Abstract
Recently, osteogenic precursor cells were isolated from human dental follicles, which differentiate into cementoblast- or osteoblast-like cells under in vitro conditions after the induction with dexamethasone or insulin. However, mechanisms for osteogenic differentiation are not understood in detail. In a previous study, real-time RT-PCR results demonstrated molecular mechanisms in dental follicle cells (DFCs) during osteogenic differentiation that are different from those in bone-marrow-derived mesenchymal stem cells. We analysed gene expression profiles in DFCs before and after osteogenic differentiation with the Affymetrix GeneChip® Human Gene 1.0 ST Array. Transcripts of 98 genes were up-regulated after differentiation. These genes could be clustered into subcategories such as cell differentiation, cell morphogenesis, and skeletal development. Osteoblast-specific transcription factors like osterix and runx2 were constitutively expressed in differentiated DFCs. In contrast, the transcription factor ZBTB16, which promotes the osteoblastic differentiation of mesenchymal stem cells as an up-stream regulator of runx2, was differentially expressed after differentiation. Transcription factors NR4A3, KLF9 and TSC22D3, involved in the regulation of cellular development, were up-regulated as well. In conclusion, we present the first transcriptome of human DFCs before and after osteogenic differentiation. This study sheds new light on the complex mechanism of osteogenic differentiation in DFCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human dental follicle is an ectomesenchymal tissue surrounding the developing tooth germ and containing progenitor cells of the periodontium [1]. As the supporting tissue of the tooth, the periodontium is composed of the periodontal ligament (PDL), alveolar bone, and the mineralized bone-like cementum covering the tooth root surfaces [2]. The PDL is a specialised connective tissue whose fibres are embedded in the cementum and the alveolar bone, providing the attachment for the tooth [1]. Recently, precursor cells were isolated from extracted human third molars, and were capable of differentiation into periodontium-like tissues [3]. These dental follicle cells (DFCs) have the capacity to differentiate into calcified tissue similar to bone-marrow-derived mesenchymal stem cells under in vitro conditions, albeit the osteogenic cell differentiation in DFCs is less complete and the mechanisms are poorly understood. Recently, we reported a differential gene expression of DLX-3 during osteogenic differentiation in DFCs, which promotes osteoblast differentiation in precursor cells. Histochemical investigations furthermore revealed that the expression of alkaline phosphatase and the formation of calcified tissues in DFCs after differentiation. BMP-2, an important indicator of osteogenic differentiation, was also up-regulated during differentiation [4]. Today, it is obvious that the osteogenic differentiation of dental follicle-derived precursors is controlled by a network of regulatory molecules including growth factors like BMP-2 and BMP-7 [3–5]. However, important factors for osteogenic differentiation such as MSX-2, DLX-5, runx-2 or osterix and osteoblast markers like osteocalcin or bone sialoprotein, were almost constitutively expressed in differentiated DFCs [6]. Here, gene expression profiles were recorded before and after osteogenic differentiation in DFCs to elucidate the mechanisms of biomineralisation in DFCs. Our analysis demonstrated 98 up-regulated and 52 down-regulated genes. They are functionally classified and their role is discussed in osteogenic differentiation.
Materials and methods
Cell culture
Dental follicle cells were isolated as described previously [3]. Briefly, an impacted human third molar was surgically removed and collected from a 20-year-old patient with informed consent. The attached dental follicle was separated from the mineralized tooth. The follicle tissues were cleaned and then digested in a solution of collagenase type I, hyaluronidase (Sigma-Aldrich, Munich, Germany) and DNAse I (Roche, Mannheim, Germany) for 1 h at 37°C. Digested tissues were seeded into T25 flasks in MesenchymStem Medium (PAA, Pasching, Austria) at 37°C in 5% CO2. Non-adherent cells were removed after medium-change. Osteogenic differentiation was induced after cultivation in alpha-MEM (PAA) supplemented with 10% foetal bovine serum (PAA), 100 µmol/L ascorbic acid 2-phosphate, 2.8 mmol/l KH2PO4, 1 × 10−7 mol/l dexamethasone sodium phosphate (Sigma-Aldrich) and HEPES (20 mmol/L). We substituted dexamethasone with insulin (5 µg/ml final concentration) for the insulin-based differentiation protocol. Stainings of differentiated cells with alizarin was described previously. Alkaline phosphatase (ALP) activity was detected with naphthol and fast red violet [4, 6].
Microarray analysis
For osteogenic differentiation, DFCs were stimulated with dexamethasone for 28 days. Total RNAs were extracted using the kit NucleoSpin® RNA II kit (Macherey Nagel, Düren, Germany) and quality-controlled using the RNA 6000 Nano LabChip (Agilent Technologies, Santa Clara, CA, USA). DNA microarray analyses were carried out with Affymetrix Human Gene 1.0 ST arrays according to the Affymetrix standard protocol. Four microarrays were performed with RNA from independent cultures of osteogenic differentiated DFCs, and two microarrays were performed with RNA from independent cultures of undifferentiated DFCs. Microarray hybridizations was carried out at the “Centre of Excellence for Fluorescent Bioanalytics” of the University of Regensburg. Data analyses were performed using the ChipInspector software (Genomatix Software GmbH, Munich, Germany), applying a significance analysis of microarrays at a false discovery rate below 0.5% and a minimum log2 ratio of 2.0 (fold change of 4.0). The database for Annotation, Visualisation, and Integrated Discovery (DAVID; http://niaid.abcc.ncifcrf.gov/) and the Bibliosphere Software tool (Genomatix) were used for annotations of significant regulated transcripts after differentiation.
Real-time RT-PCR
Total RNA was isolated from cells with NucleoSpin® RNA II. In order to digest contaminating genomic DNA, RNA was treated with DNAse I. The cDNA synthesis was performed using 400 ng total RNA and the RevertAid™ M-MuLV Reverse Transcriptase Kit (Fermentas, St. Leon-Rot, Germany). Real-time PCR was performed with TaqMan® Fast Universal PCR Master Mix (Applied Biosystems, Foster City, USA). Sequences for primers and probes can be obtained from the authors. Real-time RT-PCR (qRT-PCR) was performed with the 7900 HT Fast Real-time PCR System (Applied Biosystems). The Applied Biosystems’ RQ manager 1.2 software was used for estimation of threshold cycles (Ct-value). GAPDH gene expression was chosen for normalisation of each sample (housekeeper gene). Quantification was done with the delta/delta calculation method as described by Winer et al. [7]. For calibration, total RNA was used from cells before the induction of cell differentiation (relative gene expression = 1).
Results and discussion
Differentiation of DFCs with dexamethasone
Cultured DFCs were osteogenic differentiated using dexamethasone, a member of the glucocorticoid class of steroid hormones. To improve osteogenic differentiation the concentration of dexamethasone (1 × 10−7°M) was increased in comparison to our previous studies (1 × 10−8 M) [3, 4, 6]. The induction of osteogenic differentiation in DFCs was evaluated by gene expression of osteogenic markers and alkaline phosphatase activity (ALP) after 14 days of differentiation. We compared DFCs cultivated in two different cell culture media that were supplemented either with or without dexamethasone (Fig. 1). After 14 days of cultivation DFCs demonstrated strong alkaline phosphatase activity in cell culture medium with dexamethasone, but almost no alkaline phosphatase activity in medium without dexamethasone (Fig. 1a). In contrast to previous studies [4, 6] runx2 was higher expressed in medium with dexamethasone than without dexamethasone (Fig. 1b). Moreover the expression of the transcription factor osterix was elevated in dexamethasone-treated cells (Fig. 1b). These results demonstrate that dexamethasone has a positive effect on the initiation of osteogenic differentiation in DFCs, although markers for osteogenic differentiation are also expressed in DFCs without dexamethasone treatment. In contrast to our previous studies [4, 6], we identified an up-regulation of runx2 and osterix at day 14 of differentiation, which is probably being caused by the new concentration of dexamethasone.
DFCs after 14 days of cultivation in cell culture medium with dexamethasone (differentiation) and without dexamethasone (control). a Alkaline phosphatase activity (ALP) of DFCs with and without dexamethasone treatment. b Real-time RT-PCR with specific PCR primers for markers of osteogenic differentiation. For calibration total RNAs of DFCs were cultivated without dexamethasone (rel. gene expression = 1). PCRs were performed with three biological replicates +/−SEM. Abbreviation: runx2 runt-related transcription factor 2
Microarray analysis of osteogenic differentiated DFCs
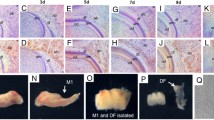
To analyse the gene expression profile of osteogenic differentiated cells, DFCs were cultivated in 1 × 10−7 M dexamethasone, l-ascorbate-2-phosphate, and KH2PO4 for 4 weeks. After 4 weeks, long-term cultures of DFCs grown in the presence of dexamethasone demonstrated the capacity to form Alizarin Red-positive areas with high levels of calcium, indicating osteogenic differentiation (Fig. 2a). No calcium accumulation was observed in long-term cultures without dexamethasone (data not shown). A high level of ALP activity was visible in dexamethasone-treated DFCs (Fig. 2a), but no ALP activity was detected in undifferentiated DFCs (data not shown). Previous studies have also demonstrated the occurrence of osteogenic differentiation in DFCs at this point in time [3, 4, 6]. The calcium deposits were scattered throughout as single mineralized zones. The formation of membrane-like structures with mineralized foci, which is a typical feature of differentiated DFCs, was also described previously [3, 4, 6]. We suppose that DFCs form membrane-like structures at an early stage of osteogenic differentiation. In previous experiments we grew long-term cultures for 5 weeks [3] and longer (own unpublished observations). Here, more complete mineralization was detectable after alizarin red staining. We therefore concluded that the differentiation process of DFCs is not completed after 4 weeks. However, this is an excellent time point to evaluate molecular mechanisms during osteogenic differentiation in DFCs, because differentiation has started but not finished. At this point in time, we would expect up-regulation of genes that are involved in the molecular process of osteogenic differentiation. In previous studies we also induced osteogenic differentiation by insulin treatment [4, 6]. In this study, we have also induced osteogenic differentiation with insulin as a control (Fig. 2). Although the gene expression of osteogenic markers was up-regulated after 4 weeks of differentiation with insulin, the osteogenic phenotype of differentiated DFCs, estimated by ALP activity and Alizarin Red staining, was similar to or weaker than that of dexamethasone-treated DFCs [4, 6]. We obtained similar results for DFCs differentiated with an insulin-based protocol (Fig. 2a) and made use of insulin-differentiated DFCs to evaluate differentially expressed genes identified with the microarray analysis by real-time RT-PCR (see below). For the microarray study, DFCs were osteogenic differentiated with dexamethasone for 4 weeks. Primary microarray data of DFCs before and after cell differentiation were compared with the ChipInspector software [8, 9]. We obtained 98 up-regulated and 52 down-regulated genes in differentiated cells (Supplemental Table). Interestingly, typical markers for osteogenic differentiation were expressed, but only alkaline phosphatase (ALP) and bone morphogenic protein (BMP)-2 were clearly up-regulated after differentiation (Fig. 2b). Although the gene expression of ALP and BMP-2 was up-regulated with microarray raw data (Fig. 2b) these genes were not significantly up-regulated after a statistical analysis. ChipInspector filtered out these genes, based on statistical criteria. In our analysis, we identified significantly regulated genes. Unlike ALP and BMP-2, some osteoblast markers were down-regulated (collagen type I) or constitutively expressed (osteocalcin, OCN) after differentiation (Fig. 2b). These gene expression results also fit with previous data [4, 6]. In Fig. 2b, we compared microarray hybridization data with the real-time RT-PCR analysis. The real-time RT-PCR data and microarray data were comparable, which demonstrates the reliability of our microarray analysis.
Differentiation potential of DFCs in vitro. a Alizarin Red staining as a measure of calcium accumulation (Alizarin) and alkaline phosphatase activity staining (ALP) in 4-week-old long-term cultures differentiated with dexamethasone and insulin. All figures have the same magnification. b Real-time RT-PCR analysis with primers for osteoblast markers after 4 weeks of osteogenic differentiation. The real-time RT-PCRs confirmed the DNA microarray data. Total RNAs from DFCs before the induction of cell differentiation (undifferentiated) were used for calibration of real-time RT-PCRs (relative gene expression = 1). Each bar of undifferentiated DFCs (light grey bar) and differentiated cells with dexamethasone (black bar) and insulin (white bar) represents the mean of three biological replicates +/−SEM. Dark grey bars represent the mean of gene expression measured by Affymetrix microarray analysis. Abbreviations ALP alkaline phosphatase, BMP-2 bone morphogenic protein 2, DLX-5 distal-less homeobox 5, MSX-2 Msh homeobox 2, runx2 runt-related transcription factor 2
Bioinformatic annotation of regulated genes
To evaluate the cellular localization of regulated genes, we used the programmes DAVID and Bibliosphere [8–10]. Gene products of both up- and down-regulated genes were predominantly localised in the extracellular region; especially in the proteinaceous extracellular matrix (Tables 1, 2). The Bibliosphere tissue filter demonstrated that transcripts present in various tissues such as the kidney, colon, nervous system, or epididymis were significantly overrepresented in up-regulated genes (data not shown). We assume that these transcripts are generally expressed in differentiated cells of various tissues. Genes that were overrepresented in down-regulated transcripts are found in immortalised cell lines or embryonic tissues (data not shown). These genes are probably involved in the maintenance of undifferentiated DFCs.
Regulated genes were further clustered with Gene Ontology (DAVID) and the functional annotation tool of DAVID. More than 50% of down-regulated genes and 30% of up-regulated genes are involved in multicellular development (Fig. 3; Tables 1 and 2). Interestingly, 10% of down-regulated genes are involved in amino acid biosynthesis and reproductive processes (Fig. 3). These genes are probably important for maintenance and proliferation of undifferentiated DFCs, and a reduction in gene expression must be important for osteogenic differentiation. Additionally, gene clusters for cell morphogenesis, skeletal development and cell differentiation were found in up-regulated genes (Fig. 3). Two small but significantly overrepresented clusters of up-regulated genes were found. These clusters are netrin-related proteins, a class of proteins involved in axon guidance, and selenium-binding proteins, which are important for multiple enzymatic reactions [11, 12]. These genes are probably involved in the differentiation of DFCs, but their functions are unknown.
Intriguingly, transcripts of complement and coagulation genes were significantly overrepresented in up-regulated genes (Table 1). Although genes of this cluster are associated with the immune response, many of them are also associated with developmental processes (Table 1). Abdallah et al. observed an up-regulation of immune response-related genes upon the inhibition of cell differentiation in mesenchymal stem cells [13]. In contrast, our investigation demonstrated the up-regulation of immune response-related genes after differentiation (Tables 2 and 3). We do not know how these genes are associated with the osteogenic differentiation process in DFCs. Interestingly, paracrine effects are suggested for the success of current clinical treatments based on mesenchymal stem cells [14, 15]. The secretion proteome of mesenchymal stem cells contains a high number of immune response-related proteins like interleukins and chemokines. These proteins are probably involved in the regulation of endogenous stem cell differentiation [15]. Additional studies will elucidate the function of immune response-related genes in cell differentiation of DFCs.
Single up-regulated genes in DFCs after osteogenic differentiation, such as IGF-2 or CD14 are known to be involved in skeletal development (Table 4), and mutations of these genes are associated with bone density diseases (Table 5). However, these genes are not typical markers for bone or dental tissues, and it is difficult to speculate on their functions in the context of our study.
The dental follicle is also involved in osteoclast recruitment, which is required for tooth eruption. Interestingly, genes involved osteoclastogenesis (like RANKL and osteoprotegerin) are not significantly regulated in differentiated DFCs. In contrast, TNFSF15, which is related to RANKL, was differentially expressed and belongs to the tumour necrosis factor (TNF) ligand family. It can activate NF-kappaB and MAP kinases, and acts as an autocrine factor to induce apoptosis in endothelial cells [16].
Transcription factors regulated by osteogenic differentiation
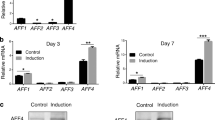
Important information about osteogenic differentiation in DFCs can be obtained from regulated transcription factors (Table 6). Six and four transcription factors were significantly up- and down-regulated, respectively, after osteogenic differentiation. A real-time RT-PCR analysis verified the obtained microarray data (Fig. 4). Interestingly, similar results were obtained for gene expression of FOXM1, ID1, ID3, KLF9, NR4A3, PRDM1 and ALF after differentiation with insulin. However, TSC22D2 and LDB2 were constitutively expressed in insulin-differentiated cells (Fig. 4). Although known osteoblast-specific transcription factors like runx2 or osterix were not regulated, known differentially expressed transcription factors are involved in cell proliferation and/or differentiation. In the following sections, we will discuss current knowledge about these identified transcription factors.
Gene expression analysis of down-regulated (LDB2, FOXM1, ID1, ID3) and up-regulated (KLF9, NR4A3, PRDM1, ALF, TSC22D3) transcription factors after osteogenic differentiation. Total RNAs from cells before the induction of cell differentiation (undifferentiated DFCs) were used for calibration (relative gene expression = 1). Insulin-differentiated DFCs were used for a comparison with dexamethasone-differentiated cells. Bars represent the mean of three biological replicates +/−SEM. Gene expression of the up-regulated transcription factor ZBTB16 is not shown, because transcripts of this transcription factor were not expressed in undifferentiated DFCs. However, they were significantly expressed after osteogenic differentiation (data not shown)
The transcription factor NR4A3 is strongly up-regulated after osteogenic differentiation of DFCs and is known to be implicated in the senescence of fibroblast cells [17]. Abrogation of NR4A3 leads to development of acute myeloid leukaemia. It has been suggested that NR4A3 is a tumour suppressor of myeloid leukaemogenesis and may functions as a homeostatic regulator of proliferation, apoptosis and differentiation [18]. However, there is no known connection between NR4A3 and osteogenic differentiation. Tsc22d3 is an up-regulated transcription factor containing a glucocorticoid-induced leucine zipper (GILZ). It was initially identified as a dexamethasone-responsive gene involved in the control of T-lymphocyte activation and apoptosis [19]. Recently, it was demonstrated that GILZ directly interacts with Ras in vitro and in vivo and that silencing of GILZ leads to an inhibition of dexamethasone antiproliferative effects [19]. We assume that the up-regulation of Tsc22d3s is due to the use of the glucocorticoid dexamethasone in the course of differentiation. This was also suggested by the real-time RT-PCR analysis of insulin-differentiated DFCs (Fig. 4). Therefore, it is highly probable that Tsc22d3 has no direct influence on the osteogenic differentiation of DFCs. Another up-regulated transcription factor is encoded by the gene PRDM1. Being a transcriptional repressor, it is considered to be a master regulator for multipotent progenitor cell populations in the posterior forelimb, caudal pharyngeal arches, secondary heart field and sensory vibrissae, maintaining key signalling centres at these diverse tissues sites [20]. However, further investigations are required to reveal a possible role of this general transcription factor in osteogenic cell differentiation. One transcription factor, the TFIIA alpha/beta-like factor (ALF), is germ cell-specific, and there is no suggestion of a possible role in somatic cell differentiation [21]. The last two up-regulated transcription factors, encoded by genes ZBTB16 und KLF9, have important functions in somatic tissue development. KLF9, for example, is an important regulator of cell migration and proliferation of intestinal cells, but the function of KLF9 in osteogenic differentiation is unknown [22]. Interestingly, ZBTB16, also known as the promyelotic leukaemia zinc finger transcription factor, promotes osteogenic differentiation of human mesenchymal stem cells as an up-stream activator of runx2 and collagen type I [23]. However, gene expression of runx2 and collagen type I was not up-regulated in DFCs after 4 weeks of differentiation. The differentially expression of BMP-2, which stimulates osteogenic differentiation in DFCs [5], can be regulated by ZBTB16 in DFCs. However, in mesenchymal stem cells the expression of BMP-2 was not regulated by ZBTB16, and BMP-2 did not induce the expression of ZBTB16 [23]. Thus further investigations of the role of ZBTB16 in DFCs will be important for our knowledge about osteogenic differentiation in DFCs.
Four different transcription factors were significantly down-regulated after differentiation, FOXM1, ID1, ID3 and LDB2. A previous study indicated that the LDB2 protein plays an important role in the homeostasis of corneal epithelium and it seems to have an important function in the maintenance of hair follicle stem cells [24]. ID1 and ID3 are transcription factors without a DNA-binding domain, but they can form heterodimers with members of the basic helix–loop–helix family of transcription factors. IDs have multiple functions. For example, they can inhibit neurogenesis, and control endothelial progenitor cell formation and the growth of vascular cells [25–27]. FoxM1 has been reported to regulate mitotic entry and prevent spindle defects in neural precursors [28]. Summarised, these down-regulated transcription factors are mainly involved in stem cell maintenance and cell proliferation. Therefore, we suppose that down-regulation of these factors supports the molecular process of osteogenic differentiation in DFCs.
Conclusion
Transcripts of 98 genes were up-regulated after differentiation in DFCs. These genes could be clustered into subcategories such as cell proliferation, cell differentiation, cell morphogenesis and skeletal development. In contrast to mesenchymal stem cells, DFCs did not up-regulate osteoblast-specific transcription factors like osterix and runx2 after 4 weeks of osteogenic differentiation. However, these markers were up-regulated after 14 days of osteogenic differentiation. However, six up-regulated transcription factors including ZBTB16, which promotes the osteoblastic differentiation of mesenchymal stem cells, were identified in our study as new top candidates for the regulation of osteogenic differentiation in DFCs. It is important to note that DFCs are also progenitors of PDL fibroblasts and that some differentially expressed genes are important for the differentiation of DFCs into PDL fibroblasts. Further studies will evaluate the role of these transcription factors in the process of osteogenic differentiation in DFCs. ZBTB16 in particular will be a target for further investigations.
References
Ten Cate AR (1997) The development of the periodontium—a largely ectomesenchymally derived unit. Periodontol 2000 13:9–19
Morsczeck C, Schmalz G, Reichert TE, Vollner F, Galler K, Driemel O (2008) Somatic stem cells for regenerative dentistry. Clin Oral Investig 12:113–118
Morsczeck C, Gotz W, Schierholz J, Zeilhofer F, Kuhn U, Mohl C, Sippel C, Hoffmann KH (2005) Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol 24:155–165
Morsczeck C, Moehl C, Gotz W, Heredia A, Schaffer TE, Eckstein N, Sippel C, Hoffmann KH (2005) In vitro differentiation of human dental follicle cells with dexamethasone and insulin. Cell Biol Int 29:567–575
Kemoun P, Laurencin-Dalicieux S, Rue J, Farges JC, Gennero I, Conte-Auriol F, Briand-Mesange F, Gadelorge M, Arzate H, Narayanan AS, Brunel G, Salles JP (2007) Human dental follicle cells acquire cementoblast features under stimulation by BMP-2/-7 and enamel matrix derivatives (EMD) in vitro. Cell Tissue Res 329:283–294
Morsczeck C (2006) Gene expression of runx2, Osterix, c-fos, DLX-3, DLX-5, and MSX-2 in dental follicle cells during osteogenic differentiation in vitro. Calcif Tissue Int 78:98–102
Winer J, Jung CK, Shackel I, Williams PM (1999) Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270:41–49
Scherf M, Epple A, Werner T (2005) The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform 6:287–297
Seifert M, Scherf M, Epple A, Werner T (2005) Multievidence microarray mining. Trends Genet 21:553–558
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:3
Bansal MP, Kaur P (2005) Selenium, a versatile trace element: current research implications. Indian J Exp Biol 43:1119–1129
Livesey FJ (1999) Netrins and netrin receptors. Cell Mol Life Sci 56:62–68
Abdallah BM, Boissy P, Tan Q, Dahlgaard J, Traustadottir GA, Kupisiewicz K, Laborda J, Delaisse JM, Kassem M (2007) dlk1/FA1 regulates the function of human bone marrow mesenchymal stem cells by modulating gene expression of pro-inflammatory cytokines and immune response-related factors. J Biol Chem 282:7339–7351
Caplan AI, Dennis JE (2006) Mesenchymal stem cells as trophic mediators. J Cell Biochem 98:1076–1084
Sze SK, de Kleijn DP, Lai RC, Khia Way TE, Zhao H, Yeo KS, Low TY, Lian Q, Lee CN, Mitchell W, El Oakley RM, Lim SK (2007) Elucidating the secretion proteome of human embryonic stem cell-derived mesenchymal stem cells. Mol Cell Proteomics 6:1680–1689
Hou W, Medynski D, Wu S, Lin X, Li LY (2005) VEGI-192, a new isoform of TNFSF15, specifically eliminates tumor vascular endothelial cells and suppresses tumor growth. Clin Cancer Res 11:5595–5602
Hardy K, Mansfield L, Mackay A, Benvenuti S, Ismail S, Arora P, O’Hare MJ, Jat PS (2005) Transcriptional networks and cellular senescence in human mammary fibroblasts. Mol Biol Cell 16:943–953
Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM (2007) Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med 13:730–735
Ayroldi E, Zollo O, Bastianelli A, Marchetti C, Agostini M, Di VR, Riccardi C (2007) GILZ mediates the antiproliferative activity of glucocorticoids by negative regulation of Ras signaling. J Clin Invest 117:1605–1615
Robertson EJ, Charatsi I, Joyner CJ, Koonce CH, Morgan M, Islam A, Paterson C, Lejsek E, Arnold SJ, Kallies A, Nutt SL, Bikoff EK (2007) Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development 134:4335–4345
Kim M, Li D, Cui Y, Mueller K, Chears WC, DeJong J (2006) Regulatory factor interactions and somatic silencing of the germ cell-specific ALF gene. J Biol Chem 281:34288–34298
Simmen FA, Xiao R, Velarde MC, Nicholson RD, Bowman MT, Fujii-Kuriyama Y, Oh SP, Simmen RC (2007) Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol 292:G1757–G1769
Ikeda R, Yoshida K, Tsukahara S, Sakamoto Y, Tanaka H, Furukawa K, Inoue I (2005) The promyelotic leukemia zinc finger promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of CBFA1. J Biol Chem 280:8523–8530
Xu X, Mannik J, Kudryavtseva E, Lin KK, Flanagan LA, Spencer J, Soto A, Wang N, Lu Z, Yu Z, Monuki ES, Andersen B (2007) Co-factors of LIM domains (Clims/Ldb/Nli) regulate corneal homeostasis and maintenance of hair follicle stem cells. Dev Biol 312:484–500
Bai G, Sheng N, Xie Z, Bian W, Yokota Y, Benezra R, Kageyama R, Guillemot F, Jing N (2007) Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev Cell 13:283–297
Ciarrocchi A, Jankovic V, Shaked Y, Nolan DJ, Mittal V, Kerbel RS, Nimer SD, Benezra R (2007) Id1 restrains p21 expression to control endothelial progenitor cell formation. PLoS ONE 2:e1338
Felty Q, Porther N (2008) Estrogen-induced redox sensitive Id3 signaling controls the growth of vascular cells. Atherosclerosis 198:12–21
Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, Rowitch DH (2007) Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol 27:8259–8270
Acknowledgement
This work was supported by the DFG (Deutsche Forschungsgemeinschaft) MO1875/2-1. We thank the “Centre of Excellence for Fluorescent Bioanalytics” of the University of Regensburg for the realisation of microarray hybridizations and Dr. Thomas Langmann for his support in the evaluation of raw-data. Finally, we would like to thank Dr. Merle Windgassen-Morsczeck for careful reading and valuable suggestions.
Conflict of interest
Authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table
Genes regulated after osteogenic differentiation (DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Morsczeck, C., Schmalz, G., Reichert, T.E. et al. Gene expression profiles of dental follicle cells before and after osteogenic differentiation in vitro. Clin Oral Invest 13, 383–391 (2009). https://doi.org/10.1007/s00784-009-0260-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-009-0260-x