Abstract
When antitumor platinum drugs react with DNA they form various types of intrastrand and interstrand cross-links (CLs). One class of new antitumor platinum compounds comprises bifunctional PtII compounds based on the dinuclear or trinuclear geometry of leaving ligands. It has been shown that the DNA-binding modes of dinuclear or trinuclear bifunctional PtII agents are distinct from those of mononuclear cisplatin, forming markedly more intramolecular interstrand CLs. However, at least two types of DNA interstrand cross-linking by bifunctional PtII complexes can be envisaged, depending on whether the platinum complex coordinates to the bases in one DNA molecule (intramolecular interstrand CLs) or in two different DNA duplexes (interduplex CLs). We hypothesized that at least some antitumor bifunctional poly(di/tri)nuclear complexes could fulfill the requirements placed on interduplex DNA cross-linkers. To test this hypothesis we studied the interduplex cross-linking capability of a representative of antitumor polynuclear agents, namely, dinuclear PtII complex [{trans-PtCl(NH3)2}2-μ-{trans-(H2N(CH2)6NH2(CH2)2NH2(CH2)6NH2)}]4+ (BBR3535). The investigations were conducted under molecular crowding conditions mimicking environmental conditions in the cellular nucleus, namely, in medium containing ethanol, which is a commonly used crowding agent. We found with the aid of native agarose gel electrophoresis that the DNA interduplex cross-linking efficiency of BBR3535 under molecular crowding conditions was remarkable: the frequency of these CLs was 54%. In contrast, the interduplex cross-linking efficiency of mononuclear cisplatin or transplatin was markedly lower (approximately 40-fold or 18-fold, respectively). We suggest that the production of interduplex CLs in addition to other DNA intramolecular adducts may provide polynuclear PtII compounds with a wider spectrum of cytotoxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
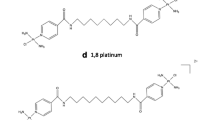
Binding to DNA and the consequences for DNA conformation, recognition, and downstream cellular effects is the mechanistic paradigm by which cytotoxic platinum complexes are believed to exert their antitumor activity [1, 2]. Consequently, altered DNA reactivity has been used as a strategy to search for new platinum antitumor compounds with improved chemotherapeutic properties in comparison with existing platinum drugs [3, 4]. One class of new antitumor active platinum compounds which emerged from this strategy comprises bifunctional PtII compounds based on the dinuclear or trinuclear geometry of leaving (reactive) ligands [5]. In these compounds, the two leaving groups are contained in the coordination spheres of the terminal platinum atoms separated by various linkers of different length. The extended distance between platinating sites is one of the factors which may account for the altered antitumor activity of dinuclear or trinuclear bifunctional PtII complexes. As a consequence of this property, the DNA-binding modes of dinuclear or trinuclear bifunctional PtII agents are distinct from those of mononuclear cisplatin (Fig. 1) and its clinically used analogues. Whereas cisplatin and its bifunctional analogues produce predominantly cross-links (CLs) between bases in the neighboring base pairs [6], dinuclear and trinuclear PtII drugs produce long-range CLs in which the sites of platination are separated by several intervening base pairs [7–9]. The properties of DNA globally modified by polynuclear complexes are also highlighted by markedly enhanced intraduplex interstrand cross-linking. For instance, dinuclear PtII complexes [{trans-PtCl(NH3)2}2-μ-{trans-(H2N(CH2)6NH2(CH2)2NH2(CH2)6NH2)}]4+ (BBR3535; Fig. 1) and [{cis-PtCl(NH3)2}2(H2N(CH2)6NH2]2+ form 57% [10] and even 90% [11], respectively, of these lesions, whereas cisplatin forms only 6% of intraduplex interstrand CLs [12].
In general, DNA interstrand cross-linking requires close proximity of binding sites in the two cross-linked DNA strands. This requirement may be easily fulfilled in the case of formation of the intraduplex DNA interstrand CLs by bifunctional PtII complexes because binding via one leaving group inevitably leaves the other close to other binding sites in the same duplex. However, if the reactive sites of the bifunctional cross-linking agents are connected by a longer linker so that the leaving groups in these agents are sufficiently distant and the linker forces those sites to point in appropriate directions, or in other words if the stereochemistry of the reactive sites of the cross-linking agent proves appropriate, such agents could also bind to adjacent duplexes [13–15]. Thus, such bifunctional PtII compounds might also be effective interduplex cross-linkers in cases when two fragments of double-helical DNA molecules are forced to lie together, for instance, during recombination, at replication forks or sites of topoisomerase action [14, 16], or more generally in cellular environmental conditions [17, 18].
Hence, at least two types of DNA interstrand cross-linking by bifunctional PtII complexes can be envisaged, depending on whether the platinum complex coordinates to the bases in one DNA molecule (intraduplex interstrand CLs) or in different DNA duplexes (interduplex interstrand CLs) (Fig. 2). We hypothesize that at least some antitumor bifunctional poly(di/tri)nuclear complexes fulfill the requirements placed on interduplex DNA cross-linkers considerably better than mononuclear cisplatin or its trans isomer (transplatin; Fig. 1) [19]. For instance, platinating sites in dinuclear BBR3535 are markedly more distant (2.7 nm [10]) than those in mononuclear cisplatin (0.28 nm [20]). Moreover, the trans geometry of leaving ligands in BBR3535 may allow the spermine-like linker to direct the reactive sites in opposite directions, which may facilitate binding to adjacent duplexes, a feature not apparently accessible by cisplatin.
Two possible types of DNA interstrand cross-linking by bifunctional PtII complexes. In intramolecular interstrand cross-links (CLs), the platinum complex coordinates the bases in one DNA molecule. In interduplex interstrand CLs, the platinum complex coordinates the bases in two different DNA duplexes. The intrastrand CL is shown as well
Therefore, to answer the question whether antitumor polynuclear PtII complexes are also efficient interduplex cross-linkers, we studied the interduplex cross-linking capability of a representative of antitumor polynuclear agents, namely, dinuclear PtII complex BBR3535 (Fig. 1). Since the entropic factor involved in bringing two DNA molecules together is so large, the investigations reported in this work were performed under molecular crowding conditions [17, 21–24] mimicking environmental conditions in the cellular nucleus. Determining this unique cross-linking capability of polynuclear PtII complexes represents a new important aspect of their DNA-binding mode and may contribute to the working mechanism of this distinct class of antitumor metallodrugs.
Materials and methods
Chemicals
Cisplatin and transplatin were obtained from Sigma (Prague, Czech Republic) (the purity was 99.9% or better based on elemental and inductively coupled plasma trace analysis). BBR3535 was synthesized and characterized by previously published procedures [25]. Stock solutions of the platinum compounds were prepared using Milli-Q sterilized water. These solutions were kept from light and allowed to hydrolyze/equilibrate for 24 h at room temperature with agitation. Calf thymus (CT) DNA [42% (G+C), mean molecular mass approximately 20 MDa] was prepared and characterized as described previously [26, 27]. The pUC19 [2,686 bp) and pSP73 (2,464 bp) plasmid DNA were purchased from Promega or MBI Fermentas, or were amplified in host Escherichia coli strain DH5a. Klenow fragment of DNA polymerase I and restriction endonucleases EcoRI and PvuI were purchased from New England Biolabs (Beverly, MA, USA). Agarose was from FMC BioProducts (Rockland, ME, USA). Ethidium bromide and NaCN were purchased from Boehringer (Mannheim, Germany). O’GeneRuler 1 kb DNA Ladder Plus was from Fermentas International (Burlington, Canada).
Platination reactions
If not stated otherwise, plasmid pUC19 DNA linearized by EcoRI or CT DNA was incubated with the platinum complex in 0.2 M sodium acetate, pH 5.5 and 75% ethanol at 37 °C in the dark. After 48 h, the samples were precipitated and resolved in the medium required for subsequent analysis by gel electrophoresis. An aliquot of these samples was used to determine the value of r b (the number of molecules of the platinum complex bound per nucleotide residue) by flameless atomic absorption spectrometry (FAAS).
Transcription mapping of DNA adducts
Transcription of the (NdeI/HpaI) restriction fragment of pSP73 DNA with T7 RNA polymerase and electrophoretic analysis of the transcripts were performed as previously described in detail [12]. The DNA concentration used in this assay was 3.9 × 10−5 M (related to the monomeric nucleotide content).
Interduplex DNA cross-linking
DNA interduplex cross-linking was examined using pUC19 DNA linearized by EcoRI (which cuts only once within this plasmid) by electrophoresis through native 1% agarose gels with 40 mM tris(hydroxymethyl)aminomethane/acetate, 1 mM EDTA, pH 7.4 running buffer. The gels were run at 20 °C in the dark, with voltages ranging between 30 and 60 V. The running time depended upon the voltage. The resultant gels were stained with ethidium bromide in water at a concentration of 0.3 μg mL−1. Bands were visualized by UV transillumination, photographed, and the electrophoretic band intensities were quantified with the aid of the AIDA image analyzer program (Raytest, Germany). The frequency of interduplex CLs (i.e., the number of interduplex CLs per adduct multiplied by 100) was calculated as follows: frequency of interduplex CLs = XL/5,372r b (pUC19 plasmid contained 5,372 nucleotide residues). XL is the number of interduplex CLs per molecule of the linearized DNA duplex and was calculated assuming a Poisson distribution of the interduplex CLs as XL = −ln A, where A is the fraction of molecules running as a band corresponding to the non-cross-linked double-stranded DNA.
Other physical methods
Absorption spectra were measured with a Beckman 7400 DU spectrophotometer and quartz cells with a thermoelectrically controlled cell holder and a path length of 1 cm. FAAS measurements were conducted with a Varian AA240Z Zeeman atomic absorption spectrometer equipped with a GTA 120 graphite tube atomizer. For FAAS analysis, DNA was precipitated with ethanol and dissolved in 0.1 M HCl. For FAAS analyses, DNA was precipitated and dissolved in 0.1 M HCl. Differential pulse polarography was performed with an EG&G Princeton Applied Research Corporation model 384B polarographic analyzer.
Results
DNA platination in aqueous ethanol solutions
To assess whether BBR3535 or cisplatin is able to form interduplex interstrand CLs in cellular especially nuclear environmental conditions, DNA was platinated under molecular crowding conditions in medium containing ethanol, which is a commonly used crowding agent [23, 24] similarly as, for instance, poly(ethylene glycol), different types of Ficoll, and dextrans [21, 22]. Therefore, first we examined DNA platination by BBR3535 or cisplatin in aqueous ethanol solutions. Solutions of double-helical CT DNA at a concentration of 0.032 mg mL−1 were incubated with cisplatin, transplatin, or BBR3535 at different r i (0.01–0.05) in 0.2 M sodium acetate plus 75% ethanol at 37 °C (r i is defined as the molar ratio of free platinum complex to nucleotide phosphates at the onset of incubation with DNA). At various time intervals an aliquot of the reaction mixture was withdrawn and assayed by differential pulse polarography for platinum complex not bound to DNA. The amount of platinum complex bound to DNA (r b) was calculated by subtracting the amount of free (unbound) platinum complex from the total amount of platinum complex present in the reaction. The amount of the free cisplatin, transplatin, or BBR3535 (not bound to DNA) decreased with time. In these binding reactions the time at which the binding of cisplatin, transplatin, or BBR3535 reached 50% was 1.3 ± 0.2, 1.3 ± 0.1, or 1.2 ± 0.2 h, respectively; 88% of cisplatin, 92% of transplatin, or 98% BBR3535 was bound after 48 h. This result indicates that all platinum compounds tested in this work are able to bind natural double-helical DNA even in ethanol solution, although the rate of binding under these conditions is significantly slower than that in commonly used 0.01 M NaClO4 (43 ± 2 min in the case of cisplatin or transplatin [28] and 2.8 ± 2 min in the case of BBR3535 [10]). Hence, the binding experiments indicate that the modification reactions in 75% ethanol resulted in the irreversible coordination of cisplatin, transplatin, or BBR3535 to polymeric double-helical DNA, which facilitates sample analysis. It was also verified using transcription mapping assay [12] that preferential DNA binding sites of all three platinum complexes tested in this work were identical if DNA was modified by these complexes in the absence or presence of 75% ethanol (not shown).
Identification of interduplex covalently cross-linked DNA
Considerable evidence suggests that the antitumor efficiency of bifunctional platinum compounds, such as cisplatin, results from the formation of both intrastrand and interstrand CLs, but the existence and specific properties of interduplex CLs have not so far been taken into account. Therefore, cisplatin, transplatin, and BBR3535 were investigated for their ability to form DNA interduplex CLs under molecular crowding conditions mimicking environmental conditions in the cellular nucleus. BBR3535, cisplatin, or transplatin at various concentrations were incubated in 75% ethanol for 48 h with 500 ng of the linearized pUC19 DNA (2,686 bp) as described in detail “in Materials and methods.” The platinated samples were precipitated, the pellets were dissolved in 40 mM tris(hydroxymethyl)aminomethane/acetate, 1 mM EDTA, pH 7.4 buffer, and samples were immediately analyzed for platinum content by FAAS and for DNA interduplex CLs by electrophoresis under nondenaturing conditions on agarose gel. Upon electrophoresis, linearized pUC19 plasmid containing no interduplex CLs migrates as a single, 2,686-bp-long duplex, whereas the interduplex cross-linked species containing two, three, and even more duplexes migrate more slowly as a higher molecular mass species (Fig. 3a). The bands corresponding to more slowly migrating fragments containing CLs formed by mononuclear cisplatin or transplatin between DNA strands belonging to different duplexes were seen for r b values of 1 × 10−3 and higher (Fig. 3a, left and middle panels, respectively). In contrast, the bands corresponding to more slowly migrating fragments containing CLs were formed by dinuclear BBR3535 at a low r b value of 1 × 10−4 (Fig. 3a, right panel, lane 2). Interestingly, the modification under identical experimental conditions by BBR3535 at an r b value of 1 × 10−3 led to such an extensive cross-linking that a major part of the cross-linked molecules did not even enter the gel (Fig. 3a, right panel, lane 4). Importantly, the supershifted bands were eliminated after treatment of the cisplatin-, transplatin-, or BBR3535-modified DNA at 45 °C overnight with 0.4 M NaCN, at pH 10–11, to remove the platinum [29, 30]. These results suggest that the species are interduplex CLs that are tethered by Pt or (Pt–Pt)–DNA coordination bonds.
The formation of interduplex CLs in the 2,686-bp linearized pUC19 DNA. a Electrophoretograms of a 1% agarose gel demonstrate DNA interduplex cross-linking by cisplatin (left), transplatin (middle), or BBR3535 (right) after incubation of DNA with the platinum complex for 48 h in medium containing 0.2 M sodium acetate plus 75% ethanol. Lanes in the left panel and the middle panel: 1 control, nonplatinated DNA (r b = 0), 2 r b = 1 × 10−3, 3 r b = 5 × 10−3, 4 r b = 1 × 10−2, and 5 r b = 5 × 10−2. Lanes in the right panel: 1 control, nonplatinated DNA (r b = 0), 2 r b = 1 × 10−4, 3 r b = 5 × 10−4, 4 r b = 1 × 10−3, and 5 r b = 5 × 10−3. b Dependences of the fraction of interduplex cross-linked DNA molecules on the level of cisplatin (left), transplatin (middle), or BBR3535 (right) modification. c Electrophoretogram of the 1% agarose gel demonstrate that cisplatin, transplatin, and BBR3535 form no interduplex CLs after incubation of DNA with the platinum complex for 48 h in medium containing 0.01 M NaClO4 (in the absence of ethanol). Lanes in c: 1 control, nonplatinated DNA (r b = 0), 2 DNA modified by cisplatin, r b = 5 × 10−2, 3 DNA modified by BBR3535, r b = 5 × 10−3, and 4 DNA modified by transplatin, r b = 5 × 10−2
The intensity of the more slowly migrating bands increased with the growing level of the modification (Fig. 3b). The intensities of the individual bands in each lane were measured to obtain estimates of the fraction of noninterduplex cross-linked or interduplex cross-linked species under each condition. To compare quantitatively the efficiency of cisplatin, transplatin, and BBR3535 to form interduplex CLs, the frequency of interduplex CLs was calculated using the Poisson distribution from the fraction of noninterduplex cross-linked double-helical DNA in combination with the r b values and the fragment size. The frequencies of DNA interduplex cross-linking of cisplatin, transplatin, and BBR3535 calculated from four independent experiments were almost independent of r b and were 1.3 ± 0.3, 3.0 ± 0.3, and 54.1 ± 7.2%, respectively. Hence, BBR3535 is markedly more effective (approximately 40-fold or 18-fold) in forming interduplex CLs in comparison with cisplatin or transplatin.
Interduplex CLs were observed in this work if cisplatin, transplatin, or BBR3535 reacted with the pUC19 plasmid linearized with restriction endonuclease EcoRI. Thus, these DNA fragments contained short, single-stranded overhangs, which might make these fragments more favorable for ligation because cisplatin or BBR3535 could more easily cross-link these overhangs. However, interduplex CLs that are tethered by platinum–DNA coordination bonds were also observed with the same frequency if cisplatin, transplatin, or BBR3535 reacted with these fragments that have blunt ends (not shown). These fragments with blunt ends were prepared using a fill-in reaction at the 3′-ends with the Klenow fragment of DNA polymerase I and an equimolar mixture of dATP, dCTP, dGTP, and dTTT. Hence, ligation of linear molecules of DNA that are stabilized by cisplatin, transplatin, or BBR3535 CLs is not likely to be responsible for interduplex cross-linking by the platinum complexes in the presence of ethanol.
The experiments performed in aqueous ethanol described above were also repeated in sodium perchlorate supplemented with 0.2 M sodium acetate, pH 5.5, i.e., in the absence of ethanol as well. In the absence of ethanol, the high salt concentration itself led to no interduplex DNA CLs (Fig. 3c). These results confirmed that the presence of ethanol was necessary to generate the conditions in which interduplex CLs could be formed upon platination.
The effect of ethanol concentration
Figure 4 shows agarose gel electrophoresis experiments when the linearized pUC19 plasmid DNA was incubated with cisplatin at r b = 0.03 or with BBR3535 at r b = 0.003, respectively, in 0.2 M sodium acetate at 37 °C at various ethanol concentrations. Whereas the platinations of DNA by both complexes in the media containing 0–30% ethanol resulted in no interduplex cross-linking, the interduplex CLs became evident in medium containing 50% ethanol and their fraction grew concomitantly with increasing concentration of this molecular crowding agent (Fig. 4).
Dependence of the interduplex CL formation by cisplatin or BBR3535 on the concentration of ethanol. Plasmid pUC19 linearized by EcoRI was incubated at 37 °C for 48 h with cisplatin at r b = 0.03 or with BBR3535 at r b = 0.003 in medium containing 0.2 M sodium acetate plus ethanol at the concentrations indicated at the top of the gels. Electrophoretograms of 1% agarose gels demonstrating cross-linking by cisplatin or BBR3535, respectively
Interduplex cross-linking of nonhomologous DNA
Previous experiments demonstrated that cisplatin and BBR3535 were able to form CLs between two or even more homologous DNA duplexes. Therefore, experiments were also performed to determine whether the homologous nucleotide sequences in the duplexes are a prerequisite for interduplex cross-linking. Formation of cisplatin- or BRR3535-induced interduplex CLs between nonhomologous DNA restriction fragments was examined using 1% agarose gel electrophoresis (Fig. 5). The 1:1 mixtures of nonhomologous 908- and 1,556-bp double-helical fragments produced by cleavage of pSP73 plasmid DNA by two restriction endonucleases (EcoRI plus PvuI) were incubated for 48 h with cisplatin or BBR3535 in the presence of 0.2 M sodium acetate and 75% ethanol at 37 °C in the dark. The binding of cisplatin or BBR3535 generated several kinds of cross-linked duplexes, those having the lowest molecular mass corresponded to cross-linked duplexes 908 + 908, 908 + 1,556, and 1,556 + 1,556 bp. Hence, the cross-linked duplexes were nonhomologous in the 908 + 1,556 bp species, which confirms that interduplex CLs can also be readily formed by cisplatin or BBR3535 under molecular crowding condition between nonhomologous duplexes.
The formation of interduplex CLs between nonhomologous DNA restriction fragments. Electrophoretogram of a 1% agarose gel demonstrate DNA interduplex cross-linking after incubation of the 1:1 mixtures of nonhomologous 908- and 1,556-bp pSP73 restriction fragments with cisplatin at r b = 0.005 and 0.01 (lanes 1 and 2, respectively), BBR3535 at r b = 0.0001 and 0.0005 (lanes 3 and 4, respectively), and in absence of platinum complexes (lane C) in medium containing 0.2 M sodium acetate and 75% ethanol at 37 °C in the dark. Lane M 1 kilobase DNA length marker; numbers indicate the length of DNA fragments in base pairs. For other details, see the text
Discussion
As illustrated in Fig. 2, two types of DNA interstrand cross-linking by bifunctional platinum complexes can exist, depending on whether the platinum complex coordinates to base residues in only one DNA molecule (intramolecular interstrand CLs) or to base residues in two separate DNA molecules (intermolecular or interduplex CLs). In the case of bifunctional polynuclear antitumor platinum drugs, formation of the intramolecular interstrand CLs under molecular crowding conditions has been assumed. To our knowledge, no direct evidence indicating the capability of antitumor platinum complexes to form interduplex CLs as major DNA adducts has been reported. However, it should be pointed out that the occurrence of such a type of DNA CLs formed by conventional mononuclear cisplatin as minor DNA adducts under conditions of mild hyperthermia has been described [31]. It has been suggested that DNA modified by cisplatin under these conditions aggregates to an extent that allows interduplex contacts sufficient for the interduplex cross-linking and that this aggregation is promoted by local premelting conformational alterations [32].
In this work interduplex contacts sufficient for the interduplex cross-linking were ensured by performing investigations under molecular crowding conditions mimicking environmental conditions in the cellular nucleus. A characteristic of the interiors of all cells is the high total concentration of macromolecules they contain. Thus, DNA in the experiments described in this work was platinated under molecular crowding conditions in medium containing ethanol, which is a commonly used crowding agent [23, 24]. The DNA interduplex cross-linking efficiency of BBR3535 under molecular crowding conditions was remarkable: the frequency of interduplex cross-linking by this dinuclear PtII complex was 54%. In contrast, the interduplex cross-linking efficiency of mononuclear cisplatin or transplatin was markedly lower (approximately 40-fold or 18-fold). It seems, therefore, reasonable to conclude that at least some antitumor bifunctional poly(di/tri)nuclear complexes fulfill the requirements placed on interduplex DNA cross-linkers considerably better than mononuclear platinum complexes. For instance platinating sites in dinuclear BBR3535 are markedly more distant (2.7 nm [10]) than those in mononuclear cisplatin (0.28 nm [20]). Thus, binding of the two reactive sites to more distant base residues in adjacent duplexes may be markedly easier in the case of dinuclear BBR3535 than in the case of mononuclear cisplatin. In other words, the geometry of the reactive sites of cisplatin or transplatin is apparently markedly less appropriate for interduplex cross-linking than the geometry of the leaving ligands in BBR3535.
It is interesting that interduplex cross-linking by BBR3535 was observed in medium containing 75% ethanol, i.e., under conditions when DNA undergoes a transition from the B conformation to the A conformation [33]. However, it has been shown [34, 35] that the binding of the platinum complexes (including dinuclear/trinuclear bifunctional compounds) to random-sequence B-DNA lowers the conformational freedom of DNA so that it cannot acquire the A conformation. Thus, interduplex cross-linking by platinum compounds seems to be connected rather with changes in the extent of hydration than with alterations in DNA conformation.
The biological consequences of the production of interduplex CLs by BBR3535 and other antitumor polynuclear PtII compounds remain to be studied. For instance, it has been shown [15] that DNA interduplex cross-linkers are very potent in inducing sister chromatid exchanges and chromosomal abnormalities in mammalian cells. It is possible that the production of interduplex CLs in addition to other DNA intramolecular CLs may provide polynuclear PtII compounds with a wider spectrum of cytotoxicity. It remains to be seen how much the interduplex CLs contribute to the remarkably high potency of polynuclear PtII compounds against a variety of experimental tumor systems. In addition, the polynuclear bifunctional PtII complexes may also be of interest because they can be used to probe the organization of DNA in three-dimensional space, especially near sites of replication, recombination, or topoisomerase action, where two duplexes must be in close proximity.
Abbreviations
- BBR3535:
-
[{trans-PtCl(NH3)2}2-μ-{trans-(H2N(CH2)6NH2(CH2)2NH2(CH2)6NH2)}]4+
- CL:
-
Cross-link
- CT:
-
Calf thymus
- FAAS:
-
Flameless atomic absorption spectroscopy
References
Wang D, Lippard SJ (2005) Nat Rev Drug Discov 4:307–320
Kelland L (2007) Nat Rev Cancer 7:573–584
Vrana O, Brabec V, Kleinwächter V (1986) Anticancer Drug Des 1:95–109
Farrell N, Qu Y, Hacker MP (1990) J Med Chem 33:2179–2184
Mangrum JB, Farrell NP (2010) Chem Commun 46:6640–6650
Fichtinger-Schepman AMJ, Van der Veer JL, Den Hartog JHJ, Lohman PHM, Reedijk J (1985) Biochemistry 24:707–713
Brabec V, Kasparkova J, Vrana O, Novakova O, Cox JW, Qu Y, Farrell N (1999) Biochemistry 38:6781–6790
Kasparkova J, Zehnulova J, Farrell N, Brabec V (2002) J Biol Chem 277:48076–48086
Farrell N (2004) In: Sigel A, Sigel H (eds) Metal ions in biological systems. Marcel Dekker, New York
McGregor TD, Hegmans A, Kasparkova J, Neplechova K, Novakova O, Penazova H, Vrana O, Brabec V, Farrell N (2002) J Biol Inorg Chem 7:397–404
Kasparkova J, Novakova O, Vrana O, Farrell N, Brabec V (1999) Biochemistry 38:10997–11005
Brabec V, Leng M (1993) Proc Natl Acad Sci USA 90:5345–5349
Mullins ST, Annan NK, Cook PR, Lowe G (1992) Biochemistry 31:842–849
Carpenter ML, Lowe G, Cook PR (1996) Nucleic Acids Res 24:1594–1601
Matsumoto L, Kurek K, Larocque K, Gustafson G, Pires R, Zhang J, Tantravahi U, Suggs JW (1999) Mutat Res 426:79–87
Huang C-H, Mirabelli CK, Mong S, Crooke ST (1983) Cancer Res 43:2718–2724
Ellis RJ, Minton AP (2003) Nature 425:27–28
Zimmerman SB, Minton AP (1993) Annu Rev Biophys Biomol Struct 22:27–65
Roberts JD, Vanhouten B, Qu Y, Farrell NP (1989) Nucleic Acids Res 17:9719–9733
Sherman SE, Lippard SJ (1987) Chem Rev 87:1153–1181
Ellis RJ (2001) Trends Biochem Sci 26:597–604
Muhuri S, Mimura K, Miyoshi D, Sugimoto N (2009) J Am Chem Soc 131:9268–9280
Renciuk D, Kejnovska I, Skolakova P, Bednarova K, Motlova J, Vorlickova M (2009) Nucleic Acids Res 37:6625–6634
Renciuk D, Zemánek M, Kejnovská I, Vorlícková M (2009) Biochimie 91:416–422
Rauter H, Di Domenico R, Menta E, Oliva A, Qu Y, Farrell N (1997) Inorg Chem 36:3919–3927
Brabec V, Palecek E (1970) Biophysik 6:290–300
Brabec V, Palecek E (1976) Biophys Chem 4:76–92
Kostrhunova H, Vrana O, Suchankova T, Gibson D, Kasparkova J, Brabec V (2010) Chem Res Toxicol 23:1833–1842
Schwartz A, Sip M, Leng M (1990) J Am Chem Soc 112:3673–3674
Lemaire MA, Schwartz A, Rahmouni AR, Leng M (1991) Proc Natl Acad Sci USA 88:1982–1985
Halamikova A, Vrana O, Kasparkova J, Brabec V (2007) ChemBioChem 8:2008–2015
Gaillard C, Flavin M, Woisard A, Strauss F (1999) Biopolymers 50:679–689
Ivanov VI, Minchenkova LE, Minyat EE, Frank-Kamenetskii MD, Schyolkina AK (1974) J Mol Biol 87:817–833
Balcarova Z, Brabec V (1986) Biochim Biophys Acta 867:31–35
McGregor TD, Balcarova Z, Qu Y, Tran MC, Zaludova R, Brabec V, Farrell N (1999) J Inorg Biochem 77:43–46
Acknowledgments
This work was supported by the Ministry of Education of the Czech Republic (grant ME10066) and the student project of Palacky University (grant PrF 2011 024).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Muchova, T., Quintal, S.M., Farrell, N.P. et al. Antitumor bifunctional dinuclear PtII complex BBR3535 forms interduplex DNA cross-links under molecular crowding conditions. J Biol Inorg Chem 17, 239–245 (2012). https://doi.org/10.1007/s00775-011-0845-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-011-0845-0