Abstract
Several nonheme iron enzymes and biomimetic model complexes catalyze a substrate halogenation reaction. Recent computational studies (Borowski et al. J Am Chem Soc 132:12887–12898, 2010) on α-ketoglutarate dependent halogenase proposed an initial isomerization reaction that is important to give halogenated products. We present here a series of density functional theory calculations on a biomimetic model complex—[FeIV(O)(TPA)Cl]+, where TPA is tris(2-pyridylmethyl)amine—and investigate the mechanisms of substrate halogenation versus hydroxylation using the reactant and its isomer where the oxo and chloro groups have changed positions. We show here that the reactions occur on a dominant quintet spin state surface, although the reactants are in a triplet state. Despite the fact that the reactants can exist in two stable isomers with the oxo group either trans or cis to the axial ligand, they react differently with substrates, where one gives dominant hydroxylation and the other gives dominant chlorination of substrates. The ligand in the cis position of the oxo group is found to be active in the reaction mechanism and donated to the substrate during the reaction. A detailed thermochemical analysis of possible reaction mechanisms reveals that the strengths of the Fe–OH and Fe–Cl bonds in the radical intermediates are the key reasons for this regioselectivity switch of hydroxylation over halogenation. This study highlights the differences between enzymatic and biomimetic halogenases, where the former only react after an essential isomerization step, which is not necessary in model complexes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Substrate halogenation is a unique chemical reaction in biology but is surprisingly widespread, with over 4,500 examples reported [1, 2]. Despite the fact that C–Cl bonds are rare in natural products, there are several enzymes that catalyze the formation of this bond in organic molecules efficiently. In particular, halogenases catalyze the addition of a halogen to a wide variety of scaffolds, ranging from aromatic and heterocyclic rings to olefinic sites and even aliphatic carbons [3]. The activated chlorination of aliphatic carbons is crucial to the biological effectiveness of the natural product, illustrated by the observed loss of function from dechlorinated syringomycin and vancomycin [4, 5]. This has huge medical significance since halogenation during drug synthesis can increase the therapeutic effectiveness of the agent by a power of between 4 and 10 [3, 6]. One particular family of enzymes responsible for halogenation reactions in nature is the α-ketoglutarate (αKG) dependent halogenases, which are highly conserved throughout evolution and have biological roles as diverse as antibiotic biosynthesis and the metabolism of toxins [2, 3, 7–10].
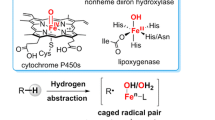
In recent years several crystal structures [11–13] of αKG-dependent halogenases were determined and show that they are structurally similar to the αKG-dependent hydroxylases and contain a nonheme iron active site, where the metal is linked to the protein via two histidine groups. In the hydroxylases there is also a linkage with a carboxylic acid group into a facial 2His–1Asp/Glu triad, but in the halogenases the acidic amino acid is missing because of an alanine amino acid in that position and the ligand position at the metal is taken in by a halide ion. Figure 1 displays the active site structure of the halogenase enzyme SyrB2 as taken from the 2FCT Protein Data Bank file [11]. This enzyme catalyzes the regioselective methyl group chlorination of l-Thr-S-SyrB1 of syringomycin E [14] on a nonheme iron center that is bound to two histidine side chains of His116 and His235 and a chloride anion in a facial triad. The chloride ion is bound in a hydrophobic pocket at hydrogen-bonding distance to a chain of crystal water molecules, whereas trans to its ligand position αKG is bound to the metal. The Protein Data Bank file does not contain a substrate but it is envisaged that it will bind near the iron center.
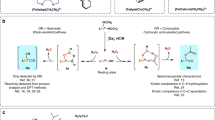
Detailed spectroscopic and kinetics studies identified several short-lived intermediates in the catalytic cycle, including an iron(IV)–oxo species that reacts with a large kinetic isotope effect with substrates [14–16]. On the basis of these studies it was concluded that the reaction proceeds with a rate-determining hydrogen abstraction step from the substrate by a high-valent iron(IV)–oxo species. These studies were followed by a series of computational studies that tried to rationalize the product distributions and explain the lack of hydroxylation products [17–19]. Owing to the absence of experimental evidence, many possible reaction mechanisms were proposed to explain the thermodynamically unfavorable halogenation over hydroxylation reaction, including CO2 trapping of the hydroxyl radical [17] and proton transfer from a nearby arginine residue [18]. In yet another study, a novel mechanism was proposed whereby the iron(IV)–oxo(halide) complex undergoes a ligand switch where the oxo and halide groups change position at the metal center prior to the hydrogen atom abstraction, as shown in Scheme 1 [19]. This ligand switch was identified as important for efficient substrate halogenation as otherwise substrate hydroxylation would have been the dominant mechanism.
Computational proposed mechanism [19] of substrate halogenation by a nonheme iron enzyme, whereby isomer A gives hydroxylation products and isomer B gives halogenation products. Labels are taken from the 2FCT Protein Data Bank file. Succ succinate
Recently there has been a positive development in substrate halogenation chemistry, and several synthetic mononuclear nonheme iron(IV)–oxo complexes were shown to react via substrate halogenation [20–23]. One particular iron ligand system known to react via halogenation of aliphatic groups is an iron with the tetradentate tripodal ligand tris(2-pyridylmethyl)amine (TPA); Fig. 2. Thus, its iron(IV)–oxo species has one vacant ligand site cis to the metal with respect to the oxo group, and with halide occupation has features resembling those of the active site of nonheme iron halogenases. To gain insight into the reactivity differences of enzymatic and biomimetic halogenases, we decided to investigate the competitive hydroxylation and halogenation of a model substrate by [FeIV(O)(TPA)X]+ as the chemical system has been well studied experimentally as well as computationally. The chemical properties and reactivities of [FeIV(O)(TPA)X]+ with X being a solvent molecule or an anion, e.g., chloride or hydroxide, have been widely studied, since the labile cis site enables studies of the equatorial (cis) ligand effect [24–32]. A combination of experimental [20] and computational [33] studies showed that [FeIV=O(Cl)(TPA)]+ is able to chlorinate cyclohexane to chlorocyclohexane via oxidative ligand transfer. Obviously, this chemical system does not discard CO2 in a previous step of the catalytic cycle, so a CO2 trapping mechanism as suggested for the enzymatic halogenase would not apply here. The question, however, is whether a ligand-switch isomerization reaction as proposed for nonheme iron halogenases would be possible for biomimetic nonheme iron complexes. To investigate this, we decided to revisit the substrate halogenation by [FeIV=O(Cl)(TPA)]+ (Re A; Fig. 2) and investigate the regioselectivity of substrate hydroxylation versus chlorination in both isomers, which we describe in this study.
Methods
The studies presented in this work used density functional theory methods as implemented in the Gaussian 03 program package [34]. Following previous experience in the field [35–37], we use the unrestricted hybrid density functional method UB3LYP for all our calculations [38, 39] in combination with a double-ζ-quality LACVP basis set for iron and a 6-311G basis set for the rest of the atoms, basis set BS1 [40]. We performed a full geometry optimization (without constraints) followed by an analytical frequency calculation for all structures and characterized local minima with a frequency calculation. Subsequently, we did single-point calculations using a triple-ζ-type LACV3P+ basis set for iron and a 6-311+G* basis set for the rest of the atoms, basis set BS2. All energies reported in this work were obtained with the latter basis set and are corrected for zero-point energies from the frequency calculations. Free energies were calculated at 1-atm pressure and a temperature of 298 K and contain entropic and thermal corrections. To test the effect of the density functional method on the energetics calculated, we performed two sets of single-point calculations, namely, using (1) the B3LYP* method, whereby the Hartree–Fock exchange was reduced to 15 %, and (2) the M06 method. The M06 results gave the same spin state ordering and the relative energies were very similar to those calculated with B3LYP and hence we will focus on the B3LYP results only. The M06 details are given in the electronic supplementary material.
Transition state searches were started by running extensive geometry scans between two local minima, whereby one degree of freedom for the reaction coordinate was fixed and all other degrees of freedom were fully optimized. This was followed by a full transition state geometry optimization starting from the maxima of these scans. Transition states were characterized by a single imaginary frequency for the correct mode.
Solvent calculations were done using the conductor polarized continuum model with dielectric constant ε of 35.688 in combination with the united atom topological model. All calculations were performed for the lowest-lying singlet, triplet, quintet, and septet spin state surfaces; details of all spin states are given in the electronic supplementary material, and we focus on the low-lying spin states only here.
The methods reported in this work have been carefully benchmarked and calibrated against experimental data and, for instance, it was shown that the experimental free energies of activation are reproduced within 3 kcal mol−1 [41–43]. Similarly, good agreement against experimental spectroscopic constants was obtained, including vibrational frequencies [44]. We are, therefore, confident that the results predict the correct trends.
Results
Recent computational studies of Borowski et al. [19] on the halogenation reaction by the nonheme iron enzyme SyrB2 highlighted an important initial isomerization reaction of the iron(IV)–oxo oxidant, whereby the oxo that was initially located trans to the axial ligand and moved to a position cis to the axial ligand, whereas the chloride ligand moved to the trans position, as shown in Scheme 1. This was found to be an important and essential isomerization in the enzyme as it affected the regioselectivity of substrate activation from hydroxylation to halogenation. The question is whether this type of isomerization is also possible in biomimetic nonheme iron(IV)–oxo species and whether the regioselectivity is influenced in a similar way. To find answers to these questions, we investigated the reaction mechanism that is displayed in Scheme 2 using ethylbenzene as the substrate (SubH). The reactant structures are the [FeIV(O)(TPA)Cl]+ isomers Re A and Re B, which have either the oxo or the chloride group trans to the amine group of TPA. The interconversion barrier between these two isomeric structures is TS AB. Both reactants attack the substrate and abstract a hydrogen atom via barrier TS H to form an iron(III)–hydroxo intermediate (Int A and Int B) with the hydroxo group trans or cis to the amine group of TPA, respectively. From each of the radical intermediates there are three subsequent pathways in the reaction: (1) isomerization, (2) hydroxyl rebinding, and (3) chloride rebinding. The isomerization barrier between the two radical intermediates is defined as TS′ AB. Finally, the radical intermediates either rebind the OH group to form alcohol products (P OH) via transition state TS OH or rebind the chloride ligand via transition state TS Cl to form halogenated products (P Cl). Because the nonheme iron(IV)–oxo complexes often react via multistate reactivity patterns [45, 46], we calculated each intermediate and transition state in the lowest singlet, triplet, quintet, and septet spin states. Before we discuss the reactivity patterns and possible regioselectivities, however, we will start with an in-depth analysis of the isomeric reactants.
Isomerization of iron(IV)–oxo complexes
We started our studies from [FeIV=O(Cl)(TPA)]+ with the oxo group trans to the amine group and the three pyridine nitrogen atoms in the cis position: Re A. We calculated Re A in the singlet, triplet, quintet, and septet spin states, and the free energy (with solvent correction included) gives a triplet spin ground state, with the quintet, septet, and singlet spin states higher in energy by 6.1, 13.4, and 30.3 kcal mol−1, respectively. The spin state ordering and relative energies are analogous to those found in the many computational and experimental studies of nonheme iron biomimetic complexes [45–52] reported before. Interestingly, our gas-phase calculations give degenerate triplet and quintet spin reactants, which are well separated from the singlet and septet spin states, although a significant triplet–quintet energy gap is found via free energies in solvent. This is probably an effect of an overall charge of +1 in the calculations that causes differences in polarization in the triplet and quintet spin states. The calculations are in good agreement with experiment: Mössbauer and extended X-ray absorption fine structure spectroscopy experiments [25] identified [FeIV(O)(TPA)Cl]+ as a triplet spin ground state with Fe–O and Fe–Cl distances of 1.65 and 2.29 Å, respectively. Electronically, the triplet spin state reactants have orbital occupation \( {\pi^{*}_{xy}}^{2}{\pi^{*}_{xz}}^{1}{\pi^{*}_{yz}}^{1}\), whereas the quintet spin state has \({\pi^{*}_{xy}}^{1}{\pi^{*}_{xz}}^{1}{\pi^{*}_{yz}}^{1}{\sigma^{*}_{z^{2}}}^{1}\) occupation. The optimized geometries of 3,5 Re A are typical for the nonheme iron(IV)–oxo complexes calculated before [45–50] with Fe–O distances around 1.65 Å. The Fe–Cl distances are long owing to the interaction of second-row elements, and the bond lengths compare well with those from previous calculations on FeCl and FeS complexes [53–56].
Next, we investigated the isomeric iron(IV)–oxo species Re B, which has the chloride trans to the amine group of TPA and the oxo in the cis position. The spin state ordering for Re B is the same as that found for Re A, but now the quintet, septet, and singlet spin states are 4.0, 12.8, and 30.3 kcal mol−1 above the triplet spin ground state at the free energy level with solvent included; hence, the spin state ordering and relative energies of Re A and Re B are the same. Note that 3 Re A is slightly more stable than 3 Re B by about 1 kcal mol−1. Since, the septet and singlet spin states are much higher in energy than the triplet and quintet spin states, we only show the latter two spin surfaces in Fig. 3; details of the other structures can be found in the electronic supplementary material. The geometries of 3,5 Re A and 3,5 Re B are very much alike and only minor differences in bond lengths are found.
Potential energy profile with energies in kilocalories per mole relative to isolated reactants in the triplet spin state for the isomerization of 3,5 Re A to 3,5 Re B. Values not in parentheses are gas-phase UB3LYP/BS2 energies with zero-point-energy (ZPE) corrections, whereas free energies in solvent are given in parentheses. Also shown are optimized geometries of critical points with bond lengths in angstroms and the imaginary frequency in the transition states in reciprocal centimeters. Values in brackets refer to B3LYP* single-point energies
Subsequently, we studied the isomerization reaction, whereby the oxo and chloride ligands swap places via transition state TS AB to form the reactant isomer Re B. Initially, a series of geometry scans starting either from Re A or from Re B were performed on the singlet, triplet, quintet, and septet spin states. These geometry scans were the result of a full geometry optimization with fixed O–Fe–Naxial angle, which was stepwise reduced from 180° to 90° in steps of 5° from Re A and the reverse for the scans starting from Re B. A detailed analysis of the individual optimized geometries confirmed that a minimum energy pathway from Re A to Re B and vice versa was found. Subsequently, the maximum points of the individual scans were subjected to a full transition state search and led to the TS AB geometries displayed in Fig. 3.
The optimized geometries of 3,5 TS AB show similar features of Fe–O and Fe–Cl bond lengths as compared with Re A and Re B, although structurally the transition states have undergone some dramatic changes. During the isomerization, the Fe–Cl and Fe–O distances remain the same, and only the Cl–Fe–Naxial and O–Fe–Naxial angles change. Thus, Re A and Re B have octahedral symmetry of the metal, where all bound atoms surrounding the metal are equally spaced and form angles close to 90°. Re A and Re B have a triplet spin ground state and spin state ordering with the same orbital occupation, which result in similarities in geometric features of the two complexes. In TS AB, by contrast, the complex is square-pyramidal with an axial ligand trans to the oxo group and three equally separated nitrogen ligands in the equatorial positions that form angles of 120°. Both chloro and oxo ligands have left the equatorial plane and are located above the metal at roughly equal angles. The analysis of the imaginary frequency in the transition states shows that the conversion of Re A into Re B via TS AB first moves the chloride out of the equatorial position, which results in reorganization of the TPA ligand into a square-pyramidal structure and a push of the iron out of the plane through the three nitrogen atoms. The imaginary frequency for this motion is very soft (53.8i cm−1 for 3 TS AB and 56.7i cm−1 for 5 TS AB) and corresponds to a flat and broad potential energy surface. The potential energy surface displayed in Fig. 3 shows that the isomerization barriers are large especially for the triplet spin state. The lowest isomerization barrier is for the quintet spin state, which is 13.1 and 13.4 kcal mol−1 in the gas phase as calculated with B3LYP and B3LYP*, respectively, whereas a free energy of activation ΔG ‡ of 36.9 kcal mol−1 is calculated with solvent effects included.
Experimental near-infrared, NMR, Mössbauer, and extended X-ray absorption fine structure studies on [FeIV(O)(TPA)Cl]+ [25] found no evidence of isomerization reactions, whereby the oxo and chloro ligands were interchanged. This further supports high energy barriers and implies that very little—if any—isomerization will occur.
Halogenation and hydroxylation
Subsequently, we investigated the mechanisms leading to substrate hydroxylation and chlorination as displayed on the left-hand side and right-hand side, respectively, of Scheme 2. Ethylbenzene was used as a model substrate since it has been shown that it has a moderate C–H bond strength [57–59]. Figure 4 gives the hydroxylation and chlorination mechanism of ethylbenzene starting from 3,5,7 Re A, whereas the mechanism starting from 3,5,7 Re B are given in Fig. 5. Both mechanisms occur stepwise and start with an initial hydrogen atom abstraction via transition state TS H that leads to an iron(III)–hydroxo complex and a nearby radical rest group (Int). Thereafter, the reactions split into two possible reaction paths: (1) rebinding of the hydroxyl group via transition state TS OH to form hydroxylated products (P OH) or (2) chloride transfer via transition state TS Cl to form chlorinated products (P Cl).
Potential energy profile with energies in kilocalories per mole relative to isolated reactants in the triplet spin state for ethylbenzene hydroxylation and chlorination by 3,5,7 Re A. Values not in parentheses are gas-phase UB3LYP/BS2 energies with ZPE corrections, whereas free energies in solvent are given in parentheses. Also shown are optimized geometries of critical points with bond lengths in angstroms and the imaginary frequency in the transition states in reciprocal centimeters. Values in brackets refer to B3LYP* results
Potential energy profile with energies in kilocalories per mole relative to isolated reactants in the triplet spin state for ethylbenzene hydroxylation and chlorination by 3,5,7 Re B. Values not in parentheses are gas-phase UB3LYP/BS2 energies with ZPE corrections, whereas free energies in solvent are given in parentheses. Also shown are optimized geometries of critical points with bond lengths in angstroms and the imaginary frequency in the transition states in reciprocal centimeters. Values in brackets refer to B3LYP* results
Both mechanisms start with a rate-determining hydrogen atom abstraction, and although the ground state of Re A and Re B is a triplet spin state, a spin state crossing is needed to a higher spin state, since 3 TS H is very high in energy: 38.8 kcal mol−1 for 3 TS H,A and 41.2 kcal mol−1 for 3 TS H,B in the gas phase. At the free energy level in solvent, the free energy of activation in the triplet spin state is well above 70 kcal mol−1, and, therefore, too high for the reaction to proceed on this surface. Much lower barriers are found for the septet and quintet spin states, which are 2.1 and 10.7 kcal mol−1 for the septet spin state and 16.3 and 6.8 kcal mol−1 for the quintet spin state for B3LYP and B3LYP*, respectively, in the gas phase. Although the septet–quintet spin state ordering changed upon replacing the B3LYP method with B3LYP*, actually at both levels of theory the lowest TS H barrier is well below that calculated for the isomerization barrier, and therefore, the conclusions remain the same. Consequently, hydrogen abstraction is an energetically lower energy process than oxidant isomerization. Both processes lead to almost degenerate septet and quintet intermediates for both pathway A and pathway B, owing to similarities in orbital occupation. Thus, 5,7 Int A and 5,7 Int B have orbital occupation \({\pi^{*}_{xy}}^{1}{\pi^{*}_{xz}}^{1}{\pi^{*}_{yz}}^{1}{\sigma^{*}_{z^{2}}}^{1}{\sigma^{*}_{{x}^{2}-{y}^{2}}}^{1}\pi_{\rm Sub}^{1}\), whereby the radical on the substrate (in the π Sub orbital) is either ferromagnetically coupled to the five 3d electrons in the septet spin state or antiferromagnetically coupled to these electrons in the quintet spin state.
The gas-phase barriers for hydrogen abstraction from ethylbenzene by Re A (Re B) of 16.3 kcal mol−1 for 5 TS H,A and 11.6 kcal mol−1 for 5 TS H,B are higher in energy than those calculated using [FeIV(O)(Por)X], where X is SH−, Cl−, or NCCH3, as oxidants using the same methods, which gave hydrogen abstraction barriers of 11.5, 9.1, and 9.9 kcal mol−1, respectively [58, 60]. The optimized geometries of the transition states identify them as early barriers with longer O–H distances than C–H distances, and hence they are closer to the geometries of the reactants than the geometries of the intermediates. In contrast to previous hydrogen abstraction studies [57–60], the imaginary frequencies are modest in size and range from 116.0i cm−1 for 5 TS H,A to 462.3i cm−1 for 3 TS H,A. Consequently, the hydrogen abstraction barrier is rather broad and the reaction is expected to proceed with a modest kinetic isotope effect. In previous work, we calculated hydrogen abstraction barriers with imaginary frequencies well over 1,500i cm−1 [54, 61–63], and found kinetic isotope effects for the replacement of the hydrogen atoms of the substrate with deuterium atoms of well over 10.
In pathway A, we calculated a rebinding barrier of 7.7 kcal mol−1 on the quintet spin state surface, whereas a chloride transfer barrier of 2.9 kcal mol−1 is found. Thus, the relative barrier heights imply there is a regioselective chlorination reaction on a dominant quintet spin state surface. The optimized geometries of 3,5 TS Cl,A give long Cl–C bonds, whereas the Fe–Cl distance is slightly elongated with respect to the value found for the connecting intermediates. The imaginary frequencies for these transition states are small and compare well with values typically found for, e.g., sulfoxidation and phosphorylation barriers [64, 65]. For the hydroxyl rebinding barriers, the Fe–O distance is significantly elongated to well over 1.9 Å, and at the same time, the substrate approaches the oxygen atom closely (to within 2.4 Å). Imaginary frequencies of 135.4i and 143.8i cm−1 are typical values for rebinding barriers [66, 67] and indicate a flat and broad potential energy surface for this reaction. In mechanism B, the order of the rebinding transition states has changed and 5 TS OH,B is much lower in energy than 5 TS Cl,B. Thus, detailed geometry scans and subsequent transition state searches gave a rebinding barrier of less than 2 kcal mol−1 for the hydroxylation mechanism, which we identify with more than −20 kcal mol−1 in Fig. 5. By contrast, a rebinding barrier of 7.0 kcal mol−1 is found for the process passing through 5 TS Cl,B. Consequently, mechanism B will proceed with dominant substrate hydroxylation rather than halogenation, which is the opposite of what is found for mechanism A. For completeness, we also investigated rebinding on the septet spin state surface but found it considerably higher in energy, probably owing to the lack of available low-lying vacant orbitals to accommodate an extra electron.
The optimized geometries of 3,5 TS Cl,B are similar to those reported for 3,5 TS Cl,A above. The substrate is somewhat closer to the chloride anion in 5 TS Cl,B at a distance of 2.787 Å, whereas the Fe–Cl and Fe–OH bond lengths have elongated to 2.592 and 1.954 Å, respectively. These structures, therefore, appear somewhat later on the potential energy surface as compared with 5 TS Cl,A. In addition, the potential energy surface around the transition state appears to be broad and flat as follows from the low value of the imaginary frequencies: 29.8i cm−1 for 5 TS Cl,B. A similar situation has occurred for the hydroxyl rebinding barrier, which also gives a soft vibrational mode of 77.0i cm−1 for the C–O bond formation. Furthermore, there is also a substantially shorted C–O distance of 1.895 Å as compared with 5 TS OH,A, which corresponds to a somewhat later barrier. At the same time, the Fe–O bond in 5 TS OH,B is significantly weakened and has grown to 2.169 Å.
Isomerization of iron(III)–hydroxo complexes
To find out whether the radical intermediates can interconvert into each other via rotational barrier TS′ AB, we performed a series of calculations on the septet and quintet spin state surfaces. We started these calculations from the optimized geometries of the radical intermediates 5,7 Int A and 5,7 Int B and did a geometry scan leading to the isomeric structure. The O–Fe–Naxial angle was varied in steps of 5° from 90° with the hydroxyl group in the cis position to 180°, where the hydroxyl group is in the trans position with respect to the amine nitrogen atom of TPA. During these geometry scans, the energy was minimized at each step and led to the potential energy surface displayed in Fig. 6. As can be seen, the structures Int A and Int B can isomerize into each other, but barriers of 14.0 and 15.8 kcal mol−1 are encountered for the septet spin state and the quintet spin state, respectively. These isomerization barriers are considerably higher in energy than the lowest hydrogen atom abstraction barriers and are also much higher in energy than the hydroxyl rebinding or chloride transfer barriers (TS OH, TS Cl) described in the previous section for mechanisms A and B. Consequently, isomerization of the radical intermediates is unlikely to occur during the lifetime of these intermediates. Therefore, the product distributions of the alcohol versus halogenated products will be dependent on the relative barrier height of TS Cl versus TS OH and the amount of isomerization that has occurred in the reactant complexes.
Discussion
We have presented results regarding the regioselectivity of ethylbenzene hydroxylation versus halogenation by a nonheme iron(IV)–oxo oxidant with a TPA ligand system and tested many possible reaction paths and mechanisms. Scheme 3 summarizes the proposed reaction mechanism as revealed by the calculations. The reactants with either the oxo or the chloride group trans to the amide group of TPA are in a triplet spin ground state, but this state is unreactive toward hydrogen atom abstraction and isomerization. There are, however, close-lying quintet spin states, where the isomerization is accessible with a gas phase barrier of 13.1 kcal mol−1. This barrier is much higher in energy than the hydrogen abstraction barriers from Re A and Re B. Although it cannot be ruled out that some degree of isomerization will occur prior to the reaction, the calculations predict fast hydrogen atom abstraction from substrates as the dominant pathway. The hydrogen abstraction barriers lead to radical intermediates, where the isomerization barriers are high in energy, and consequently 5 TS′ AB will not contribute to the reaction mechanism. As summarized in Table 1, the rebinding process along pathway A has significantly lower barriers for chloride transfer than for hydroxyl transfer and hence will lead to chlorination of the product. On the other hand, pathway B gives dominant hydroxylation instead because of significantly lower hydroxylation rebinding barriers than chloride transfer barriers. It appears, therefore, that the ligand in the cis position is transferred to the substrate radical in the rebinding process.
Reaction mechanism of chlorination versus hydroxylation of ethylbenzene (SubH) by [FeIV(O)(TPA)Cl]+ isomers Re A and Re B. The energies reported were obtained in the gas phase at the UB3LYP/B2 level with zero-point-energy corrections and were calculated relative to isolated reactants in the triplet spin state
Experimental studies [18] of cyclohexane activation by [FeIV(O)(TPA)Cl]+ gave only halogenated products and no alcohols. Indeed our calculations imply there is little isomerization between Re A and Re B owing to high isomerization barriers and consequently a dominant pathway via Re A leading to halogenated products. To understand this process, we performed a detailed thermochemical analysis of the reactions involved.
The first step of the reaction mechanism involves hydrogen atom transfer from the substrate to the oxidant, which can be described by the difference in energy of the bond dissociation energy of the C–H bond of the substrate (BDECH, Eq. 1) and the bond dissociation energy of the O–H bond of the iron(III)–hydroxo structure (BDEOH, Eq. 2) [68–72]. The difference in energy between BDECH and BDEOH reflects the energy of hydrogen atom abstraction from ethylbenzene by [FeIV(O)(TPA)Cl]+, ΔH HA (Eqs. 3, 4).
We calculated a BDECH value of 81.6 kcal mol−1 in the gas phase and 82.8 kcal mol−1 in solvent for removal of a hydrogen atom from the benzyl position of ethylbenzene, which is in good agreement with previous calculations [48]. The BDEOH values for Re A are 98.4 kcal mol−1 in the gas phase and 95.6 kcal mol−1 in solvent, whereas those for Re B are 100.1 kcal mol−1 in the gas phase and 98.4 kcal mol−1 in solvent, and hence lead to ΔH HA,A values of −16.8 kcal mol−1 in the gas phase and −12.8 kcal mol−1 in solvent for mechanism A and ΔH HA,B = −18.5 kcal mol−1 in the gas phase and ΔH HA,B = −15.6 kcal mol−1 in solvent for mechanism B. The values for ΔH HA are in good agreement with the energy differences between reactants and Int A and Int B and show that both isomers are equally potent oxidants.
To understand the origins of the regioselectivity of the reaction, we performed a thorough thermodynamic analysis of the rebinding mechanisms, and the results are shown in Scheme 4. Firstly, we calculated the adiabatic strength of the Fe–Cl bond in Int A and Int B by calculating the energy difference between [FeIII(OH)(TPA)Cl]+ and the optimized geometries of [FeII(OH)(TPA)]+ and a chloride ion. We found virtually identical adiabatic Fe–Cl bond strengths (ΔH Fe–Cl) for isomers Int A and Int B, although it should be mentioned that geometry optimization with the chloride ion removed from either Int A or Int B converges to the same structure with trigonal bipyramidal symmetry and the hydroxo group trans to the amine nitrogen of TPA. Consequently, the adiabatic bond dissociation energies are similar for the chloride in the cis and trans positions; however, in the pentacoordinated structure in both cases the optimized geometry converged to a geometry with the hydroxo group trans to the amide of TPA, which should incur a considerable geometric and energetic change for the intermediate with chloride in the cis position. We, therefore, decided to calculate the diabatic bond dissociation energy of the Fe–Cl bond in Int A and Int B. This was done by taking the Int A and Int B optimized geometries and running a single-point calculation with chloride or hydroxide removed. These values in combination with the energies of the optimized geometries of [FeIII(OH)(Cl)(TPA)]+ and either Cl/OH give the diabatic bond dissociation energies. As follows from the diabatic bond dissociation energies in Scheme 4, the weakest bond in Int A is the Fe–Cl bond by almost 10 kcal mol−1 over the Fe–OH bond. These values contrast with the situation for Int B, where the Fe–OH bond is the weaker of the two by more than 12 kcal mol−1. The diabatic bond dissociation energies, therefore, imply that in Int A the chloride will be transferred to the substrate, whereas in Int B it is more likely that the hydroxyl group rebinds. Indeed the potential energy profiles shown in Figs. 4 and 5 give chlorination via pathway A and hydroxylation via pathway B in support of the diabatic bond dissociation energies. These studies also highlight the fact that the cis ligand is always the source of the rebinding process.
Finally, we calculated the energy to form 1-chloro-1-phenylethane and 1-phenylethanol products from the 1-phenylethyl radical (bottom of Scheme 4). The reaction to form alcohol products is 10.5 kcal mol−1 and 6.1 kcal mol−1, respectively, more exothermic than the reaction leading to chlorination products, which means that the driving force for the reaction to give alcohols is larger. On the basis of the Polanyi principle, therefore, one would expect dominant hydroxylation products, but in these halogenation oxidants the driving force is not the dominant factor that determines the reaction products. In halogenases, stereochemical interactions between the oxidant and the substrate determine the relative barrier heights of halogenation versus hydroxylation and consequently the product distributions.
Conclusions
We have described the regioselectivity of substrate hydroxylation versus chlorination using a nonheme iron(IV)–oxo oxidant. We have shown that the regioselectivity is dependent on the stereochemistry of the oxidant, whereby the system with the oxo group trans to the amine group of TPA reacts via hydroxylation and the system with the oxo group in the cis position reacts via chlorination. The reaction starts with a rate-determining hydrogen abstraction barrier, which is much lower in energy than the competitive isomerization barrier between the two isomeric oxidants. This implies that little equilibration between the two oxidants will occur prior to the hydrogen atom abstraction. A detailed thermochemical analysis of bond strengths shows a change in Fe–Cl and Fe–OH bond strengths between the two isomers, whereby the weaker ligand is placed in the cis position. Because of this, the removal of the cis ligand in the rebinding phase of the reaction mechanism is more likely to happen. The work also highlights the differences between enzymatic halogenases, on the one hand, and biomimetic models, on the other hand. Thus, a suggested mechanism for enzymatic halogenation includes an essential isomerization that regulates the differences between hydroxylases and halogenases. By contrast, in biomimetic model complexes, such as the one described in this work, the isomerization is not a prerequisite for the reaction as the weakest bond is the iron–cis ligand bond and hence the cis ligand will be transferred in the rebinding phase. The work further emphasizes that the catalytic mechanism proposed for nonheme iron halogenases is far from being resolved and further testing on the enzymatic system may be required.
References
Gribble GW (1998) Acc Chem Res 31:141–152
Butler A, Sandy M (2009) Nature 460:848–854
Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT (2006) Chem Rev 106:3364–3378
Grgurina I, Barca A, Cervigni S, Gallo M, Scaloni A, Pucci P (1994) Experientia 50:130–133
Harris CM, Kannan R, Kopecka H, Harris TM (1985) J Am Chem Soc 107:6652–6658
Neumann CS, Galonić Fujimori D, Walsh CT (2008) Chem Biol 15:99–109
Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee S-K, Lehnert N, Neese F, Skulan AJ, Yang Y-S, Zhou J (2000) Chem Rev 100:235–349
Costas M, Mehn MP, Jensen MP, Que L Jr (2004) Chem Rev 104:939–986
Blasiak LC, Drennan CL (2009) Acc Chem Res 42:147–155
de Visser SP, Kumar D (eds) (2011) Iron-containing enzymes: versatile catalysts of hydroxylation reactions in nature. RSC Publishing, Cambridge
Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL (2006) Nature 440:368–371
Wong C, Galonić Fujimori D, Walsh CT, Drennan CL (2009) J Am Chem Soc 131:4872–4879
Khare D, Wang B, Gu L, Razelun J, Sherman DH, Gerwick WH, Håkansson K, Smith JL (2010) Proc Natl Acad Sci USA 32:14099–14104
Galonić Fujimori D, Barr EW, Walsh CT, Bollinger JM Jr, Krebs C (2007) Nat Chem Biol 3:113–116
Matthews ML, Krest CM, Barr EW, Vaillancourt FH, Walsh CT, Green MT, Krebs C, Bollinger JM Jr (2009) Biochemistry 48:4331–4343
Matthews ML, Neumann CS, Miles LA, Grove TL, Booker SJ, Krebs C, Walsh CT, Bollinger JM Jr (2009) Proc Natl Acad Sci USA 106:17723–17728
de Visser SP, Latifi R (2009) J Phys Chem B 113:12–14
Pandian S, Vincent MA, Hillier IA, Burton NA (2009) Dalton Trans 6201–6207
Borowski T, Noack H, Radoń M, Zych K, Siegbahn PEM (2010) J Am Chem Soc 132:12887–12898
Kojima T, Leising RA, Yan S, Que L Jr (1993) J Am Chem Soc 115:11328–11335
Podgoršek A, Zupan M, Iskra J (2009) Angew Chem Int Ed 48:8424–8450
Comba P, Wunderlich S (2010) Chem Eur J 16:7293–7299
Liu W, Groves JT (2010) J Am Chem Soc 132:12847–12849
Rohde J-U, In J-H, Lim MH, Brennessel WW, Bukowski MR, Stubna A, Münck E, Nam W, Que L Jr (2003) Science 299:1037–1039
Rohde J-U, Stubna A, Bominaar EL, Münck E, Nam W, Que L Jr (2006) Inorg Chem 45:6435–6445
Sastri CV, Lee J, Oh K, Lee YJ, Lee J, Jackson TA, Ray K, Hirao H, Shin W, Halfen JA, Kim J, Que L Jr, Shaik S, Nam W (2007) Proc Natl Acad Sci USA 104:19181–19186
Company A, Feng Y, Güell M, Ribas X, Luis JM, Que L Jr, Costas M (2009) Chem Eur J 15:3359–3362
Makhlynets OV, Das P, Taktak S, Flook M, Mas-Ballesté R, Rybak-Akimova EV, Que L Jr (2009) Chem Eur J 15:13171–13180
Lyakin OY, Bryliakov KP, Britovsek GJP, Talsi EP (2009) J Am Chem Soc 131:10798–10799
Paine TK, Paria S, Que L Jr (2010) Chem Commun 46:1830–1832
Das P, Que L Jr (2010) Inorg Chem 49:9479–9485
Lyakin OY, Bryliakov KP, Talsi EP (2011) Inorg Chem 50:5526–5538
Noack H, Siegbahn PEM (2007) J Biol Inorg Chem 12:1151–1162
Frisch MJ et al (2004) Gaussian 03, revision C.02. Gaussian, Wallingford
Aluri S, de Visser SP (2007) J Am Chem Soc 129:14846–14847
Kumar D, Karamzadeh B, Sastry GN, de Visser SP (2010) J Am Chem Soc 132:7656–7667
Kumar D, Thiel W, de Visser SP (2011) J Am Chem Soc 133:3869–3882
Becke AD (1993) J Chem Phys 98:5648–5652
Lee C, Yang W, Parr RG (1988) Phys Rev B 37:785–789
Hay PJ, Wadt WR (1985) J Chem Phys 82:270–283
Kumar D, de Visser SP, Shaik S (2005) Chem Eur J 11:2825–2835
de Visser SP, Oh K, Han A-R, Nam W (2007) Inorg Chem 46:4632–4641
Vardhaman AK, Sastri CV, Kumar D, de Visser SP (2011) Chem Commun 47:11044–11047
Karamzadeh B, Kumar D, Sastry GN, de Visser SP (2010) J Phys Chem A 114:13234–13243
Hirao H, Kumar D, Que L Jr, Shaik S (2006) J Am Chem Soc 128:8590–8606
de Visser SP (2006) J Am Chem Soc 128:9813–9824
de Visser SP (2006) J Am Chem Soc 128:15809–15818
Latifi R, Bagherzadeh M, de Visser SP (2009) Chem Eur J 15:6651–6662
Seo MS, Kim NH, Cho K-B, So JE, Park SK, Clémancey M, Garcia-Serres R, Latour J-M, Shaik S, Nam W (2011) Chem Sci 2:1039–1045
Ye S, Neese F (2011) Proc Natl Acad Sci USA 108:1228–1233
de Visser SP, Nam W (2008) J Phys Chem A 112:12887–12895
Cheng L, Wang J, Wang M, Wu Z (2010) Phys Chem Chem Phys 12:4092–4103
de Visser SP, Shaik S, Sharma PK, Kumar D, Thiel W (2003) J Am Chem Soc 125:15779–15788
de Visser SP (2006) Chem Eur J 12:8168–8177
Green MT (1999) J Am Chem Soc 121:7939–7940
de Visser SP, Latifi R, Tahsini L, Nam W (2011) Chem Asian J 6:493–504
de Visser SP, Kumar D, Cohen S, Shacham R, Shaik S (2004) J Am Chem Soc 126:8362–8363
Shaik S, Kumar D, de Visser SP (2008) J Am Chem Soc 130:10128–10140
de Visser SP (2010) J Am Chem Soc 132:1087–1097
de Visser SP, Tahsini L, Nam W (2009) Chem Eur J 15:5577–5587
Kumar D, de Visser SP, Shaik S (2003) J Am Chem Soc 125:13024–13025
Kumar D, de Visser SP, Sharma PK, Cohen S, Shaik S (2004) J Am Chem Soc 126:1907–1920
Kumar D, de Visser SP, Shaik S (2004) J Am Chem Soc 126:5072–5073
Kumar D, Sastry GN, de Visser SP (2011) Chem Eur J 17:6196–6205
Latifi R, Tahsini L, Kumar D, Sastry GN, Nam W, de Visser SP (2011) Chem Commun 47:10674–10676
de Visser SP, Ogliaro F, Shaik S (2001) Angew Chem Int Ed 40:2871–2874
Shaik S, Cohen S, de Visser SP, Sharma PK, Kumar D, Kozuch S, Ogliaro F, Danovich D (2004) Eur J Inorg Chem: 207–226
Bordwell FG, Cheng J-P (1991) J Am Chem Soc 113:1736–1743
Mayer JM (1998) Acc Chem Res 31:441–450
Prokop KA, de Visser SP, Goldberg DP (2010) Angew Chem Int Ed 49:5091–5095
Prokop KA, Neu HM, de Visser SP, Goldberg DP (2011) J Am Chem Soc 133:15874–15877
de Visser SP (2012) Adv Inorg Chem 64:1–31
Acknowledgments
M.G.Q. thanks the BBSRC for a studentship. The National Service for Computational Chemistry Software is acknowledged for generous CPU time.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Detailed energies, group spin densities, and charges as well as structural information for all local minima and transition states described in this work are available.
Rights and permissions
About this article
Cite this article
Quesne, M.G., de Visser, S.P. Regioselectivity of substrate hydroxylation versus halogenation by a nonheme iron(IV)–oxo complex: possibility of rearrangement pathways. J Biol Inorg Chem 17, 841–852 (2012). https://doi.org/10.1007/s00775-012-0901-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-012-0901-4