillus ferrivorans. Although they have many physiological traits in common, At. ferrivorans and At. ferrooxidans display differences in terms of their pH and temperature characteristics, suggesting that the two bacteria would tend to dominate different environmental niches. In addition, preliminary evidence suggests a possible different biochemical mechanism for ferrous iron oxidation in these two acidophiles.
Materials and methods
Isolation and cultivation of bacteria
Four strains of iron- and sulfur-oxidizing acidophilic bacteria (referred to subsequently as the “NO-37 group”) were selected for study: (i) NO-37 isolated from copper mine spoil drainage in Norway (Johnson et al. 2001), (ii) CF27 isolated from mine water draining an abandoned cobalt/copper mine in Cobalt, ID, USA, a strain which was noted for its propensity to aggregate pyrite particles when grown in liquid media (Blake and Johnson 2000); (iii) Peru6 obtained from mine water draining an abandoned mine in the Andes (ca. 3000 m elevation) Peru (D. B. Johnson, unpublished); and (iv) OP14, isolated in the 1980s from the abandoned Cae Coch pyrite mine, North Wales (Ghauri et al. 2007). Each strain was isolated and purified by repeated streaking on iron/tetrathionate overlay solid medium (Johnson 1995), and individual colonies transferred into liquid medium (see below). The type strain of At. ferrooxidans (NCIMB 11820, which is equivalent to ATCC 23270) was also grown alongside the new isolates for comparative purposes.
Bacteria were routinely grown in heterotrophic basal salts (“HBS”) supplemented with a trace elements mix (Wakeman et al. 2008) with ferrous iron (at 10–25 mM) as electron donor. For improved growth on tetrathionate and thiosulfate as electron donor, a variant of the basal salts mixture was used (“UBS”; Hallberg and Lindström 1994). Isolates were maintained by storage of pyrite supplemented cultures at 4°C, and these were subcultured periodically (approximately every 6 months). Cultures were also concentrated 100-fold by centrifugation, amended with dimethylsulfoxide to 7% (v/v) and frozen at −80°C for long-term storage.
Cell morphology
Bacteria were viewed routinely by phase contrast microscopy (Leitz Labolux) at ×400 magnification. To determine cell size, cultures were grown in liquid medium with either 25 mM ferrous iron or elemental sulfur as electron donor, fixed with glutaraldehyde, critical point-dried and gold-coated ahead of viewing with a Hitachi S-520 scanning electron microscope. The lengths and widths of ten cells were measured and mean dimensions (and the standard deviations) were recorded.
Determination of effects of temperature and pH on growth
To determine the optimum temperature and pH for growth, isolate NO-37 (selected as the type strain of the new species) was grown in a 2-l bioreactor (Electrolab Ltd., UK) with a working volume of 1 l of a liquid medium containing 25 mM ferrous sulfate in HBS. Cultures were aerated (1 l/min) and stirred at 150 rpm. The bioreactor temperature was set at varying temperatures (20–35°C) at a constant pH of 2.2, and growth was determined by measuring the ferric iron produced as a yellow-colored chloride complex (Schnaitman et al. 1969). When the pH was varied (pH 2.1–3.5), the temperature was held at 25°C and growth was measured as ferrous iron oxidized by titration of residual ferrous iron with 1 mM potassium permanganate, due to the poor solubility of ferric iron at pH > 2.5. Semi-logarithmic plots of ferric iron produced or ferrous iron oxidized against time was used to identify exponential growth phases, and from them specific growth rates were calculated.
To determine the lower pH limit for growth, At. ferrooxidans T and the NO-37 group isolates were grown in 20 mM ferrous sulfate medium at pH 2.0, 1.9, 1.8, 1.7, and 1.5. The complete media were prepared, pH adjusted (with sulfuric acid), and then filter-sterilized (though 0.2 μm pore size cellulose nitrate membranes) into sterile conical flasks. Cultures were inoculated with small volumes (0.5%, v/v) in order not to change the pH values of the media, and flasks were incubated, shaken at 30°C, for up to 10 days.
The lower limit of temperature for growth was determined in shake flask cultures containing liquid medium containing 25 mM ferrous iron (pH 2.2) that were inoculated with isolate NO-37 or At. ferrooxidans T. These were incubated, shaken at 10°C, and growth was monitored by measuring changes in ferric iron concentration. All four of the NO-37 groups were also grown in shake flasks at 4°C (the lowest temperature tested).
Specific rates of ferrous iron oxidation
Isolates NO-37, CF27, and Peru6, and At. ferrooxidans T and strain IESL32 (an isolate from the Escondida copper mine, Chile, that shares >99% 16S rRNA gene sequence identity with At. ferrooxidans T, Pedro Galleguillos, unpublished) were grown in shake flasks containing 25 mM ferrous sulfate, adjusted to an initial pH of 1.9. When cultures had oxidized all of the available iron, cells were harvested by centrifugation, washed, and resuspended in acidified basal salts. The protein contents of the cell suspensions were determined using the Bradford assay (Bradford 1976), and aliquots dispensed into shake flasks containing 20 ml of 1 mM ferrous sulfate/basal salts (pH 2.0) medium (in triplicate), and iron oxidation monitored by measuring concentrations of ferrous iron (using the ferrozine assay; Lovley and Phillips 1987) over a 2-h time period.
Growth on reduced sulfur and other electron donors
Potential electron donors, other than ferrous iron, tested for being able to support the growth of the novel isolates included pyrite and elemental sulfur (both at 1%, w/v), tetrathionate (2 mM), and thiosulfate (5 mM) in liquid media (the latter two using UBS) adjusted initially to pH 2.0 (pH 4.0 in the case of acid-labile thiosulfate). Growth on pyrite was determined by measuring the increase in cell numbers microscopically, whereas growth on reduced sulfur was measured as an increase in culture turbidity. The ability of the new isolates to use sulfide was assessed by inoculating gradient tubes of semi-solid HBS, pH 2.5, using a modified technique of that described previously by Drobner et al. (1992), with agarose as gelling agent in place of agar.
To screen for aerobic growth on hydrogen, colonies of the novel isolates and At. ferrooxidans T grown on ferrous iron/tetrathionate overlay plates were streak-inoculated onto a variety ferrous iron overlaid solid media (Johnson and Hallberg 2007). Plates were incubated (at 30°C) in 3.7 l sealed jars in which a hydrogen- and carbon dioxide-enriched atmosphere was created using gas generating kits (Oxoid, UK). Since no catalyst was used, the atmosphere in the sealed jars also contained oxygen. Control plates were incubated in jars in which atmospheric CO2 concentrations were enriched (using acidified sodium bicarbonate). Colonies were compared after 2 weeks incubation.
Assimilation of carbon
No organic carbon was provided in all of the experimental work described above, indicating that the NO-37 group isolates were autotrophic, as are all other members of the genus Acidithiobacillus. Portions (618 or 505 bp) of the genes coding for the large subunit of forms I and II RuBisCO (cbbL and cbbM, respectively) were amplified by PCR as described previously (Johnson et al. 2009) and sequenced (GenBank accession numbers FJ467341 and FJ467342).
To test whether the novel isolates were able to grow as mixotrophs, using an inorganic (ferrous iron) electron donor and organic carbon, isolates were grown in 5 mM ferrous sulfate medium, pH 2.2, supplemented with glucose, glycerol, glutamic acid, citric acid (at 2.5 mM), or yeast extract (at 0.01%, w/v). Cultures were incubated for 10 days (at 30°C) and cell numbers recorded using a Helber counting chamber marked with Thoma ruling (Hawksley, UK) relative to organic carbon-free control cultures.
Potential for diazotrophy
To assess whether isolate NO-37 could fix nitrogen (N2), a washed cell suspension (grown on ferrous iron) was inoculated into HBS containing 20 mM ferrous iron, and into N-free medium, where the (NH4)2SO4 in the HBS was replaced with an equimolar amount of K2SO4. These cultures were incubated at 30°C for 2 weeks in a sealed chamber that contained a flask of 50% (v/v) H2SO4 to absorb any trace amounts of ammonia in the atmosphere. At. ferrooxidans T was also inoculated into the same media as a positive control, and At. caldus T was used as a negative control (with tetrathionate as electron donor in place of ferrous iron). Growth was scored as an increase in ferric iron concentration (or decrease in pH for At. caldus cultures) and also by microscopic observation of cultures. Genetic evidence for the capacity of NO-37 to fix nitrogen was obtained by amplifying a portion (408 bp) of the nifH gene using PCR (Ueda et al. 2005), and confirmation by sequencing (GenBank accession number FJ467343).
Ferric iron respiration
Isolates NO-37, CF27, and Peru6 and At. ferrooxidans T were grown aerobically on elemental sulfur medium (initial pH 3.0) and active cultures inoculated into de-oxygenated sterile liquid medium containing 1% (w/v) elemental sulfur and 20 mM ferric iron (pH ~ 2.2) in sterile 100 ml serum bottles (in triplicate). These were sealed and incubated at 30°C for up to 13 days. Changes in ferrous iron concentrations were measured using the ferrozine assay.
Tolerance of selected transition metals and anions
The NO-37 group isolates and At. ferrooxidans T were grown at 30°C for 2 weeks in ferrous iron liquid medium containing varying concentrations of added transition metals (added as sulfate salts from stock solutions at pH 2.0) or anions (listed in Table 2). Growth of the bacteria was assessed by measuring increases in ferric iron concentrations. To confirm that any inhibitory effects were due to toxicity of the transition metals rather than elevated osmotic potentials, cultures were also grown in the presence of equimolar concentrations of MgSO4.
Chemotaxonomy
Isolate NO-37 was grown in 16 l of 2 mM tetrathionate/10 mM Fe2+ to obtain sufficient biomass to facilitate DNA–DNA hybridization analysis with the type strain of At. ferrooxidans, which had previously been grown using the same medium. Portions of each were washed twice with 10 mM sodium citrate (pH 4.0) before a single wash in TE buffer (10 mM Tris, 1 mM EDTA at pH 8). DNA was extracted by incubating the cell suspensions (in TE) with proteinase K and SDS at 55°C for 3 h, followed by two extractions with phenol:chloroform:isoamyl alcohol (25:24:1) and three extractions with chloroform:isoamyl alcohol (24:1). The DNA was further purified by cesium chloride gradient centrifugation. The G + C content of purified chromosomal DNA was determined by melting point analysis as described previously (Okibe et al. 2003). The remaining biomass (of both strains) was preserved in 50% (v/v) isopropanol and was sent to the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany) for DNA–DNA hybridization analysis.
Analysis of the respiratory quinones of isolate NO-37 grown with ferrous iron was carried out as previously described (Johnson et al. 2009).
Phylogeny
Sequences were analyzed using BLAST at the NCBI database (http://ncbi.nlm.nih.gov/BLAST) and added to a database of over 200,000 prokaryotic 16S rRNA gene sequences by using the aligning tool of the ARB software package (Ludwig et al. 2004). The rRNA alignments were corrected manually and alignment uncertainties were omitted in the phylogenetic analysis, resulting in a final alignment of 1,400 nucleotides. Phylogenetic trees were generated using DNA parsimony, neighbor-joining, and maximum likelihood analyses with a subset of 200 nearly full-length sequences (>1400 bp). Filters, which excluded highly variable positions, were used. In all cases, general tree topology and clusters were stable, and reliability of the tree topologies was confirmed by bootstrap analysis using 1,000 replicate alignments. A consensus tree was generated.
PCR amplification of rusticyanin coding genes
Primers were designed to amplify portions of the genes coding for the two isoforms of rusticyanin (RusA and RusB) found previously in strains considered to be At. ferrooxidans (Sasaki et al. 2003). The primers rusAF (5′-TTGGTGCGGCTGCGGCTCTG-3′) and rusAR (5′-GGTCACGTCCACCGTTGCC-3′) were synthesized to target the rusA gene, and rusB1F (5′-TCGCCGCAGGACTTTCCACC-3′) and rusB1R (5′-TCCGAACCCCCCACTCTTGG-3′) the rusB gene, to yield expected PCR products of 284 and 422 bp, respectively. PCR was carried out in 20 μl reactions (GoTaq, Promega, Southampton, UK) with MgCl2 added to 1.5 mM for 35 cycles, after denaturation of template DNA for 5 min at 95°C, of 30 s at 95°C, 30 s at 65°C, and 30 s at 72°C. Final extension was carried out for 5 min at 72°C before the temperature was reduced to 4°C. DNA extracted from the NO-37 group isolates was used as template, along with DNA extracted from At. ferrooxidans T and strains ATCC 19859 and DSM 9465. The identity of the PCR product from isolate NO-37 was confirmed by sequence analysis (GenBank accession number FJ467344).
Results
Isolation and morphology of novel strains of Acidithiobacillus
All isolates formed relatively large colonies (>1 mm) on iron/tetrathionate overlay plates. Colonies were typically initially ferric iron-stained, but matured with protracted incubation into opaque/translucent colonies with cream/yellow centers (considered to be due to deposition of sulfur). All four isolates were rod-shaped bacteria that were highly motile (especially in the early stages of growth). The mean size of NO-37 cells grown with iron as electron donor was 2.4 × 0.5 (±0.3 × 0.03) μm, much larger than the 1.0 × 0.5 μm reported for cells of At. ferrooxidans T. Cells of NO-37 were shorter, 1.6 × 0.5 (±0.1 × 0.04) μm, when grown with sulfur as electron donor.
16S rRNA gene sequence analysis
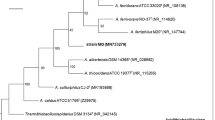
Phylogenetic analysis based on 16S rRNA gene sequences of several iron-oxidizing isolates from acid mine drainage in Norway (including strain NO-37) clearly identified two distinct groupings for the iron-oxidizing acidithiobacilli (Johnson et al. 2001), though a limited number of gene sequences were used in that study. Inclusion of the other iron-oxidizing isolates described here (the NO-37 group isolates), along with many sequences for more recent isolates or acidophiles detected as cloned genes from various acidic sites, providing much stronger support for the placement of iron-oxidizing bacteria of the genus Acidithiobacillus into more than one distinct phylogenetic group (Fig. 1). Group I circumscribes those bacteria related to At. ferrooxidans T, whereas Group II includes the NO-37 group isolates.
A consensus phylogenetic tree derived from 16S rRNA gene sequence data showing the two groups of iron-oxidizing acidithiobacilli (NO-37 group isolates in bold), and their relationship to the other species of the genus Acidithiobacillus, as well as to related clones detected in acidic environments. Bootstrap values of each major division are given at the respective nodes, and the scale bar represents 1% sequence divergence. Accession numbers for the sequences used to make this tree are given in parentheses, and the tree was rooted with the sequence from Thermithiobacillus tepidarius T (AJ459801)
Chemotaxonomy
The G + C content for NO-37 was determined as 55.5 ± 0.5 mol% (n = 3), whereas that of At. ferrooxidans T was found to be 59.2 mol% (n = 1), which is close to the 58.7 mol% calculated from genome sequence data of this acidophile. The G + C contents of Peru6, OP14, and CF27 were 55.9 ± 0.3, 55.4 ± 0.3, and 55.2 ± 0.4 mol%, respectively. The DNA–DNA hybridization value between the chromosomal DNAs of isolate NO-37 and At. ferrooxidans T was 37%. Like At. ferrooxidans T, the ubiquinone detected in isolate NO-37 was Q8.
Temperature and pH ranges for growth of NO-37 group isolates
From the phylogenetic and genotypic data, it appeared that the NO-37 group isolates belong to a different and novel species of the genus Acidithiobacillus, for which we propose isolate NO-37 as the type strain. Strain NO-37 exhibited a broad temperature range at which optimal growth occurred, between 25 and 32°C (Fig. 2). The highest temperature tested at which growth of NO-37 occurred was 37°C. The optimum pH for growth of NO-37 was found to be 2.5 (Fig. 2). At higher pH values, growth was moderately slower, whereas at lower culture medium pH the growth rate rapidly decreased. The culture doubling time of isolate NO-37 at its optimum pH (2.5) and temperature (30°C) was 3.2 h, which is significantly faster than the maximum growth rates of ca. 0.1 h−1 (equivalent to culture doubling times of about 6 h) that have been reported for At. ferrooxidans T (e.g., Tuovinen and Kelly 1974). However, while the growth rate of NO-37 under optimum growth conditions was seemingly more rapid than that of At. ferrooxidans T, the specific rates of ferrous iron oxidation by the NO-37 group isolates were significantly smaller (at pH 2.0) than those of both At. ferrooxidans T and the closely related isolate IESL32 (Table 1).
Growth rates of isolate NO-37 slowed rapidly as pH decreased below 2.5, suggesting that the NO-37 group might be less tolerant of extremely low pH than At. ferrooxidans. This was confirmed in shake flask cultures where At. ferrooxidans T was able to oxidize all of the available ferrous iron in cultures adjusted to initial pH values of 1.5–2.0, whereas the four NO-37 group isolates oxidized iron only in cultures set at pH 1.9 and 2.0.
Since phylogenetic analysis revealed that the NO-37 group clade included an At. ferrooxidans-like isolate (SS3) described as psychrotolerant (Kupka et al. 2007), the lower temperature limits of growth for the NO-37 group was determined. At 10°C, isolate NO-37 was able to oxidize ferrous iron in liquid medium while At. ferrooxidans T did not. When all four novel isolates were grown in shake flasks at 4°C, ferric iron concentrations increased exponentially and culture doubling times for all four bacteria were found to be about 100 h. These experiments confirmed that, like isolate SS3, the four NO-37 group isolates are psychrotolerant, in contrast to At. ferrooxidans T.
Utilization of other sources of electron donors
In addition to growth with ferrous iron, all of the isolates were able to grow by oxidation of tetrathionate and thiosulfate, though growth was initially preceded by an extended lag phase (on the order of weeks). Upon subculture into fresh medium with either electron donor, growth proceeded with a much reduced (or no) lag phase. When isolate NO-37 was then inoculated into ferrous iron medium, growth and oxidation of iron occurred immediately; however, this culture again exhibited a long lag before growth with tetrathionate. All four of the NO-37 group isolates were also able to grow by oxidation of elemental sulfur and sulfide with little lag. These four isolates were also able to grow by the oxidative dissolution of pyrite.
Growth of At. ferrooxidans T on hydrogen was obtained on solid media, as evidenced by far more luxurious growth of colonies grown in a hydrogen-containing atmosphere. Evidence for oxidation of hydrogen by the NO-37 group isolates was far more variable, with colonies of isolates Peru6 and CF27 also being larger when grown in the presence of hydrogen, whereas those of isolates NO-37 and OP14 were of very similar size to those on control plates.
Ferric iron respiration
Like At. ferrooxidans T, all four of the NO-37 group isolates could grow in anoxic media through ferric iron respiration, using elemental sulfur as electron donor. Figure 3 shows data for iron reduction by isolates NO-37, Peru6, and CF27 in comparison with At. ferrooxidans T. Iron reduction by the latter was more rapid (possibly due to more conducive culture pH) than by any isolates of the NO-37 group, which displayed variable lag times before iron reduction occurred.
Reduction of ferric iron by three strains of the NO-37 group compared with At. ferrooxidans T. Anaerobic cultures were set up (in triplicate) in which elemental sulfur was provided as electron donor. Data show mean values and standard deviations. Filled square NO-37; filled triangle Peru6; filled inverted triangle CF27; filled circle At. ferrooxidans T; circle non-inoculated control
Carbon metabolism of the NO-37 group
All known acidithiobacilli fix CO2 using the CO2-fixing enzyme RuBisCO. Portions of the gene coding for both forms I (cbbL) and II (cbbM) of the CO2-fixing enzyme RuBisCO were amplified by PCR using DNA from isolate NO-37 as template. The cbbL gene of isolate NO-37 was 91.6% identical to the cbbL-2 gene of At. ferrooxidans T (translated gene products shared 97% amino acid similarity) and only 76.5% identical to cbbL-1 (94% amino acid similarity) from At. ferrooxidans T. It is presently unclear if isolate NO-37 harbors more than one cbbL gene as does At. ferrooxidans T. The cbbM gene fragment from isolate NO-37 was 91.1% identical (98% similarity of amino acid sequence) to that of At. ferrooxidans T. Both cbbL and cbbM genes were also amplified from the other three isolates of the NO-37 group.
Supplementing culture media with organic carbon compounds did not inhibit ferrous iron oxidation or enhance cell yields of any isolate of the NO-37 group, implying that they are obligately chemo-autotrophic bacteria.
Nitrogen fixation by NO-37 group isolates
Both isolate NO-37 and At. ferrooxidans T grew by ferrous iron oxidation in both ammonium-amended and nitrogen-free liquid medium, whereas At. caldus T grew and oxidized sulfur only when ammonium was provided, which is consistent with an earlier report by Norris et al. (1995). The ability of isolate NO-37 to fix nitrogen was corroborated by PCR amplification of a portion of the nifH gene (coding for the Fe protein of the nitrogenase enzyme), which was 85.0% identical to nifH from At. ferrooxidans T. A PCR product of the same size as that from isolate NO-37 was also obtained from isolates CF27, Peru6, and OP14 (data not shown).
Tolerance to transition metals and anions
In general, the NO-37 group isolates were more sensitive to the transition metals and anions tested than the type strain of At. ferrooxidans (Table 2). At. ferrooxidans T grew well in medium containing up to 400 mM Fe2+ while the NO-37 group grew in medium containing 200 mM Fe2+ but not 400 mM. In contrast, Fe3+ completely inhibited growth of NO-37 at 100 mM, whereas At. ferrooxidans T grew in medium containing 200 mM but not 400 mM Fe3+. Both the NO-37 group isolates and At. ferrooxidans T were more tolerant of Zn than Cu (Table 2), and none of the former were able to grow in medium containing 50 mM Cu (the lowest concentration tested). All of the bacteria were able to grow in medium containing up to 400 mM MgSO4, confirming that the metals themselves were toxic rather than inhibition being due to elevated solute potentials. The NO-37 group isolates were more sensitive to chloride than At. ferrooxidans T, and all of these iron-oxidizing acidophiles were highly sensitive to molybdate (Table 2).
Analysis of rusticyanin gene variations in iron-oxidizing acidithiobacilli
Previously, it has been shown that different strains of iron- and sulfur-oxidizing acidophiles described as At. ferrooxidans harbor either one or two variants of a gene encoding for rusticyanin (Sasaki et al. 2003). The type strain and some others harbor the archetypal gene called rusA, whereas other strains considered at the time to be At. ferrooxidans were shown by Southern hybridization to harbor both the rusA gene and also a second gene called rusB. These two genes share 81.1% identity, and the gene products share 75.9% identity at the amino acid level. PCR was used to determine if the new isolates harbored both the rusA and rusB genes. As expected, only the rusA gene was amplified from the three At. ferrooxidans strains (Fig. 4). Unexpectedly, however, only the rusB gene could be amplified from three of the NO-37 group isolates (Fig. 4), and no PCR product was obtained from isolate CF27 even when lower annealing temperatures were tested.
Discussion
The first iron-oxidizing acidophile to be isolated and characterized was given the name Thiobacillus ferrooxidans (Temple and Colmer 1951). In the following years, other isolates that appeared to lack the ability to oxidize thiosulfate or elemental sulfur were named as Ferrobacillus ferrooxidans (Leathen et al. 1956) and Ferrobacillus sulfooxidans (Kinsel 1960), though Kelly and Tuovinen (1972) reclassified these two species as T. ferrooxidans following the demonstration of growth of both on sulfur and thiosulfate. Interestingly, the bacterium that was eventually designated as the type strain of this species was not the original isolate, but rather the isolate of Leathen et al. (“F. ferrooxidans”). The need to revisit the classification of bacteria labeled as At. ferrooxidans has been apparent for some time. We have elected to classify one particular phylogenetic sub-group of Acidithiobacillus that includes closely related isolates from different continents (North and South America, and two areas within Europe) as a novel species. In addition to the indication by 16S rRNA gene sequence analysis that these isolates were distinct from At. ferrooxidans T, other supporting data for the delineation of these as separate species came from differences (ca. 4%) in G + C content of their chromosomal DNA and, crucially, DNA–DNA hybridization (37%) which was far below the minimum figure of 70% that serves as a boundary to differentiate species. To reflect the propensity of the isolates of this new species to oxidize iron, we propose to name this new species At. ferrivorans (“iron eating”).
Comparison of At. ferrivorans and At. ferrooxidans showed that the two acidophiles have several major phenotypic characteristics in common (Table 3). These include being obligately chemoautotrophic acidophiles that can grow using ferrous iron, elemental sulfur, thiosulfate, or tetrathionate as sole energy sources and that fix both carbon dioxide (through the Calvin–Benson–Bassham cycle, implied for At. ferrivorans by the presence of cbbL and cbbM genes), and nitrogen gas. Both iron-oxidizers can also grow anaerobically by coupling the oxidation of sulfur to the reduction of ferric iron. Differences between the two species were more apparent with their responses to temperature and pH (Table 3). One of the most significant of these is that, whereas At. ferrooxidans T is mesophilic, At. ferrivorans is a psychrotolerant mesophile that can grow at <5°C, though both bacteria have similar temperature optima for growth. Also, At. ferrivorans is less tolerant of extremely low pH than At. ferrooxidans, though its pH optimum (2.5) is similar to that reported for At. ferrooxidans (Kelly and Wood 2005). While the specific rates of ferrous iron oxidation determined in this study were significantly smaller for strains of At. ferrivorans than for At. ferrooxidans T, this was probably greatly influenced by the fact that these were determined at pH 2.0, which is close to the pH minimum (1.9) for At. ferrivorans. When grown under optimum growth conditions, the culture doubling time (~3.2 h) determined for At. ferrivorans strain NO-37 was significantly less than that (~6 h) for At. ferrooxidans T when using ferrous iron as electron donor. These differences imply that At. ferrivorans would tend to dominate in iron-rich environments (e.g., acid mine drainage streams) that are cool and have a pH value of >2.3, while At. ferrooxidans would have competitive advantage in warmer and more acidic sites.
Another differentiating characteristic between the two iron-oxidizing acidithiobacilli appears to be their motility. Although being described initially as a motile rod (Leathen et al. 1956), we have never observed motility in the cultures of At. ferrooxidans T that have been confirmed as being axenic, regardless of growth substrate (ferrous iron, sulfur, or pyrite), whereas the four strains of At. ferrivorans described are all highly motile rods. Interestingly, examination of the genome of At. ferrooxidans T has failed to reveal the presence of genes coding for flagella (Valdés et al. 2008). The apparent discrepancy for motility in At. ferrooxidans T could be explained by the initial descriptions being made of enrichment cultures that contained heterotrophic acidophiles (and also possibly At. ferrivorans) in addition to At. ferrooxidans. This hypothesis is supported by the fact that the type strain culture that was deposited at the National Collection of Industrial and Marine Bacteria, UK (and from which the American Type Culture Collection, ATCC 23270, was derived) was shown by Johnson and Kelso (1983) to be contaminated with Acidiphilium-like heterotrophic acidophiles, most species of which are motile (Wichlacz et al. 1986).
While limited information is currently available on the biochemistry of iron oxidation by At. ferrivorans, it does appear that this may be different to that of At. ferrooxidans T, where the periplasmic copper-containing protein rusticyanin is considered to play an important role in electron transfer (Holmes and Bonnefoy 2007). None of the At. ferrivorans strains examined contained the gene coding for rusticyanin A (the sole form present in At. ferrooxidans T), though three out of the four strains examined contained a gene coding for the isozyme rusticyanin B. Attempts to amplify rusA and rusB genes in one strain of At. ferrivorans (CF27) failed, confirming an earlier report that this isolate does not synthesize rusticyanin when grow on ferrous iron (Blake and Johnson 2000), a feature that has previously been considered to be diagnostic for At. ferrooxidans. The biochemical pathway of ferrous iron oxidation must therefore be different in At. ferrivorans CF27 to that previously described for iron-oxidizing acidithiobacilli, and whether this is also the case for other strains of At. ferrivorans (i.e., there is no direct role for rusticyanin B in iron oxidation) is an intriguing question that will be addressed in future work.
Emended description of the genus Acidithiobacillus (Kelly and Wood 2000)
The description of the genus Acidithiobacillus (Kelly and Wood 2000) is emended to reflect the novel phenotypic traits described here and previously for At. ferrooxidans (Pronk et al. 1992; Mackintosh 1978). Some species are facultative anaerobes, capable of respiration with ferric iron as terminal electron acceptor. Some species are diazotrophic.
Description of At. ferrivorans, sp. nov.
Acidithiobacillus ferrivorans (fer.ri.vo’rans. L. n. ferrum, iron; L. part. adj. vorans, devouring; N.L. part. adj. ferrivorans, iron-devouring).
Cells are motile, Gram-negative, straight rods (1.6–2.4 μm long and 0.5 μm wide), and do not form endospores. They form small, “fried-egg”-like colonies (round orange-centered colonies with off-white margins) on acidic iron/tetrathionate overlay medium, which become opaque/translucent with cream/yellow centers with protracted incubation. Obligate autotrophs capable of growth with ferrous iron, S°, sulfide, tetrathionate, thiosulfate, and sulfide minerals as electron donors. Facultative anaerobes are capable of anaerobic growth with ferric iron as electron acceptor and elemental sulfur as electron donor. Diazotrophic. Psychrotolerant; optimum temperature for growth is 27–32°C, and growth occurs from 4°C (lowest temperature tested) to 37°C. Acidophilic; the optimum pH for growth is 2.5 and the pH range for growth is from 1.9 to 3.4 (highest pH tested). The G + C content of genomic DNA is 55.5 ± 0.5 mol%.
The type strain, NO-37T (=JCM 15606T = DSM 22755T), was isolated from copper mine spoil drainage in Norway. Other isolates of Acidithiobacillus ferrivorans were obtained from acid mine drainage sites worldwide that were typically characterized by low temperatures and pH of between 2 and 3.
References
Blake R, Johnson DB (2000) Phylogenetic and biochemical diversity among acidophilic bacteria that respire on iron. In: Lovley DR (ed) Environmental microbe-metal interactions. American Society of Microbiology Press, Washington, DC, pp 53–78
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Drobner E, Huber H, Stetter KO (1990) Thiobacillus ferrooxidans, a facultative hydrogen oxidizer. Appl Environ Microbiol 56:2922–2923
Drobner E, Huber H, Rachel R, Stetter KO (1992) Thiobacillus plumbophilus sp. nov., a novel galena and hydrogen oxidizer. Arch Microbiol 157:213–217
Ghauri MA, Okibe N, Johnson DB (2007) Attachment of acidophilic bacteria to solid surfaces: the significance of species and strain variations. Hydrometallurgy 85:72–80
Hallberg KB, Lindström EB (1994) Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140:3451–3456
Hallberg KB, Thomson HEC, Boeselt I, Johnson DB (2001) Aerobic and anaerobic sulfur metabolism by acidophilic bacteria. In: Ciminelli VST, Garcia O Jr (eds) Biohydrometallurgy: fundamentals, technology and sustainable development. Elsevier, Amsterdam, pp 423–431
Harrison AP Jr (1982) Genomic and physiological diversity amongst strains of Thiobacillus ferrooxidans, and genomic comparison with Thiobacillus thiooxidans. Arch Microbiol 131:68–76
Holmes DS, Bonnefoy V (2007) Genetic and bioinformatic insights into iron and sulfur oxidation mechanisms of bioleaching organisms. In: Rawlings DE, Johnson DB (eds) Biomining. Springer-Verlag, Berlin, pp 281–307
Johnson DB (1995) Selective solid media for isolating and enumerating acidophilic bacteria. J Microbiol Methods 23:205–218
Johnson DB, Hallberg KB (2007) Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms. In: Rawlings DE, Johnson DB (eds) Biomining. Springer-Verlag, Berlin, pp 237–261
Johnson DB, Hallberg KB (2008) Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv Microb Physiol 54:202–256
Johnson DB, Kelso WI (1983) Detection of heterotrophic contaminants in cultures of Thiobacillus ferrooxidans and their elimination by subculturing in media containing copper sulphate. J Gen Microbiol 123:2969–2972
Johnson DB, Rolfe S, Hallberg KB, Iversen E (2001) Isolation and phylogenetic characterisation of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine. Environ Microbiol 3:630–637
Johnson DB, Bacelar-Nicolau P, Okibe N, Thomas A, Hallberg KB (2009) Characteristics of Ferrimicrobium acidiphilum gen. nov., sp. nov., and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic iron-oxidizing, extremely acidophilic actinobacteria. Int J Syst Evol Microbiol 59:1082–1089
Karavaiko GI, Tourova TP, Kondrat’eva TF, Lysenko AM, Kolganova TV, Ageeva SN, Muntyan LN, Pivovarova TA (2003) Phylogenetic heterogeneity of the species Acidithiobacillus ferrooxidans. Int J Syst Evol Microbiol 53:113–119
Kelly DP, Tuovinen OH (1972) Recommendation that the names Ferrobacillus ferrooxidans Leathen and Braley and Ferrobacillus sulfooxidans Kinsel be recognised as synonyms of Thiobacillus ferrooxidans Temple and Colmer. Int J Syst Bacteriol 22:170–172
Kelly DP, Wood AP (2000) Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int J Syst Evol Microbiol 50:511–516
Kelly DP, Wood AP (2005) Genus Acidithiobacillus (Kelly and Wood 2000). In: Brenner DJ, Krieg NR, Staley JT (eds) Bergey’s manual of systematic bacteriology, second edition, volume two the Proteobacteria. Part B. The Gammaproteobacteria. Bergey’s Manual Trust, Michigan, pp 60–62
Kinsel N (1960) New sulfur oxidizing iron bacterium: Ferrobacillus sulfooxidans. J Bacteriol 80:628–632
Kupka D, Rzhepishevska OI, Dopson M, Lindström EB, Karnachuk OV, Tuovinen OH (2007) Bacterial oxidation of ferrous iron at low temperatures. Biotechnol Bioeng 97:1470–1478
Leathen WW, Kinsel NA, Braley SA (1956) Ferrobacillus ferrooxidans: a chemosynthetic autotrophic bacterium. J Bacteriol 72:700–704
Lovley DR, Phillips EJP (1987) Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl Environ Microbiol 53:1536–1540
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Mackintosh MEJ (1978) Nitrogen fixation by Thiobacillus ferrooxidans. J Gen Microbiol 105:215–218
Ni YQ, He KY, Bao JT, Wan DS, Li HY (2008) Genomic and phenotypic heterogeneity of Acidithiobacillus spp. strains isolated from diverse habitats in China. FEMS Microbiol Ecol 64:248–259
Nordstrom DK (2000) Advances in the hydrogeochemistry and microbiology of acid mine waters. Int Geol Rev 42:499–515
Norris PR, Murrell JC, Hinson D (1995) The potential for diazotrophy in iron- and sulfur-oxidizing acidophilic bacteria. Arch Microbiol 16:294–300
Okibe N, Gericke M, Hallberg KB, Johnson DB (2003) Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred tank bioleaching operation. Appl Environ Microbiol 69:1936–1943
Pronk JT, Meijer WM, Hazeu W, van Dijken JP, Bos P, Kuenen JG (1991) Growth of Thiobacillus ferrooxidans on formic acid. Appl Environ Microbiol 57:2057–2062
Pronk JT, de Bruyn JC, Bos P, Kuenen JG (1992) Anaerobic growth of Thiobacillus ferrooxidans. Appl Environ Microbiol 58:2227–2230
Rawlings DE, Johnson DB (eds) (2007) Biomining. Springer-Verlag, Berlin
Sasaki K, Ida C, Ando A, Matsumoto N, Saiki H, Ohmura N (2003) Respiratory isozyme, two types of rusticyanin of Acidithiobacillus ferrooxidans. Biosci Biotechnol Biochem 67:1039–1047
Schnaitman CA, Korczynski MS, Lundgren DG (1969) Kinetic studies of iron oxidation by whole cells of Ferrobacillus ferrooxidans. J Bacteriol 99:552–557
Temple KL, Colmer AR (1951) The autotrophic oxidation of iron by a new bacterium: Thiobacillus ferrooxidans. J Bacteriol 62:605–611
Tuovinen OH, Kelly DP (1974) Studies on the growth of Thiobacillus ferrooxidans. V. Factors affecting growth in liquid culture and the development of colonies on solid media containing inorganic sulfur compounds. Arch Microbiol 98:351–364
Ueda T, Suga Y, Yahiro N, Matsuguchi T (2005) Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol 177:1414–1417
Valdés J, Pedroso I, Quatrini R, Holmes DS (2008) Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94:180–184
Wakeman K, Auvinen H, Johnson DB (2008) Microbiological and geochemical dynamics in simulated heap leaching of a polymetallic sulfide ore. Biotechnol Bioeng 101:739–750
Wichlacz PL, Unz RF, Langworthy TA (1986) Acidiphilium angustum sp. nov., Acidiphilium facilis sp. nov., and Acidiphilium rubrum sp. nov.: acidophilic heterotrophic bacteria isolated from acid coal mine drainage. Int J Syst Bacteriol 36:197–201
Acknowledgments
This study was carried out in the frame of BioMinE Project, supported by the European Commission under the Sixth Framework Programme for Research and Development (Contract NMP1-CT-500329-1). We wish to thank our various partners on the project for their contributions to the work reported in this paper. The authors would also like to thank Professor Jean Euzéby for his expert advice on bacterial nomenclature, Dr. Eleanor Jameson for conducting experiments on anaerobic growth of At. ferrivorans, and Mr. Pedro Galleguillos and Dr. Tadayoshi Kanao for measurement of specific iron oxidation rates. D.B.J. is grateful to the Royal Society (UK) for the award of an Industrial Fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Matsunaga.
Rights and permissions
About this article
Cite this article
Hallberg, K.B., González-Toril, E. & Johnson, D.B. Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 14, 9–19 (2010). https://doi.org/10.1007/s00792-009-0282-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-009-0282-y