Abstract
Lake Tebenquiche is one of the largest saline water bodies in the Salar de Atacama at 2,500 m above sea level in northeastern Chile. Bacteria inhabiting there have to deal with extreme changes in salinity, temperature and UV dose (i.e., high environmental dissimilarity in the physical landscape). We analyzed the bacterioplankton structure of this lake by 16S rRNA gene analyses along a spatio–temporal survey. The bacterial assemblage within the lake was quite heterogeneous both in space and time. Salinity changed both in space and time ranging between 1 and 30% (w/v), and total abundances of planktonic prokaryotes in the different sampling points within the lake ranged between two and nine times 106 cells mL−1. Community composition changed accordingly to the particular salinity of each point as depicted by genetic fingerprinting analyses (denaturing gradient gel electrophoresis), showing a high level of variation in species composition from place to place (beta-diversity). Three selected sites were analyzed in more detail by clone libraries. We observed a predominance of Bacteroidetes (about one third of the clones) and Gammaproteobacteria (another third) with respect to all the other bacterial groups. The diversity of Bacteroidetes sequences was large and showed a remarkable degree of novelty. Bacteroidetes formed at least four clusters with no cultured relatives in databases and rather distantly related to any known 16S rRNA sequence. Within this phylum, a rich and diverse presence of Salinibacter relatives was found in the saltiest part of the lake. Lake Tebenquiche included several novel microorganisms of environmental importance and appeared as a large unexplored reservoir of unknown bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diversity of microbial communities is believed to be very large and poorly characterized (see overview in Pedrós-Alió 2006). Yet, its knowledge is of interest for several reasons, both practical and theoretical. Bacterial diversity is a reservoir of potentially interesting genes for biotechnology and medicine, and the large seed-bank of bacterial taxa hidden in natural communities should be of interest to better delineate both the taxonomy and the evolutionary relationships among microorganisms (Baldauf 2003; Pedrós-Alió 2007). In this respect, the ocean and freshwater lakes have received a lot of attention (Glöckner et al. 1999; Giovannoni and Rappé 2000). However, bacterial diversity in saline lakes have been studied sparsely (Bowman et al. 2000; Humayoun et al. 2003; Jiang et al. 2006; Wu et al. 2006), despite the fact that they are numerous and widespread (Williams 1996). Microorganisms from such environments have potentially interesting enzymes (Oren 2002), and some authors have claimed that the physiology and ecology of microorganisms in hypersaline environments may be relevant for a better understanding of both the early stages of life on Earth (Kunte et al. 2002) and potential life in Mars evaporitic environments (Mancinelli et al. 2004).
Lake Tebenquiche is one of the largest hyperhaline high-altitude water bodies in the Salar de Atacama (Chile). A preliminary limnological characterization of this lake was carried out by Zúñiga et al. (Zúñiga et al. 1991). Next, a large collection of bacteria and archaea were isolated in pure culture. Results have been reported for moderately halophilic Gram-negative rods (Prado et al. 1991) and Gram-positive cocci (Valderrama et al. 1991), heterotrophic halophilic microorganisms (Prado et al. 1993) and extreme halophilic Archaea (Lizama et al. 2001, 2002). Most of the bacteria isolated belonged to the Gammaproteobacteria, especially members of the genera Vibrio, Halomonas (including Deleya and Volcaniella), Acinetobacter, Alteromonas, Psychrobacter and Marinococcus. The only other groups that were recovered with some frequency were the High and Low GC Gram positives. No Bacteroidetes were recovered. The real extent of bacterial diversity within the system remains still unexplored, because it is well known that isolation in pure culture selects some of the microorganisms present in the sample and that those able to grow in culture are in many occasions, not the most abundant ones in nature (Staley and Konopka 1985; Amann et al. 1995; Pedrós-Alió 2006).
Here, we present a detailed study of Lake Tebenquiche covering spatial heterogeneity and changes in time of the bacterioplankton composition by genetic fingerprinting on the environmental 16S rRNA gene pool. We also constructed clone libraries from selected sampling sites to obtain a more precise description of the bacterial diversity. In a previous paper (Demergasso et al. 2004), we carried out a general fingerprinting survey of the bacterial and archaeal diversity in other undersampled and remote athalassohaline environments from the Atacama Desert. Thus, it was of additional interest to compare the sequences retrieved by molecular methods and check whether any of the isolates could be found among them. Our goal was to use Lake Tebenquiche as a model to determine the degree of spatial heterogeneity in this kind of shallow lakes and to explore whether bacterial taxa specific to systems with intermediate salinities existed.
Materials and methods
Description of Lake Tebenquiche
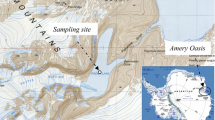
The Salar de Atacama is a huge system (about 2,900 km2) with several different water bodies in its interior. Lake Tebenquiche is one of the largest and it is located in the northern part of the Salar (Fig. 1). A summary of the geographical coordinates and other parameters of the locations sampled can be found in Table 1 and in Demergasso et al. (2004). Information on the geochemistry of this system can be found in Risacher et al. (1999) and in Zúñiga et al. (1991).
The hydrochemistry of salt lakes and marshes within the Salar de Atacama basin shows significant differences (Carmona et al. 2000). Water inputs have a wide range of compositions and flows, draining very different geological formations surrounding the Salar. In addition, the evaporation along the flow path and within the Salar itself, contributes to the heterogeneity. A spatial distribution of pore brines within the Salar nucleus in two zones has been proposed (Risacher and Alonso 1996): calcium-rich brines (of Na–Ca–(Mg)–Cl type) are present in its south-western part, while sulfate-rich brines (of Na–(Mg)–SO4–Cl type) are found towards the eastern part. This distribution agrees with the 87Sr/86Sr data, suggesting different water sources feeding each area (Carmona et al. 2000).
Sampling and measurements
Lake Tebenquiche was visited in August 1999 (winter) and March 2000 (summer). In October 2000 (spring), an intensive sampling expedition was carried out, and six different points in the lake were sampled to investigate the spatial heterogeneity of the microbial assemblage (Fig. 1). For comparison, additional samples were taken at two nearby dolines (small sinkholes) and at Burro Muerto, a shallow lake south of Lake Tebenquiche but within the Salar de Atacama. The environments sampled showed a variety of salinities and other physicochemical conditions (Table 1). An Orion model 290 pH meter was used to measure temperature and pH. Salinity was measured using an Orion model 115 conductivity meter.
Water samples were transferred to plastic bottles and kept in an icebox with ice until further processing. Samples for chlorophyll analysis were filtered through 25-mm-diameter Whatman GF/F glass fiber filters. The filters were placed in aluminum foil and kept frozen. Chlorophyll a concentration was determined by fluorescence of acetone extracts (Yentsch and Menzel 1963) with a Turner Designs Fluorometer.
Total bacterial number was determined by flow cytometry in 1.8 ml samples fixed with 200 μl of paraformaldehyde:glutaraldehyde (1 and 0.05% final concentration, respectively) in criovials. Vials were frozen until processing in the laboratory. The protocol followed was that of Gasol and del Giorgio (2000) and Gasol et al. (2004). Briefly, 100-μl aliquots were stained with Syto13 (Molecular Probes, Eugene, OR, USA), a suspension of fluorescently labeled beads was added at a known concentration and the samples were counted in a FACScalibur flow cytometer (Becton and Dickinson, San Jose, CA, USA). In some cases, bacteria were counted by epifluorescence microscopy using the DNA-specific dye 4',6-diamidino-2-phenylindole (DAPI) with a Leica DMLS epifluorescence microscope.
Nucleic acid analyses: DGGE, clone libraries and 16S rRNA sequences analyses
Between 20 and 650 ml of water was filtered through 0.2-μm polycarbonate membranes (Nuclepore Millipore, Bedford, MA, USA) and stored at −70°C. Filters were incubated with lysozyme, proteinase K and sodium dodecyl sulfate (SDS) in lysis buffer as described previously (Schauer et al. 2000). DNA was extracted with phenol–chloroform–isoamyl alcohol (25:24:1, vol/vol/vol) and precipitated with ethanol. The extracted genomic DNA was used as target in the PCR to amplify 16S rRNA genes. Bacterial fragments suitable for subsequent denaturing gradient gel electrophoresis (DGGE) analysis were amplified with the primer combinations 358fGC-907r as described previously (Dumestre et al. 2002). A 6% polyacrylamide gel was obtained with a gradient of DNA-denaturant agent 40–80% (100% denaturant agent is defined as 7 M urea and 40% deionized formamide). Around 800 ng of PCR product was loaded for each sample and the gels were run at 100 V, 60°C for 16 h in a CBS DGGE-2000 system (CBS Scientific Company, Del Mar, CA, USA). The gels were stained with the nucleic acid dye SybrGold for 45 min, and visualized with UV in a Fluor-S MultiImager (Bio-Rad, Hercules, CA, USA) with the Multi-Analyst software (Bio-Rad, Hercules, CA, USA). High-resolution images (1,312 × 1,034 pixels, 12-bits dynamic range) were saved as computer files. Then the picture was analyzed using the gel plotting macro tool of the NIH-Image software package version 1.62 (National Institute of Health, USA). After background subtracting, the intensity of each band was measured integrating the area under the peak and was expressed as percent of the total intensity in the lane. The error measured among replicates was less than 4%. Bands were excised from the gels, reamplified and purified for sequencing as reported (Casamayor et al. 2001). Bands that provided sequence between 450 and 540 bp length were submitted to GenBank with accession numbers AJ487523 to AJ487534 and AJ568004 to AJ568014.
Cloning and RFLP analysis were performed as previously described (Ferrera et al. 2004). 16S rRNA genes were amplified by PCR with the universal primers 27f and 1492r. PCR amplifications were digested with the restriction enzyme HaeIII (Invitrogen Corporation, Madison, WI, USA), and the RFLP patterns of the clones were compared. Chimeric sequences were identified by using the CHECK_CHIMERA (Maidak et al. 2000) and by visual inspection of the BLAST search outputs.
Sequences were sent to BLAST search (www.ncbi.nlm.nih.gov) to determine the closest relative in the database. A similarity matrix was built with the ARB software package (Technical University of Munich, Munich, Germany; www.arb-home.de). Partial sequences were inserted into the optimized and validated tree available in ARB (derived from complete sequence data), by using the maximum-parsimony criterion and a special ARB parsimony tool that did not affect the initial tree topology. The respective ARB tools were used to perform maximum parsimony (MP), neighbor-joining (NJ) and maximum likelihood (ML) analyses for full sequences. The calculation methods were combined with different filters, and the resulting phylogenetic trees were compared manually to obtain a final consensus tree. Results from the three types of analyses were essentially identical and only the maximum parsimony trees are shown. The sequence data have been submitted to the EMBL database under accession numbers AY862726 to AY862797.
Results
Table 1 provides the geographical location, physicochemical and biological parameters for all the samples used in the present study. The different sampling sites in Lake Tebenquiche have also been indicated in Fig. 1. Despite the different seasons at which samples were collected, the temperature range was moderate, between 14°C in winter and 27.6°C in one spring sample. On the other hand, salinity of the samples ranged between 1 and 30% (w/v) even within the same sampling date. Likewise, salinity also changed considerably between sampling dates at the same sampling site (see different visits to sites 12 and 18 for example). Chlorophyll a was generally low and ranged by one order of magnitude between 0.03 and 0.6 μg l−1. In contrast, chlorophyll a was much higher in the very shallow Lake Burro Muerto, where resuspension from the sediments is an important factor. Bacterial numbers changed by a factor of two (3–6 × 106 cells ml−1) with only two exceptions. There was no correlation between temperature, salinity, chlorophyll a and bacterial numbers.
Heterogeneity in space and time
Six different sampling sites on the lake were chosen as representative of the different water-inundated areas, and depth of water was less than 50 cm at all sampling sites. We analyze the possible heterogeneity within the system in time and space by DGGE (Fig. 2), and identification of the excised and sequenced DGGE bands is shown in Table 2. We observed major differences in the composition of the bacterial assemblage among the different sampling locations within Lake Tebenquiche (see lanes 3–8, all taken on the same date, i.e., 13 October 2000). The general grouping of the DGGE lanes was in agreement with the local salinity at each place (see Table 1). There were at least three types of assemblages. Lanes 3 and 4 were identical. This made sense, since the samples had been collected very closely in space and their salinity was the same. Lane 5 was from a close sampling location, but the salinity was three times lower, and finally lanes 6–8 formed a third cluster. Therefore, we observed change in the bacterial assemblages among places because of the environmental variability and topographic complexity, within the lake.
Negative image of a denaturing gradient gel electrophoresis (DGGE). Bands that were cut off from the gel are labeled with the same number as in Table 2, and in Figs. 4 and 5. When bands across several lanes could be identified as being the same, they all have the same number. Sample from the freshwater Lake Miscanti is added for comparison (see also Demergasso et al. 2004)
We also analyzed some samples from other water bodies in the Salar de Atacama to see whether the same assemblage would be found all over the complete system. Sample in lane 14 corresponds to Lake Cejas, north of Lake Tebenquiche; samples in lanes 12 and 13 correspond to two dolines (West and East dolines located in the sampling point 13 of our general survey in this area), a few hundred meters east from Lake Tebenquiche (Fig. 1); and samples in lanes 9 and 10 correspond to Burro Muerto (sampling points 14 and 15), south of Tebenquiche. As could be expected, the band patterns from these water bodies were completely different from those in Lake Tebenquiche.
Finally, we examined samples taken on the same site (Lake Tebenquiche) but at different seasons to evaluate changes with time. Samples in lanes 2 and 6 were taken at site 12 in fall and spring, respectively. Most bands were different on the two sampling dates. Samples in lanes 1 and 3, in turn, were taken in winter and spring at site 18, east of the previous one. In this case, although a few bands did not appear in both sampling dates, the patterns were relatively similar and the most intense bands were the same (bands 17, 19 and 20). This is coherent with the salinities of the samples. The two samples from site 12 differed in salinity by almost six times, while the two samples from site 18 only differed by a factor of two. Comparing lanes corresponding to the dolines (12 and 13) and lanes corresponding to Burro Muerto (9–11) taken at different times of the year, it appears that the assemblages in these two areas were less variable with time. Thus, changes with time were quite important in Lake Tebenquiche, but the differences were associated to the changes in salinity caused by the variable hydrographic regime and did not conform to a seasonal succession.
Diversity in Lake Tebenquiche
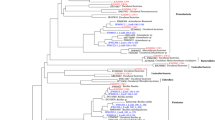
Three samples were chosen from the previous ones for a more in depth analysis of the community by clone libraries. On the one hand, those for At12Oct and At18Oct were representative of two of the DGGE patterns found in Lake Tebenquiche (Fig. 2). On the other hand, At18Aug was intended to provide a comparison with At18Oct at a different time of the year. Based on results from both partial and complete sequences, a similarity level of ≥97% was chosen to define operational taxonomic units (OTUs). The resultant OTUs are shown in Table 3 with the number of clones belonging to each OTU found in each library, the clones sequenced completely and their closest relative as identified with BLAST. The assemblage was clearly dominated by Bacteroidetes and Gammaproteobacteria, whereas the remaining phylogenetic groups were represented only by a few clones. A general tree constructed with full sequences is shown in Figs. 3 and 4, and more details are provided in Table 3.
Bacteroidetes grouped consistently into four clusters (Fig. 4). The best-represented cluster was Atacama-I, and it was very distantly related to the genus Psychroflexus (at the 87% level). Atacama-I also included three DGGE bands from Lake Tebenquiche and six more bands from other aquatic saline systems in Northern Chile (Fig. 5), showing that this cluster could be abundant and widely distributed in the area. Two clones from a survey of a hypersaline endoevaporitic microbial mat in Eilat (Israel) (Sorensen et al. 2005) showed similarities between 95 and 97% to sequences in cluster Atacama-I as well. Even though we sampled environments with salinities ranging between freshwater and 36.4%, sequences from this cluster were only retrieved from samples within the range of 3–15%, suggesting that it may contain bacteria adapted to intermediate salinities. This cluster had sequences from both libraries from site 18 and none from site 12. Since the salinity of the latter at the time of sampling was 29.6% (Table 1), this again suggests that members of the Atacama-I cluster could not survive at high salinities. We split this cluster into three OTUs at a similarity ≥97% (Fig. 5). OTU-1, only found in Northern Chile up to the present, was the most numerous in the sites of intermediate salinity and also included sequences from Ascotán (Nch31 and 34-AS6), a different salt flat system located at 4,000 m a.s.l. in Northern Chile (Demergasso et al. 2004). The Bacteroidetes cluster Atacama-I, therefore, showed a rich diversity at different levels of similarity and appeared to be a novel and phylogenetically complex group of bacteria inhabiting intermediate salinity environments. Cluster Atacama-II and Cluster Atacama-III again were not related to any cultivated bacterium and distantly related to any sequence in the databases (90% similarity) with the only closer clone relatives (99–95% similarity) obtained from the mat in Eilat. Finally, the last Bacteroidetes cluster (Atacama-IV) was retrieved from the At12Oct library exclusively and from the most intense DGGE band at sampling sites 12, 21 and 22 (Fig. 2, Table 2), and comprised some sequences related to Salinibacter spp. (95–96% similarity) (Fig. 4), and other group of sequences distantly related to Salinibacter (85% similarity) or to clones from the microbial mat in Eilat (91–95% similarity). These sequences were only found in the highest salinity sample, indicating a rich and diverse presence of Salinibacter relatives in Lake Tebenquiche. Altogether, the diversity of Bacteroidetes sequences in Lake Tebenquiche was large and showed a remarkable degree of novelty.
Phylogenetic tree including partial sequences from DGGE bands and clones for the cluster Atacama-I. Scale bar 0.10 mutations per nucleotide position. The code includes the band number in Fig. 2 and Table 2, and the code for the natural environment from which the 16S rRNA gene sequence was originally obtained. Bands with site codes -AS and -LL were obtained from previous work (Demergasso et al. 2004)
Proteobacteria were the second most abundant group of sequences. The beta subdivision was represented by only one DGGE sequence (Table 2), but no clones were retrieved. Conversely, the gamma subdivision was the best-represented group, followed by the alpha subdivision. The latter were all within the Roseobacter group, but with low similarity to all cultivated strains reported so far. Finally, the epsilon subdivision was represented by only two clones. A tree based on full 16S rRNA gene sequences is shown in Fig. 3, and Fig. 6 includes all the partial sequences obtained from DGGE. Most sequences were included within three clusters in the gamma-Proteobacteria, distantly related (92–93%) to Nitrococcus mobilis, Halorhodospira halophila and Thiomicrospira sp. (Fig. 6, Table 3). Cluster Atacama-V contained sequences from Lake Tebenquiche from sampling site 18, with salinity ranging between 8 and 15%, from Lake Cejas (another shallow lake on the Salar de Atacama, relatively close to Lake Tebenquiche, Table 1), and from Mono Lake, another highly saline lake in California, USA (Figs. 3, 6), but the clones were not closely related to any other bacterium or environmental sequence. Judging from the number of clones and the intensity of the DGGE bands, this cluster appeared to be important in the lake. The Alphaproteobacteria sequences found were related with environmental clones retrieved from relatively high salinity environments (Fig. 3) and, again, had no close relatives isolated in pure cultures.
Phylogenetic tree including partial sequences from DGGE bands and clones for the Gammaproteobacterial clusters. Scale bar 0.10 mutations per nucleotide position. The code includes the band number in Fig. 2 and Table 2, and the code for the natural environment from which the 16S rRNA gene sequence was originally obtained. See also previous work (Demergasso et al. 2004)
Finally, the remaining divisions of bacteria were represented by very few sequences. One clone and two bands belonged to the High G+C Gram-positive bacteria, one clone (related to some Mono Lake clones) was affiliated to the Low G+C Gram-positive bacteria and a series of clones was associated with yet poorly defined lineages: two clones could be affiliated with the candidate division OD1; three more clones associated with uncultivated clones CS_B020 and BD1-5 from marine sediments; and one clone was related to sequences of the KB1 group, from sediments in hypersaline brines (Fig. 3).
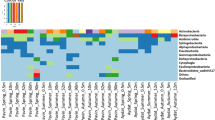
The percentage contribution to the total assemblage of the different groups is shown in Fig. 7, together with the percentage contribution to total band intensity in DGGE gels for the same samples. The recovered groups were the same in both the techniques: Psychroflexus-like (Atacama-I), Salinibacter-like (Atacama-IV), and DG890 cluster (Atacama-II) among the Bacteroidetes, and Mono Lake cluster (Atacama-V) and Thiomicrospira-like (Atacama-VII) cluster for the Gammaproteobacteria (Fig. 7). Since primers used for cloning and DGGE are very different, the good coincidence between techniques gives support to the idea that the targeted bacterial assemblage was considerably well sampled. The least abundant groups were usually absent from the DGGE results (Fig. 7) due to the fact that faint bands in DGGE gels are very difficult to sequence (Sánchez et al 2007).
Discussion
At a global level, the biodiversity of the Earth’s aquatic systems can be approached by sampling different ecosystems, each with a different diversity. In this respect, both low and high salinity systems have received considerable attention. At the seawater end, many studies have been performed in the last 15 years (Giovannoni et al. 1990; Giovannoni and Rappé 2000; Pommier et al. 2007). At the other end, crystallizer ponds from solar salterns have also been studied extensively (Benlloch et al. 1996, 2001, 2002; Rodríguez-Valera et al. 1999; Casamayor et al. 2000, 2002; Oren 2002; Estrada et al. 2004; Pedrós-Alió 2005; Maturrano et al. 2006). Aquatic systems with intermediate salinities, however, have not received much attention. In the case of solar salterns, ponds with intermediate salinities show relatively high levels of heterotrophic activities (Gasol et al. 2004). The case of Lake Tebenquiche shows that such systems hold the potential to reveal a considerable degree of phylogenetic novelty at different levels. Thus, we found sequences that are candidates for new branches in the bacterial phylogenetic tree, novel clusters within the well-characterized bacterial groups and a large microdiversity within these novel clusters.
Consistently, all three libraries were dominated by the same two large phylogenetic groups: Bacteroidetes and Gammaproteobacteria. All the other groups represented minor components. The identity of the particular OTUs within these large groups, however, was very different between library At12Oct (the highest salinity point) and the other two, and substantially different between At18Aug and AT18Oct (the same site with intermediate salinities in winter and spring, respectively). Bacteroidetes accounted for 39% of the clones in the At12Oct library, mostly distantly associated (similarities ranged between 91 and 95%) to the halophilic bacterium Salinibacter ruber isolated from crystallizer ponds (salinities up to 35%) in coastal solar salterns (Antón et al. 2002). Since this library came from the most saline sample in Lake Tebenquiche (29.6%), it makes sense that Salinibacter-like sequences were retrieved in abundance. Certainly, the potential for new extremely halophilic members of the Bacteria exists within this cluster. In clone libraries At18Aug and At18Oct, Bacteroidetes were the most abundant groups followed by Gammaproteobacteria. Salinity was about twice in October (14.8%) than in August (7.9%), and most Bacteroidetes clones were distantly related to Psychroflexus spp. (similarities ranged between 87 and 92%), mainly in October. The most abundant cluster of Gammaproteobacteria was associated with uncultured bacteria from Mono Lake (Humayoun et al. 2003), a saline alkaline lake in the USA, as well as the few clones belonging to the Low and High G+C groups of bacteria were also associated to sequences from Mono Lake. As indicated above, aquatic systems with intermediate salinities have not received much attention, and our data will help for future comparisons.
A noteworthy finding is the consistent predominance of Bacteroidetes. A similar predominance was found in Salar de Huasco (C. Dorador, K.P. Witzel, C. Vargas, I. Vila, J.F. Imhoff, unpublished data) where both DGGE and clones libraries were done. Salar de Huasco is north of the areas studied here and it is found at 3,800 m a.s.l. Bacteroidetes were also the most abundant groups of bacteria in clone libraries from the water column of Lake Chaka (Jiang et al. 2006) with a salinity of 32.5%. In contrast, a study of several lakes in the Tibetan plateau (Wu et al. 2006) found Bacteroidetes to account for only 5–10% of the total cell count by fluorescent in situ hybridization, in the most saline lakes. When DGGE was carried out, however, Bacteroidetes were the most represented group accounting for almost half of the sequences retrieved. Preliminary FISH counts in Lake Tebenquiche indicated that Bacteroidetes accounted for 3–17% of the total count (average 8%). This discrepancy between PCR-related methods and FISH deserves further research, because in marine systems FISH usually gives higher representation of Bacteroidetes than cloning or DGGE (Cottrell and Kirchman 2000). Obviously, the high salinity Bacteroidetes are very different from the marine representatives of this group and better FISH probes have to be designed for them. Alternatively, most of the Bacteroidetes would have low ribosomal content in the case where they were inactive under in situ conditions being difficult to detect by FISH but still recovered by DNA amplification methods.
A large number of bacteria have been isolated in pure culture from Lake Tebenquiche in the past (Prado et al. 1991, 1993, 2006; Valderrama et al. 1991), but none of them could be retrieved in our molecular study of the same lake. The difference between microorganisms retrieved from natural systems by molecular and pure culture methods is a well-known phenomenon (Amann et al. 1995; Pedrós-Alió 2007). Usually, it is attributed to inability of microbiologists to find suitable conditions to tame those microbes that are abundant in nature. Microorganisms isolated from a given system but absent from clone libraries strongly indicate that they were at low abundances but can potentially become dominant. Thus, the topographic complexity and environmental heterogeneity of Lake Tebenquiche make “visible” for the molecular methods different fractions of the whole collection of microorganisms present in the system, presumably the ones best adapted to each particular environment.
Conclusions
We found that the bacterial community composition in Lake Tebenquiche was very heterogeneous both in space and time, in close relationship to the salinity of the water. This system is highly variable depending on the groundwater inputs in different zones of the lake basin, and thus, water with very different salinities can coexist in the lake. In addition, hypersaline places within the lake can be diluted by dynamic freshwater inputs and bacteria have to adapt locally to the new conditions. These differences in salinity override any seasonal changes that would be mild anyway in this tropical region. Overall, this intermediate salinity lake showed a remarkable degree of novelty in its bacterial assemblage, both in terms of deep lineages and of microdiversity within known clusters. All these novel sequences belong to the diversity of the system and, therefore, to microorganisms that are relevant for the ecosystem functioning. The challenge now is to isolate them in pure culture to analyze in detail their specific adaptations to the dynamic perturbations found in Lake Tebenquiche.
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R (2002) Salinibacter ruber gen. nov., sp nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol 52:485–491
Baldauf S (2003) The deep roots of eukaryotes. Science 300:1703–1706
Benlloch S, Acinas SG, Antón J, López-López A, Luz SP, Rodríguez-Valera F (2001) Archaeal Biodiversity in Crystallizer Ponds from a Solar Saltern: Culture versus PCR. Microb Ecol 41:12–19
Benlloch S, Acinas SG, Martínez-Murcia A, Rodríguez-Valera F (1996) Description of prokaryotic biodiversity along the salinity gradient of a multipond solar saltern by direct PCR amplification of 16S rDNA. Hydrobiologia 329:19–31
Benlloch S, López-López A, Casamayor EO, Øvreas L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedrós-Alió C, Rodríguez-Valera F (2002) Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol 4:349–360
Bowman JP, McCammon SA, Rea SM, McMeekin TA (2000) The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett 183:81–88
Carmona V, Pueyo JJ, Taberner C, Chong G, Thirlwall M (2000) Solute inputs in the Salar de Atacama (N. Chile). J Geochem Explor 69:449–452
Casamayor E, Muyzer G, Pedrós-Alió C (2001) Composition and temporal dynamics of planktonic archaeal assemblages from anaerobic sulfurous environments studied by 16S rDNA Denaturing Gradient Gel Electrophoresis and sequencing. Aquat Microb Ecol 25:237–246
Casamayor EO, Calderón-Paz JI, Pedrós-Alió C (2000) 5S rRNA fingerprints of marine bacteria, halophilic archaea and natural prokaryotic assemblages along a salinity gradient. FEMS Microbiol Ecol 34:113–119
Casamayor EO, Massana R, Benlloch S, Øvreas L, Díez B, Goddard VJ, Gasol JM, Joint I, Rodríguez-Valera F, Pedrós-Alió C (2002) Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol 4:338–348
Cottrell MT, Kirchman DL (2000) Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol 66:5116–5122
Demergasso C, Casamayor EO, Chong G, Galleguillos P, Escudero L, Pedrós-Alió C (2004) Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiol Ecol 48:57–69
Dumestre JF, Casamayor EO, Massana R, Pedrós-Alió C (2002) Changes in bacterial and archaeal assemblages in an equatorial river induced by the water eutrophication of Petit Saut dam reservoir (French Guiana). Aquat Microb Ecol 26:209–221
Estrada M, Hendriksen P, Gasol JM, Casamayor EO, Pedrós-Alió C (2004) Diversity of planktonic photoautotrophic microorganisms along a salinity gradient as depicted by microscopy, flow cytometry, pigment analysis and DNA-based methods. FEMS Microbiol Ecol 49:281–293
Ferrera I, Massana R, Casamayor EO, Balagué V, Sánchez O, Pedrós-Alió C, Mas J (2004) High-diversity biofilm for the oxidation of sulfide-containing effluents. Appl Microbiol Biotechnol 64:726–734
Gasol J, del Giorgio P (2000) Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar 64:197–224
Gasol J, Casamayor EO, Join I, Garde K, Gustavson K, Benlloch S, Díez B, Schauer M, Massana R, Pedrós-Alió C (2004) Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol 34:193–206
Giovannoni S, Rappé M (2000) Evolution, diversity, and molecular ecology of marine prokaryotes. In: Kirchman DL (ed) Microbial ecology of the oceans. Wiley Interscience, New York, pp 47–84
Giovannoni SJ, Britschgi TB, Moyer CL, Field KG (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60–63
Glöckner FO, Fuchs BM, Amann R (1999) Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol 65:3721–3726
Humayoun SB, Bano N, Hollibaugh JT (2003) Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol 69:1030–1042
Jiang H, Dong H, Zhang G, Yu B, Chapman L, Fields M (2006) Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China. Appl Environ Microbiol 72:3832–3845
Kunte H, Trüper H, Stan-Lotter H (2002) Halophilic microorganisms. In: Horneck G, Baumstark-Khan C (eds) Astrobiology, the quest for the conditions of life. Springer, Koln, pp 185–200
Lizama C, Monteoliva-Sanchez M, Prado B, Ramos-Cormenzana A, Weckesser J, Campos V (2001) Taxonomic study of extreme halophilic archaea isolated from the “Salar de Atacama”, Chile. Syst Appl Microbiol 24:464–474
Lizama C, Monteoliva-Sánchez M, Suárez-García A, Roselló-Mora R, Aguilera M, Campos V, Ramos-Cormenzana A (2002) Halorubrum tebenquichense sp. nov., a novel halophilic archaeon isolated from the Atacama Saltern, Chile. Int J Syst Evol Microbiol 52:149–155
Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR, Stredwick JM, Garrity GM, Li B, Olsen GJ, Pramanik S, Schmidt TM, Tiedje JM (2000) The RDP (Ribosomal Database Project) continues. Nucleic Acids Res 28:173–174
Mancinelli R, Fahlen T, Landheim R, Klovstad M (2004) Brines and evaporates: analogs for Martian life. Adv Space Res 33:1244–1246
Maturrano L, Santos F, Rosselló-Mora R, Antón J (2006) Microbial diversity in Maras salterns, a hypersaline environment in the peruvian andes. Appl Environ Microbiol 72:3887–3895
Oren A (2002) Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biot 28:56–63
Pedrós-Alió C (2005) Diversity of microbial communities: the case of solar salterns. In: Gunde-Cimerman N, Plemenitas A, Oren A (eds) Adaptations to life at high salt concentrations in archaea, bacteria, and eukarya. Cellular origins, life in extreme habitats and astrobiology (COLE) (Seckbach J, series editor), vol 9. Springer, Dordrecht, pp 71–90
Pedrós-Alió C (2006) Marine microbial diversity: can it be determined? Trends Microbiol 14:257–263
Pedrós-Alió C (2007) Dipping into the rare biosphere. Perspect Sci 315:192–193
Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P, Tunlid A, Hagstrom A (2007) Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16:867–880
Prado B, del Moral A, Quesada E, Ríos R, Monteoliva-Sánchez M, Campos V, Ramos-Cormezana A (1991) Numerical taxonomy of moderately halophilic Gram negative rods isolated from the Salar de Atacama, Chile. Syst Appl Microbiol 14:275–281
Prado B, del Moral A, Campos V (1993) Distribution and Types of Heterotropic Halophilic Flora from Salar De Atacama, Chile. Toxicol Environ Chem 38:163–166
Prado B, Lizama C, Aguilera M, Ramos-Cormenzana A, Fuentes S, Campos V, Monteoliva-Sánchez M (2006) Chromohalobacter nigrandesensis sp. nov., a moderately halophilic, Gram-negative bacterium isolated from Lake Tebenquiche on the Atacama Saltern, Chile. Int J Syst Evol Microbiol 56:647–651
Risacher F, Alonso H (1996) Geochemistry of the Salar de Atacama. 2. Water evolution. Rev Geol Chile 23:123–134
Risacher F, Alonso H, Salazar C (1999) Geoquímica de aguas en cuencas cerradas: I, II y III Regiones—Chile (Technical Report S.I.T. No. 51). Convenio de Cooperación DGA, UCN, IRD, Santiago, Chile
Rodríguez-Valera F, Acinas S, Antón J (1999) Contribution of molecular techniques to the study of microbial diversity in hypersaline environments. In: Oren A (ed) Microbiology and Biogeochemistry of Hypersaline Environments. CRC Press, Boca Raton, pp 27–38
Sánchez O, Gasol J, Massana R, Mas J, Pedrós-Alió C (2007) Comparison of different denaturing gradient gel electrophoresis primer sets for the study of marine bacterioplankton communities. Appl Environ Microbiol 73:5962–5967
Schauer M, Massana R, Pedrós-Alió C (2000) Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol Ecol 33:51–59
Sorensen KB, Canfield DE, Teske AP, Oren A (2005) Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol 71:7352–7365
Staley J, Konopka A (1985) Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 39:321–346
Valderrama MJ, Prado B, del Moral A, Ríos R, Ramos-Cormenzana A, Campos V (1991) Numerical taxonomy of moderately halophilic gram-positive cocci isolated from the Salar de Atacama (Chile). Microbiologia 7:35–41
Williams W (1996) The largest, highest and lowest lakes of the world: saline lakes. Verh Int Verein Limnol 26:61–79
Wu Q, Zwart G, Schauer M, Kamst-van Agterveld M, Hahn M (2006) Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl Environ Microbiol 72:5478–5485
Yentsch C, Menzel D (1963) A method for the determination of phytoplankton chlorophyll and phaeophytin by fluorescence. Deep-Sea Res 10:221–231
Zúñiga L, Campos V, Pinochet H, Prado B (1991) A limnological reconnaissance of Lake Tebenquiche, Salar de Atacama, Chile. Hydrobiologia 210:1–2
Acknowledgments
Sampling and measurements carried out in Chile were funded by grants FONDECYT 1030441 and FONDEF D99I1026. Measurements carried out in Barcelona were funded by grant “ATACAMA-2002” (CICYT, BOS2002-10258-E). Grant “BIOARSENICO” from Fundación BBVA funds current work in both laboratories. EOC was supported by the Programa Ramón y Cajal and REN2003-08333, and VB by grant GENμMAR (CTM2004-02586/MAR), both from the Spanish Ministerio de Educación y Ciencia. LEG was supported by the Centro de Investigación Científica y Tecnológica para la Minería, Antofagasta, Chile. We thank Juan José Pueyo for suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi.
Rights and permissions
About this article
Cite this article
Demergasso, C., Escudero, L., Casamayor, E.O. et al. Novelty and spatio–temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama). Extremophiles 12, 491–504 (2008). https://doi.org/10.1007/s00792-008-0153-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-008-0153-y