Abstract
The proteolytic processing of collagen I by three matrix metalloproteinases (MMPs), a collagenase (MMP-1), a gelatinase (MMP-2), and the ectodomain of a membrane-type metalloproteinase (MMP-14), has been investigated at 37 °C between pH 6.0 and 9.2, a pH range reflecting conditions found in different body compartments under various physiopathological processes. In the proteolytic degradation the native collagen triple helix must be partially unwound to allow the binding of α chains to the protease’s active-site cleft. We have found that MMP-1 interacts with the two types of collagen I α chains in a similar fashion, whereas both MMP-2 and MMP-14 bind the two α chains in a different way. The overall enzymatic activity is higher on the α-2 chain for both MMP-1 and MMP-2, whereas the MMP-14 ectodomain preferentially cleaves the α-1 chain. In MMP-2 a marked difference for substrate affinity (higher for the α-1 chain) is overwhelmed by an even more marked propensity to cleave the α-2 chain. As a whole, the three classes of MMPs investigated appear to process collagen I in a significantly different fashion, so various MMPs play different roles in the collagen homeostasis in various compartments (such as bloodstream, synovial fluid, normal and tumoral tissues), where different pH values are observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Matrix metalloproteinases (MMPs) are a family of Zn2+-dependent endopeptidases which are involved in the enzymatic processing of several components of the extracellular matrix (ECM), such as collagens, proteoglycans and fibronectin [1–3]. They are implicated in several physiological and pathological processes, such as skeletal growth and remodeling, wound healing, cancer, arthritis and multiple sclerosis [4–8]. MMPs are virtually capable of proteolytically processing all molecules present in the ECM, though they display differing propensity for various substrates [1–3]. From the structural viewpoint, all MMPs share a common catalytic site, where a Zn2+ ion is coordinated by three histidyl residues, whereas they differ regarding the presence and the structural arrangement of additional domains, such as the hemopexin-like domain and the fibronectin-like domain, which are important for the recognition of macromolecular substrates [9–13].
Collagen I is the most abundant macromolecular substrate for MMPs present in the ECM; it is usually found as fibrils, originating from the self-association of monomers, which gives rise to a supramolecular organization of 67-nm D periodic fibrils [14, 15]. Each monomer displays a particular quaternary structure (called a triple helix), made of three left-handed α chains (two α-1 chains and one α-2 chain) that are coiled around each other into a right-handed triple helix [16].
On the basis of their structural features and of their propensity to proteolytically process collagen I, MMPs are classified into five main groups, namely, (1) collagenases (i.e., MMP-1, MMP-8, and MMP-13), which preferentially cleave collagen I, recognizing the substrate through the hemopexin-like domain [10, 17, 18], (2) gelatinases (i.e., MMP-2 and MMP-9), which are able to cleave both collagen I and collagen IV [19, 20], recognizing the substrates preferentially through the fibronectin-like domain [9, 11, 13, 20], a fibronectin II-like domain inserted only in the catalytic domain of gelatinases, (3) stromelysins (i.e., MMP-3, MMP-10, and MMP-11), which enzymatically process collagen IV, but not collagen I [3], (4) matrilysins (i.e., MMP-7 and MMP-26), which are unable to cleave collagen I [21, 22], and (5) membrane-type MMPs (i.e., MMP-14, MMP-15, MMP-16, MMP-17, and MMP-24), which display at the C-terminal a transmembrane domain with a short cytoplasmic tail and a long extracellular ectodomain; among these, only MMP-14 is able to enzymatically process collagen I [23]. Hence, only collagenases, gelatinases, and MMP-14 are able to cleave collagen I.
As previously reported, the substrate recognition occurs in a differing fashion for these MMPs [13, 24], even though no structural information is available for the interaction of any of the MMPs with the natural substrate collagen I. Further, a detailed comparison of the different proteolytic processing of a natural substrate by the three types of MMPs has never been carried out before under different pH conditions. This pH range can be of a major pathophysiological relevance, since it is known that almost all mammalian tumors are characterized by a significant variation of pH values both at the intracellular and at the extracellular level (ranging from 5.85 to 7.68) [25]. Furthermore, pH variations may also occur under inflammatory conditions and even different compartments can physiologically display drastic differences in pH values. As an example, whereas in the bloodstream the pH is maintained around 7.4 ± 0.1, in the synovial liquid of joints the pH is much higher, being 8.1 ± 0.1 under normal conditions [26]; in both cases, inflammation (such as in osteoarthritis) brings about a lowering of the pH to 7.1 ± 0.1 and 7.6 ± 0.1, respectively [26]. Up to now, investigation of the pH dependence of enzymatic properties of MMPs has been carried out for synthetic substrates on porcine collagenases and gelatinases [27, 28], on stromelysin MMP-3 [29, 30], and on MMP-8, MMP-2, and MMP-9 [31], whereas for a macromolecular substrate an investigation of the pH dependence has been undertaken only for the enzymatic activity of MMP-8 toward collagen I [24]. In this paper we have extended the investigation of collagen I to the pH dependence of the enzymatic activity of MMP-1 (a collagenase), MMP-2 (a gelatinase), and the ectodomain of MMP-14 (ect-MMP-14; a membrane-type MMP). Historically, studies on collagen degradation have been performed at 25 °C, because synthetic substrates are not stable at temperatures higher than 25 °C. However, in the work reported here, as well as in previous work [10, 24], we carried out an investigation at 37 °C, since the partial unwinding of collagen I at the physiological temperature [32] is fully reversible (Fig. 1b), clearly demonstrating that it is not due to denaturation (as it occurs for gelatin), but it is a fully physiological conformational change of a macromolecular substrate. Therefore, to acquire physiologically relevant information (in the absence of structural information for the complex between collagen I and MMPs) on the different aspects of the proteolytic mechanism of various MMPs regarding the main natural macromolecular substrate present in the ECM, we carried out this analysis at 37 °C, even though it is known that at this physiological temperature the structural triple-helix arrangement of collagen I is less tight [32].
a pH dependence of circular dichroism (CD) spectra of collagen I at 37 °C in agarose gels. Only selected spectra at the pH values indicated are shown to illustrate the proton-linked spectral changes. b Circular dichroism spectra of collagen I at pH 7.3 and 37 °C before the pH-dependence investigation (continuous line), at pH 9.5 and back to pH 7.3 (dashed line) after the exposure to pH 9.5. c pH dependence of ellipticity at 224 nm. The continuous line in c represents the nonlinear least-squares fitting of data according to \( {\text{CD}}_{\text{obs}} = {\text{CD}}_{0} + \Updelta {\text{CD}}_{1} \times {\frac{{K_{1} [{\text{H}}^{ + } ]}}{P}} + \Updelta {\text{CD}}_{2} \times {\frac{{K_{1} K_{2} [{\text{H}}^{ + } ]^{2} }}{P}} + \Updelta {\text{CD}}_{3} \times {\frac{{K_{1} K_{2} K_{3} [{\text{H}}^{ + } ]^{3} }}{P}} \) where \( P = 1 + K_{1} [{\text{H}}^{ + } ] + K_{1} K_{2} [{\text{H}}^{ + } ]^{2} + K_{1} K_{2} K_{3} [{\text{H}}^{ + } ]^{3} \) employing the following parameters CD0 = +1.29 ± 0.15, ΔCD1 = +2.55 ± 0.29, ΔCD2 = +36.57 ± 4.34, ΔCD3 = −38.48 ± 4.46, K1 = 1.04 (±0.21)×109 M−1 (corresponding to pK1 = 9.02 ± 0.11), K2 = 1.55 (±0.34)×106 M−1 (corresponding to pK2 = 6.19 ± 0.12), and K3 = 2.27 (±0.53)×106 M−1 (corresponding to pK3 = 6.36 ± 0.12)
Materials and methods
Materials
The purity of MMP-1, MMP-2, and MMP-14 proenzymes (R&D System, London, UK) was measured by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) according to the method of Laemmli [33]. After the gels had been run, they were stained using a silver staining kit (Bio-Rad, Hercules, CA, USA). The broad-spectrum protein markers (Bio-Rad, Hercules, CA, USA) were used as molecular weight standards.
The stability of each MMP was checked by zymography (employing as the substrate either gelatin or collagen). All three MMPs are perfectly stable (without undergoing any autocatalytic activity in the presence of the collagen solution at 37 °C) for well over 60 min, which is the time period needed to obtain kinetic data for the calculation of the catalytic parameters. The stability experiment was performed as follows: 2 μl of MMP in collagen solution was mixed with a fivefold excess of sample buffer [0.25 M tris(hydroxymethyl)aminomethane (Tris)–HCl, 0.8% SDS, 10% glycerol, and 0.05% bromophenol blue] and run on 12% SDS–polyacrylamide gels containing either 1 mg/ml gelatin or bovine type I collagen as described previously [34].
Trypsintpch and soybean trypsin inhibitor were purchased from Sigma (St. Louis, MO, USA). The quenched fluorogenic substrates employed for activity assays were (7-methoxycoumarin-4-yl)acetyl-Pro-cyclohexylalanine-Gly-norvaline-His-Ala-(N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl)-NH2 (MCA-1) and (7-methoxycoumarin-4-yl)acetyl–Pro-Leu-Gly-Leu-(N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl)-Ala-Arg-NH2 (MCA-omni) (Calbiochem, La Jolla, CA, USA). Batimastat (BB-94), a peptidomimetic inhibitor, which stoichiometrically inhibits MMPs, was kindly provided by British Biotech Pharmaceutical (Cowley, Oxford, UK).
We used rat-tail tendon type I collagen, isolated by standard ammonium sulfate precipitation procedures from tail tendons of 8-week-old Wistar rats, as previously described by others in detail [35, 36]. The amount of collagen was quantified as previously described [37]. The triple-helix nature of collagen was checked by circular dichroism (CD) measurements and further confirmed by the trypsin degradation assay. This assay validates the quality of collagen triple-helix structure, verifying the absence of collagen degradation at 1:10 trypsin to collagen ratio over 3 h at 28 °C, as described previously [11]. Lyophilized collagen was stored at −80 °C, and stock solutions were prepared as needed. No significant difference in the triple-helix content and in the stability was detected between the two preparations of collagen upon solubilization.
Methods
Enzymatic assays
Recombinant human MMP-2 proenzyme was activated by incubating 0.1 mg/ml progelatinase solution with 0.25 mM aminophenyl mercuric acid (Sigma, St. Louis, MO, USA) at 37 °C for 30 min. The activation of the pro-ect-MMP-14 was performed using 5 μg/ml trypsintpch at 25 °C. Trypsin activity was quenched using 50 μg/ml soybean trypsin inhibitor. The concentration of active MMP was determined by the classic fluorimetric assay, as described by others [38], following the progressive decrease of hydrolysis [upon addition of Batimastat (BB-94)] of the quenched fluorogenic substrate MCA-1 (λ exc = 325 nm, λ em = 398 nm) for MMP-1 and MCA-omni (λ exc = 325 nm, λ em = 398 nm), which is resistant to the trypsin action, for MMP-2 and ect-MMP-14. All measurements were performed at 37 °C using a solution of 50 mM Tris–HCl, 0.1 M NaCl, 10 mM CaCl2 plus 0.05% Brij 35 buffered at pH 7.3. The same synthetic substrates were used for the investigation of the pH dependence (between pH 6.4 and 9.55) of the catalytic parameters for MMP-1 and ect-MMP-14 to be compared with those obtained with collagen chains (see later).
Determination of kinetic and thermodynamic parameters
Kinetic analyses of α chain degradation by MMP-1, MMP-2, and ect-MMP-14 were performed in 50 mM Tris–HCl, 0.1 M NaCl, 10 mM CaCl2 at several pH values (ranging between pH 6.0 and 9.5), employing different concentrations of collagen I (spanning from 0.3 to 3.2 μM); active MMP was added to the solutions (10 pM final concentration). For each collagen concentration, the mixtures were kept at 37 °C and small aliquots were harvested at different time intervals. The reactions were stopped by the addition of SDS-PAGE loading buffer containing 20 mM EDTA and frozen to −80 °C until they were used. The aliquots in the reducing sample buffer were separated on 7.5 or 10% SDS-PAGE gels and they were stained using either (1) the silver staining method or (2) 0.5% Commassie blue, followed by destaining until substrate bands were clearly visible. The Coomassie blue electrophoretic spots, corresponding to different aliquots at different time intervals, were analyzed by image analysis software (Image Quant TL, Amersham Biosciences) and their intensity was calibrated (to obtain concentration values) using standard substrate solutions. For each α chain (i.e., α-1 and α-2 chains) the substrate disappearance rates were derived at each collagen concentration employed. The initial velocities were measured within 1 h, which is the time period during which the rate was constant and less than 10% of the substrate was degraded. This ensured a steady-state condition for the first cleavage step, and it was a prerequisite for the subsequent analysis step, which was based on the observation of an inverse linear correlation between velocity and substrate concentration according to the Lineweaver–Burk equation (Eq. 1) and Eadie–Hofstee linear regression (Eq. 2):
where E 0 is the total enzyme concentration, v is the actual rate (expressed as moles per second), K M is the Michaelis–Menten equilibrium constant (expressed as moles), k cat is the rate-limiting-step kinetic constant (expressed as per second), S is the substrate concentration, and V MAX is the ratio between E 0 and v. Linear regression plots were constructed from the velocity data and the catalytic parameters k cat and K M were extracted. Investigation of the kinetics with synthetic substrates of MMP-1 and ect-MMP-14 was carried out at different pHs (within the range 6.4–9.5 pH units) under the same experimental conditions. Enzymatic processing of fluorogenic synthetic substrates was followed by spectrofluorimetry (Eclypse, Varian), employing 325 nm as the excitation wavelength and following the fluorescence at 393 nm (for MCA-1) and 400 nm (for MCA-MMP14). The data were analyzed according to Eqs. 1 and 2.
CD experiments
CD spectra were recorded with a JASCO J-710 spectropolarimeter equipped with a thermostated cell holder and connected to a data station for signal averaging and processing. All spectra are averages of four scans and were recorded employing quartz cells of 2-mm path length. For the spectroscopic observations, collagen I was entrapped in agarose gel by mixing the solubilized protein with a 1% low-melting-point agarose solution at pH 3.0. The temperature of the agarose solution when collagen was added was 37 °C. After rapid stirring the mixture was poured on a simple gel casting (Mini-protean II, Bio-Rad) and immediately cooled to obtain the gel. The final concentrations were 0.5% agarose and 1 μM collagen. The thickness of the homogeneous gel was 1 mm. For the kinetic experiments, slices of suitable size were cut off from the homogeneous gel and kept overnight in buffer solution to reach the desired pH (without the formation of aggregates) and then used for CD measurements [24]. Since collagen I has been reported to be somewhat thermally unstable in solution [32], gel-entrapped collagen samples of each proteolytic assay were monitored for a time as long as 3 days at 37 °C and no significant change in the CD spectra was detected in the absence of the enzyme, as previously reported [13]. The CD spectra so obtained are in agreement with those obtained for collagen-like peptides showing the characteristic triple-helix maximum in the 223–225-nm range [39]. Therefore, we achieved a ‘quasi-native’ model for evaluating structural arrangements during proteolytic processes on type I collagen [24]. The kinetics of the unwinding of the triple helix was followed in 0.2 M Tris–HCl, 1 M NaCl, 10 mM CaCl2 solution at 37 °C and at different pHs (spanning from 6.0 to 9.0) by recording all the spectroscopic range (200–250 nm) and reporting the values of molar ellipticity at 223 nm of CD spectra recorded at different pHs.
Data analysis
The pH dependence of the catalytic parameters was fitted according to the following equations:
where obs k cat/K M, obs k cat, and obs K M are the observed parameters as a function of pH, i k cat/K M, i k cat, and i K M are the parameters for the ith-protonated forms, i K Ua are the ith protonation constants for the free enzyme, and i K La are the ith protonation constants for the substrate-bound enzyme when it is involved in the cleavage event at the rate-limiting step. The global fitting procedure, employing simultaneously Eqs. 3, 4, and 5 to describe the pH dependence of all three catalytic parameters (i.e., k cat/K M, k cat, and K M) of a MMP, is also able to furnish information on different parameters at various protonation levels (see Eqs. 3, 4, 5), which is reported in Tables 2 and 3 for the two chains.
Results
The investigation on the pH dependence of the enzymatic activity of MMP-1, MMP-2, and ect-MMP-14 toward collagen I involves not only the proton-linked effect on the specific activity of the enzyme(s), but also the pH effect on the structural arrangement of the macromolecular substrate (i.e., collagen I). This obviously represents an additional difficulty for the quantitative analysis, since (unlike synthetic small substrates [31]) our observations are influenced by pH-dependent structural changes of both the enzyme(s) and the macromolecular substrate.
pH dependence on the collagen triple-helix structure
Collagen I shows a pH dependence of the triple-helix assembly (Fig. 1a), which is characterized by a variation of the ellipticity in the 223–225-nm spectral region, such that at pH ≈ 6.3 the positive ellipticity is maximal and it decreases as the pH is raised, becoming very small at pH ≥ 9.0 (Fig. 1a). The spectroscopic features at different pH values are very stable and time-independent for at least 48 h at 37 °C (data not shown), and their proton-linked behavior is fully reversible (Fig. 1b). This clearly indicates that the ellipticity changes are not due to denaturation of the collagen molecule toward a gelatine-like state, but they reflect instead a pH-dependent conformational change characterized by a different packing of the triple-helix arrangement. This fully reversible proton-linked behavior can be accounted for by three protonation events, characterized by pK a1 = 9.02 ± 0.15, pK a2 = 6.36 ± 0.18, and pK a3 = 6.16 ± 0.19 (Fig. 1c). The last two protonation processes have opposite effects on the ellipticity of collagen I, since the second protonation (i.e., characterized by pK a2) induces a drastic increase of the ellipticity, likely related to a proton-linked structural change of the triple helix, whereas the third protonation (i.e., characterized by pK a3) brings about a decrease of the positive ellipticity, possibly due to a partial unwinding of the triple helix. The pH range of these two last processes almost overlaps, suggesting that they could be somehow correlated, such that the protonation of the second group (responsible for pK a2) might affect the protonation of the third group (responsible for pK a3).
This behavior clearly indicates that collagen I cannot be considered simply a “passive” substrate whose enzymatic processing is only modulated by the pH-dependent activity of the MMPs (as for small synthetic substrates); it actively participates in modulating its own proteolytic processing.
pH dependence of the MMP-1 collagenolytic activity
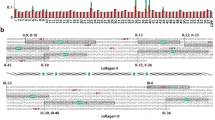
Figure 2 reports the SDS-PAGE electrophoretic pattern of the proteolytic cleavage of the two chains from collagen I upon incubation with MMP-1 at 37 °C at pH 8.0 for different time intervals; the pattern is very similar at all pH values investigated. The the formation of the two three-quarter fragments as a function of time is evident; however, their formation kinetics was not analyzed, since the significance of its apparent rate is not straightforward to determine, being affected by different cleavage events. We only analyzed the rate of disappearance of the two intact chains, to which the overall catalytic activity refers. Figures 3a and 4a display the pH dependence of the enzymatic efficiency of MMP-1 (i.e., k cat/K M) on the two chains of collagen I; Fig. 5a shows the pH dependence of the enzymatic activity against the synthetic substrate MCA-1.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis of the kinetics of fragmentation of collagen I by 18 nM wild-type matrix metalloproteinase (MMP) 1 at 37 °C. Left column collagen I sample in the absence of the enzyme. Right column molecular weight markers. Intermediate columns samples of collagen I incubated with wild type MMP-1 for a given interval, as indicated
pH dependence of processing at 37 °C of the α-1 chain of collagen I by MMP-1 (circles), MMP-2 (crosses), and the ectodomain of MMP-14 (asterisks), as expressed by the pH effect on k cat/K M (a), k cat (b), and K M (c). The continuous lines are the nonlinear least-squares fitting of experimental data according to Eq. 3 (a), Eq. 4 (b), and Eq. 5 (c), employing parameters reported in Tables 1, 2, and 3. In a the arrows marked a and b indicate the inflammation-dependent pH decrease in the bloodstream and in the synovial fluid, respectively. For further details, see the text
pH dependence of processing at 37 °C of the α-2 chain of collagen I by MMP-1 (circles), MMP-2 (crosses), and the ectodomain of MMP-14 (asterisks), as expressed by the pH effect on k cat/K M (a), k cat (b), and K M (c). The continuous lines are the nonlinear least-squares fitting of experimental data according to Eq. 3 (a), Eq. 4 (b), and Eq. 5 (c), employing parameters reported in Tables 1, 2, and 3. In a the arrows marked a and b indicate the inflammation-dependent pH decrease in the bloodstream and in the synovial fluid, respectively. For further details, see the text
pH dependence of processing at 37 °C of synthetic substrate [(7-methoxycoumarin-4-yl)acetyl-Pro-cyclohexylalanine-Gly-norvaline-His-Ala-(N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl)-NH2 (MCA-1) for MMP-1 and MCA-MMP14 for the ectodomain of MMP-14] by MMP-1 (circles) and the ectodomain of MMP-14 (crosses), as expressed by the pH effect on k cat/K M (a), k cat (b), and K M (c). The continuous lines are the nonlinear least-squares fitting of experimental data according to Eq. 3 (a), Eq. 4 (b), and Eq. 5 (c), giving the pK a values reported in Table 1
It appears immediately evident that over the whole pH range investigated the proteolytic activity on the α-2 chains is higher than for the α-1 chain. This difference is fairly large at alkaline pH (being about 40-fold at pH 9.2) and it decreases as the pH decreases toward the physiological value (i.e., pH 7.3), where the enzymatic processing of the α-2 chain is only 4–5 times higher than for the α-1 chain. As the pH is further lowered, the difference remains essentially unchanged down to pH 6.3.
This difference is almost completely attributable to a difference in k cat for the rate-limiting cleavage step of the two chains (see Figs. 3b, 4b), whereas the K M values are fairly similar for the two chains (see Figs. 3c, 4c), the difference always being less than threefold over the pH 6.5–9.5 range. This clearly indicates that the higher susceptibility of the α-2 chains to the proteolytic attack is not due to a preferential interaction of the enzyme with the substrate, but it is instead attributable to easier accessibility of the active site to the cleavage site and/or to a higher tendency of the α-2 chain to undergo the partial unwinding transition enzymatic processing. It is very important to underline that the proteolytic activity of MMP-1 toward the synthetic substrate is essentially pH-independent for pH ≤ 9.0, increasing only at very alkaline pH values (see Fig. 5a). This behavior reflects the very limited pH dependence of both k cat and K M (see Fig. 5b, c).
Table 1 reports the values of pK a required to quantitatively describe the pH dependence of all three catalytic parameters. Even though the pH dependence of individual parameters could have been fitted with fewer protonating groups, the overall description of the enzymatic process could only be accounted for by three groups (i.e., n = 3 in Eqs. 3, 4, 5) which display a substrate-linked variation of their pK a values.
A first point to be outlined from the values reported in Table 1 is that pK Ua values are the same for the α-1 and for the α-2 chains, clearly indicating that the same residues of MMP-1 are involved in the interaction with both types of chains. However, the effect of substrate binding is different for the two chains, since the pK La values display significant variations for the two substrates. In particular, the second protonating group (characterized by pK U2 and pK L2 in Table 1) shows a marked decrease for pK a upon binding of MMP-1 to the α-1 chains, whereas it remains substantially unaffected upon interaction of the enzyme with the α-2 chains (see Table 1). On the other hand, the first and the third protonating groups (characterized in Table 1 by pK U1 and pK L1 and by pK U3 and pK L3, respectively) show an increase of pK a (the first one) and a marked decrease (the third one) upon formation of the enzyme–substrate complex, which then carries out the rate-limiting cleavage step. Completely different values of pK U and pK L are observed for the synthetic substrate MCA-1 (see Table 1), clearly indicating that in the case of the synthetic substrate the interaction area with MMP-1 is drastically different from that involved in the interaction with collagen chains. This is particularly relevant for the pK U values, which refer to the enzyme in the absence of substrate, and which give some information on the portion of the enzyme involved in the interaction with the substrate, whereas the pK L values refer to the effect of the substrate interaction and are obviously dependent on the type of substrate.
However, we cannot attribute the observed pH dependence exclusively to a proton-linked effect on the enzyme activity, since, as already outlined, collagen I also undergoes a pH-induced structural change (Fig. 1a), which may modulate the observed proteolytic process.
A closer look at the effect of protonation can obtained from the catalytic parameters for the different protonation levels (see Table 2). In this respect, we find that before the first protonation occurs MMP-1 has a much lower enzymatic efficiency toward the α-1 chain, and this is due to a much slower rate for the rate-limiting cleavage step (about 80-fold slower than for the α-2 chain, see Table 2), which overwhelms the slightly higher affinity of MMP-1 for the α-1 chain.
The first protonation brings about a dramatic enhancement of the enzymatic activity toward the α-1 chain (mostly due to a sevenfold binding affinity increase, see Table 2), whereas the activity toward the α-2 chain remains essentially unchanged, showing a compensation between an increased substrate affinity and a slower rate-limiting step. Therefore, this group, which somewhat raises its pK a upon formation of the enzyme–substrate complex, seems to play a major role in facilitating the interaction of MMP-1 with the α-1 chain, thus decreasing the difference between the two chains for the interaction with MMP-1.
The second protonation process induces a fivefold decrease of the enzymatic activity of MMP-1 toward both chains, but this behavior has a completely different origin for the two chains, since in the case of the α-1 chain it is completely due to a decrease of substrate affinity (only partially compensated by an increase of k cat, see Table 2), whereas in the case of the α-2 chain it is wholly attributable to a decrease of the rate constant for the rate-limiting cleavage step (see Table 2). This difference is reflected in the fact that the pK a of the groups is markedly decreased upon interaction of the enzyme with the α-1 chain, whereas it does not seem affected by the interaction with the α-2 chains (see Table 1); this clearly demonstrates that (1) the collagen groups interacting with MMP-1 are different in α-1 and in α-2 chains and (2) the binding of the α-1 chain by MMP-1 brings about the deprotonation of a group (not observed for the α-2 chain, see Table 1) and this is accompanied by a decrease of the binding affinity for the α-1 chain. This second protonation event, although not affecting the substrate binding affinity for the α-2 chain, has a dramatic effect on k cat of MMP-1 on the two chains, which becomes much faster for the α-1 chain, whereas it is markedly reduced for the α-2 chain (see Table 2). Indeed, it suggests that the substrate-linked deprotonation of this group is required for an efficient cleavage process by MMP-1 (as observed for the α-1 chain) and the fact that it remains protonated even after the interaction with the substrate (as observed for the α-2 chain) induces a relevant decrease of its cleavage efficiency.
The third protonation event is accompanied by a marked decrease of the enzymatic activity for both chains, being associated with a decrease of substrate affinity and of k cat (see Table 2), thus suggesting that this phenomenon, which occurs at acid pH, is likely accompanied by the inactivation of the enzymatic activity of MMP-1.
pH dependence of the MMP-2 collagenolytic activity
Figures 3a and 4a show the pH dependence of the enzymatic efficiency of MMP-2 (i.e., k cat/K M) on the two chains of collagen I. Over the whole pH range investigated the enzymatic activity toward the α-2 chain is significantly higher than that toward the α-1 chain. The difference is maximal at alkaline pH (being about 20-fold at pH 9.2) and it somewhat decreases at neutral pH, such that at pH 7.3 the enzymatic processing of the α-2 chain is only about fivefold higher than for the α-1 chain. The difference at alkaline pH is only due to a slower k cat (which is about 25-fold slower for the α-1 chain, see Figs. 3b, 4b), whereas K M is very similar for the two chains (see Figs. 3c, 4c). However, as the pH decreases the difference for k cat between the two chains becomes even more pronounced, being about 300-fold at pH 7.3 (Figs. 3b, 4b); this effect is, however, almost completely compensated by the opposite trend for K M, which decreases for the α-1 chain and increases for the α-2 chain (see Figs. 3c, 4c), such that at pH 7.3 the affinity of MMP-2 for the α-1 chain is about 60-fold higher than for the α-2 chain. Further decrease down to pH 6.0 brings about a reduced difference between the k cat values for the two chains (still amounting to a 30-fold higher value for the α-2 chain, see Figs. 3b, 4b) accompanied by the annihilation of the difference for the K M values (see Figs. 3c, 4c), resulting in a marked difference in favor of the overall enzymatic processing of the α-2 chain (Figs. 3a, 4a).
Unlike MMP-1, MMP-2 shows different pK Ua values for the interaction of the free enzyme with the two types of collagen chains (see Table 1). Since it is known that MMP-2 binds collagen I preferentially by the fibronectin-like domain [11, 13], this difference might underlie the possibility that the α-1 chain interaction involves a different portion of the collagen binding domain of MMP-2 with respect to that binding the α-2 chain. In this respect, it is very important to underline that the pK U values for the α-1 chain are very similar to those reported for the interaction of MMP-2 with synthetic substrates [31]. Although a comparison between data on synthetic macromolecular substrates is not straightforward, since the enzyme–substrate interface is obviously different, this evidence suggests that in MMP-2 similar groups are possibly involved in the binding both to the synthetic substrate and to the α-1 chain of collagen I. However, it must be remarked that this is limited to the α-1 chain, since the pK U2 and pK U3 values for the interaction of MMP-2 with the α-2 chain are clearly different, strengthening the idea that either (1) distinct portions of the fibronectin-like domain of MMP-2 bind the two chains of collagen I or else (2) a different domain(s) (such as the hemopexin-like domain) is (are) involved in the interaction with the two chains.
The interaction of MMP-2 with the α-1 chain is then characterized by an increase of pK 1 and by a decrease for the other two protonating groups, whereas the interaction with the α-2 chain brings about a decrease of the first two proton-linked groups and an increase of pK 3 (see Table 1), supporting the idea that the two collagen chains interact with a somewhat different portion of MMP-2.
Obviously, in view of the pH effect on the structure of collagen I (see Fig. 1a), we cannot rule out at this stage the possibility that some of these pK a values are also related to the proton-linked conformational change of collagen I.
The behavior observed for the catalytic parameters upon protonation confirms the drastic difference for the two chains (see Tables 2, 3). Before the first protonation takes place, MMP-2 displays very similar affinity for the two collagen I chains, whereas a dramatic 20-fold difference can be observed for the speed of the rate-limiting step (with k cat much faster for the α-2 chain, see Tables 2, 3), resulting in a faster enzymatic activity (i.e., k cat/K M) toward the α-2 chain (see Tables 2, 3). However, the first protonation is accompanied by a tenfold increase of affinity for the α-1 chain, whereas the interaction with the α-2 chain is only very slightly affected. In both cases, after the first protonation event a reduction of k cat is observed, even though this is much more marked for the α-1 chain (see Tables 2, 3); for both chains the net result is a slight reduction of the enzymatic activity. On the other hand, whereas the second protonation does not change significantly the affinity of the enzyme for the α-1 chains, the affinity for the α-2 chain is dramatically decreased, being accompanied by a marked increase of k cat for both collagen I chains, with an overall increase of the enzymatic activity, which for the α-1 chain attains the highest value over the whole pH range investigated (see Table 2). The third protonation brings about a drastic decrease of the enzymatic activity of MMP-2 toward the α-1 chain, which is mostly due to a dramatic decrease of affinity (see Table 2). Conversely, after the third protonation the enzymatic activity toward the α-2 chain is kept constant by a marked increase of affinity, which counterbalances the relevant decrease of k cat (see Table 3).
pH dependence of the ect-MMP-14 collagenolytic activity
Figures 3a and 4a show the pH dependence of the overall enzymatic activity (i.e., k cat/K M) of ect-MMP-14 toward the collagen chains, whereas Fig. 5a displays the pH dependence of the proteolytic activity of ect-MMP-14 toward the synthetic substrate MCA-MMP14. The observed proton-linked behavior is fairly similar for the two chains of collagen I, though displaying a somewhat different pH dependence, such that at pH 7.5 a tenfold higher activity is observed toward the α-2 chain, whereas a very similar value for the two chains is detected at pH 7.0. This behavior can be attributed to the difference reported for the pK U2 values of the two chains (see Table 1), in spite of the close similarity of the k cat/K M for the two chains after the first and the second protonation process (see Tables 2, 3). Further, at pH < 7.0 the overall enzymatic activity toward the α-1 chain increases, whereas the processing of the α-2 chain remains essentially constant between pH 7.3 and 6.0 (Figs. 3a, 4a). This different proton-linked effect can be instead related to the much lower value of pK U3 for the α-1 chain associated with a somewhat faster enzymatic activity against the α-1 chain after the third protonation occurs.
The decrease of the overall enzymatic activity of ect-MMP-14 after the first and the second protonation event is fully attributable to a drastic reduction of the velocity of rate-limiting step for both chains (i.e., k cat, see Figs. 3b, 4b, Tables 2, 3). This decrease is more marked for the α-2 chain but it is fully compensated by the higher affinity of ect-MMP-14 for this chain (Fig. 4c), being especially evident after the second protonation (see Tables 2, 3). On the other hand, after the third protonation, ect-MMP-14 displays a somewhat higher activity toward the α-1 chain, which is only due to a faster rate-limiting step (i.e., k cat, see Fig. 3b, Tables 2, 3), since the substrate affinity is very similar for the two chains.
In the case of the synthetic substrate the overall enzymatic activity of ect-MMP-14 displays a pH dependence characterized by maximum efficiency at pH 7.0, which decreases upon both pH increase and pH decrease (see Fig. 5a). This effect is almost completely attributable to the proton-linked behavior of the substrate affinity (see Fig. 5c), whereas the rate-limiting-step k cat shows a very limited pH dependence (see Fig. 5b).
Unlike MMP-1 and similarly to MMP-2, ect-MMP-14 displays different pK U values for the two types of collagen chains (see Table 1), underliying also in this case the possibility that the interaction of ect-MMP-14 is different with the two chains. In particular, the interaction with the α-1 chains brings about a marked increase of pK 1 and a decrease of pK 2, leaving pK 3 essentially unchanged (see Table 1). Conversely, the interaction with the α-2 chain shows a marked increase of pK 1 and a decrease for pK 3, whereas the protonating group characterized by pK 2 undergoes only a moderate pK a decrease (see Table 1). Like for MMP-1, also in the case of ect-MMP-14 the pK U values obtained for the enzymatic processing of the synthetic substrate are different from those observed for the activity toward the collagen chains (see Table 1), even though pK U3 for the synthetic substrate is similar to the value of pK U2 obtained for the collagen α-1 chain, suggesting that this might be attributable to the same group.
A closer look at the catalytic parameters for different protonation levels shows that the similar enzymatic activity for the two types of chains derives from a different balancing of k cat and K M (see Tables 2, 3). Before the first protonation, the much faster k cat of ect-MMP-14 for the α-2 chains is fully compensated by the much lower affinity with respect to the α-1 chain (see Tables 2, 3), whereas for both chains the decrease of the enzymatic activity after the first protonation event is associated with a decrease of k cat (though to a different extent) compensated by a similar increase of the substrate affinity (i.e., a decrease of K M, see Figs. 3c, 4c, Tables 2, 3). On the other hand, the 100-fold lower enzymatic activity observed for both chains after the second protonation process is only due to a decrease of affinity of ect-MMP-14 for the α-1 chain, whereas for the α-2 chain the same effect results from a combination of a fivefold decrease for k cat and a 20-fold decrease of substrate affinity (i.e., an increase of K M, see Figs. 3c, 4c, Tables 2, 3). Finally, the third protonation event brings about a 40-fold increase of the enzymatic activity toward the α-1 chain, which is only due to a 40-fold increase of k cat (see Figs. 3b, 4b, Table 2), whereas the tenfold enhancement of the enzymatic activity toward the α-2 chain is attributable to a 300-fold increase of k cat, partially compensated by a 30-fold decrease of substrate affinity (i.e., an increase of K M, see Figs. 3c, 4c, Table 3).
Discussion
MMP-1, MMP-2, and MMP-14 are valid representatives of three classes of MMPs (namely, of collagenases, gelatinases, and membrane-type metalloproteinases, respectively) and the detailed comparison of their enzymatic activity toward the major natural substrate present in the ECM represents an important contribution to the elucidation of different aspects of the proteolytic mechanism exerted by these MMPs. This is particularly true since no structural information is available on the interaction between collagen I and any of the MMPs
In particular, from the physiological point of view, pH may turn out to be very important for the modulation of enzyme activity under different physiopathological conditions, which can be accompanied by relevant pH changes. It is well known that during pathological processes and in particular in tumor growth there is a significant remodeling of the interstitial space and of the ECM surrounding a tumor, favoring in most cases the tumor growth itself. This process is characterized by a unbalanced degradation of ECM components (such as collagen, laminin, and fibronectin) and it is mediated by MMPs and other matrix-degrading enzymes, secreted by tumor cells and other cell types. The activity of these enzymes is strictly connected to physicochemical parameters, such as pH, which can dramatically change under various conditions. In this sense it is well known that the tumor microenvironment is characterized by a more acid pH (ranging between 5.85 and 7.68 depending on the tumor) than under physiological conditions. These pH changes may also affect the radiation response and the effectiveness of chemotherapy on mammalian cells [25].
However, also under physiological conditions, different body compartments can display drastic pH differences. As an example, it is worth mentioning that in the bloodstream, where many MMPs are secreted by circulating cells (such as neutrophils), and in the ECM (where most MMPs are physiologically operative) the average pH is 7.4 ± 0.1, whereas in joints (where collagen I and collagen II are present) the synovial fluid displays an average pH of 8.1 ± 0.1 [26]. This difference can have a relevant effect for the action of MMPs depending on whether they are functioning in the bloodstream (and/or the ECM) or in specific microenvironments.
In general terms, we can outline that over the pH range investigated (i.e., between pH 6.0 and 9.5) both MMP-1 and MMP-2 appear to enzymatically process preferentially the α-2 chain, whereas MMP-14 does not show any preferential activity toward either of the two types of chains (see Figs. 3a, 4a, Tables 2, 3). However, a closer look at the catalytic parameters of the three MMPs allows one to extract more information on the different molecular aspects of the proteolytic mechanism operative for the three MMPs. The preferential cleavage of the α-2 chain by MMP-1 and MMP-2 is in both cases completely attributable to a much faster rate-limiting cleavage step (i.e., a faster k cat, see Figs. 3b, 4b, Tables 2, 3), since a higher affinity for the α-1 chain is observed for both MMPs (i.e., a lower K M, see Figs. 3c, 4c, Tables 2, 3). This behavior, which is observed for the zero-, one-, and two-protonation levels in the case of MMP-1 and MMP-2, is not detected in the case of ect-MMP-14, where, for the one- and two-protonation levels, a higher affinity for the α-2 chain and a faster k cat for the α-1 chain is instead observed (see Figs. 3, 4, Tables 2, 3). This behavior clearly suggests that binding of the three MMPs involves different topological regions of the collagen I triple helix and occurs through a different interaction mode for the two types of chains. In this respect, it is important to remark as for MMP-2 a close similarity is observed for the values of the three pK U with the α-1 chain of collagen I (see Table 1) and those obtained with synthetic substrates [31], indicating that these proton-linked groups belong unequivocally to the free enzyme and no contribution comes from the interacting substrate. On the other hand, the drastically different values of pK U between the synthetic substrates and the collagen chains observed for MMP-1 and ect-MMP-14 (see Table 1) clearly indicate that in these enzymes the mode of interaction with collagen chains is not regulated by the same residues involved in the reactivity modulation of the enzymatic activity toward synthetic substrates. Since in the case of synthetic substrates the active site of MMP-1 and ect-MP-14 is likely to be the main area responsible for the observed pK a values, this result indeed suggests that the enzymatic activity of MMP-1 and ect-MMP-14 is mostly regulated by exosites, such as the substrate interaction with the hemopexin-like domain and/or additional regions of the catalytic domain and of the hinge portion [40]. Unfortunately, the lack of any detailed structural information on the interaction between macromolecular substrates and this region of both MMP-1 and MMP-14 rules out the possibility of any detailed structural interpretation of this behavior.
The significantly faster rate for the cleavage step in MMP-1 and MMP-2 suggests that these two enzymes interact with regions where the α-2 chain might be more susceptible to being cleaved off (possibly because of a more pronounced unwinding of the triple-helix assembly either intrinsically or as a consequence of the interaction with the enzyme) [13, 34]. On the other hand, the binding of ect-MMP-14 probably involves a region of the triple helix, which, though displaying a fairly high affinity for the enzyme (see Tables 2, 3), does not alter the structural assembly of the triple helix of both the α-1 and the α-2 chain (as also indicated by the lack of CD signal change upon binding of the fully inhibited MMP-14) [13].
An important aspect to be considered is also the conformation of the triple helix of collagen I at different pH values. At pH > 7.5 a relevant decrease is observed for the ellipticity (see Fig. 1a, c), suggesting a progressive relaxation of the triple-helix arrangement of the molecule at alkaline pH values. Interestingly, this proton-linked conformational transition of collagen I is associated in most cases with a relevant decrease of affinity for the enzyme(s), as represented by the increase of K M (see Figs. 3c, 4c). This behavior clearly suggests that in most cases the partial unwinding of the collagen triple helix decreases the affinity for the enzyme, likely because the structural loosening distorts the interaction pocket, rendering less efficient the molecular recognition by the enzyme binding domain. The only exception among the MMPs investigated concerns the interaction between MMP-2 and the α-2 chain of collagen I (see Fig. 4c), where a decrease of K M is observed instead between pH 7.0 and 8.5, underlying the possibility that this interaction has features that are clearly distinct from those of the other substrate binding processes, as also demonstrated by the fact that the pK U values for MMP-2 with the α-2 chain are different from those operative in the interaction with the α-1 chain (see Table 1) and with synthetic substrates [31].
In line with the MMP-2 behavior observed at 25 °C [19], our results indicate that also at 37 °C MMP-2 is a very efficient collagenase. If we compare the three MMPs investigated, we can observe that for the enzymatic processing of the α-1 chain at physiological pH (i.e., pH 7.3) the faster overall rate observed for MMP-2 (only slightly higher than for MMP-1 and about tenfold higher than for MMP-14, see Fig. 3a) is only due to the higher affinity of the interacting domain (likely the fibronectin-like one) of MMP-2 (somewhat higher than that of the hemopexin-like domain of MMP-1 and about 70-fold higher than for MMP-14, see Fig. 3c). On the other hand, k cat for the cleavage of the α-1 chain is significantly higher for MMP-14 (about twice that of MMP-1 and sevenfold faster than for MMP-2, see Fig. 3b), partially compensating for the different substrate affinity.
In the case of the α-2 chain, at pH 7.3 the much higher enzymatic activity of MMP-2 (only slightly faster than for MMP-1, but almost 100-fold higher than for MMP-14, see Fig. 4a) is predominantly related to a faster k cat of MMP-2 (about 100-fold faster than that displayed by MMP-14 and tenfold faster than what we observed for MMP-1, see Fig. 4b), whereas the substrate affinity is very similar for MMP-2 and MMP-14 and only about fivefold higher for MMP-1 (Fig. 4c).
As a whole, we can summarize these data at physiological pH (i.e., at pH 7.4), stating as a working hypothesis that the enzymatic processing of the α-1 chain appears to be mainly regulated by K M, through a modulation of the binding with different types of MMPs, whereas the enzymatic processing of the α-2 chain is mostly controlled by variations of k cat. This different mode of controlling the enzymatic activity on the different components of the collagen I triple helix implies that (1) the α-1 chain discriminates among various MMPs through the binding region and/or the binding mode, keeping unchanged the overall tertiary structure of the chain (and thus its susceptibility to cleavage by various MMPs), and (2) the α-2 chain interacts quite similarly with different MMPs, though undergoing different ligand-linked conformational changes, which bring about a MMP-dependent unwinding of the helical arrangement, exposing to a different extent the chain to the proteolytic cleavage. As previously reported, the reciprocal structural flexibility between the hemopexin-like domain and the catalytic domain seems to be a general property of all MMPs [41]. In line with these findings, different levels of conformational freedom seem to be very important, as indicated by different effects on K M (mostly regulated by the interaction with exosites) and on k cat (mostly regulated by the catalytic domain) of the proteolytic processing of different chains of collagen I. As also observed for MMP-9 [42], we might postulate, as a working hypothesis at this stage, that the two types of chains of collagen I have different flexibility and this feature, associated with the different intramolecular flexibility of various domains, modulates the MMP-specific enzymatic properties against collagen I.
Although the lack of structural information on the interaction between collagen I and MMPs impairs a detailed identification of residues involved in the recognition mechanism, this proton-linked behavior may turn out very useful to predict the enzymatic activity of MMPs in different environmental conditions. Therefore, at physiological pH values (i.e., 7.4 in the bloodstream and 8.1 in the synovial fluid) the overall proteolytic activity of the three MMPs is significantly different, being (1) on the α-1 chain much higher for MMP-2 with respect to the other MMPs in the bloodstream and very similar in the synovial fluid (see Fig. 3a) and (2) on the α-2 chain similar for MMP-1 and MMP-2 both in the bloodstream and in the synovial fluid, but much lower for ect-MMP-14 (see Fig. 4a). Furthermore, on the basis of the pH dependence of the proteolytic function and of the fact that inflammatory processes induce an acidification, lowering the pH of the bloodstream to 7.1 ± 0.1 and of the synovial fluid to 7.5 ± 0.1 [26], we can predict that under inflammatory conditions (1) the MMP-1 collagenolytic activity is decreased on both collagen I chains in the bloodstream, but it prefers the α-1 chain degradation in the synovial fluid (see Figs. 3a, 4a), (2) MMP-2 preferentially degrades the α-1 chain both in the bloodstream and in the synovial fluid (see Figs. 3a, 4a), and (3) the overall activity of ect-MMP-14 is decreased on both chains in the bloodstream as well as in the synovial fluid (see Figs. 3a, 4a).
In conclusion, we can say that these MMPs are modulated in a different fashion by pH compartmentalization, envisaging the possibility that their enzymatic activity toward the two α chains can be finely tuned in various body compartments, and allowing them to play differing roles in different environmental conditions. All these data confirm the complexity of the system, where different environmental conditions (which may occur in vivo) affect the functional behavior of these MMPs toward an important natural substrate, such as collagen I. In this sense, both the substrate and the enzyme(s) show a different way of interacting, strictly dependent on where and when this interaction happens. Therefore, this complexity justifies a deeper analysis of the biochemical parameters, as attempted in this work, and this information should also be helpful for understanding why it is often difficult to achieve a successful therapeutic strategy based on the inhibition of the catalytic activity of MMPs.
Abbreviations
- CD:
-
Circular dichroism
- ECM:
-
Extracellular matrix
- ect-MMP-14:
-
Ectodomain of matrix metalloproteinase 14
- MCA-1:
-
(7-Methoxycoumarin-4-yl)acetyl-Pro-cyclohexylalanine-Gly-norvaline-His-Ala-(N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl)-NH2
- MCA-MMP14:
-
(7-Methoxycoumarin-4-yl)acetyl-Pro-Leu-Ala-Cys(p-OMeBz)-Trp-Ala-Arg(N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl)-NH2
- MCA-omni:
-
(7-Methoxycoumarin-4-yl)acetyl–Pro-Leu-Gly-Leu-(N-3-(2,4-dinitrophenyl)-l-2,3-diaminopropionyl)-Ala-Arg-NH2
- MMP:
-
Matrix metalloproteinase
- PAGE:
-
Polyacrylamide gel electrophoresis
- SDS:
-
Sodium dodecyl sulfate
- Tris:
-
Tris(hydroxymethyl)aminomethane
References
Nagase H, Woessner JF Jr (1999) J Biol Chem 274:21491–21494
Sternlicht MD, Werb Z (2001) Annu Rev Cell Dev Biol 17:463–516
Aureli L, Gioia M, Cerbara I, Monaco S, Fasciglione GF, Marini S, Ascenzi P, Topai A, Coletta M (2008) Curr Med Chem 15:2192–2222
Matrisian LM, Bowden GT, Krieg P, Furstenberger G, Briand JP, Leroy P, Breathnach R (1986) Proc Natl Acad Sci USA 83:9413–9417
Hirose T, Riefe RA, GN Smith Jr, Stevens RM, Mainardi CL, Hasty KA (1992) J Rheumatol 19:593–599
Bafetti LM, Young TN, Itoh Y, Stack MS (1998) J Biol Chem 273:143–149
Vu TH, Shipley JM, Bergers G, Berger JE, Helms JA, Hanahan D, Shapiro SD, Senior RM, Werb Z (1998) Cell 93:411–422
Opdenakker G, Nelissen I, van Damme J (2003) Lancet Neurol 2:747–756
Patterson ML, Atkinson SJ, Knäuper V, Murphy G (2001) FEBS Lett 503:158–162
Gioia M, Fasciglione GF, Marini S, D’Alessio S, De Sanctis G, Diekmann O, Pieper M, Politi V, Tschesche H, Coletta M (2002) J Biol Chem 277:23123–23130
Tam ER, Moore TD, Butler GS, Overall CM (2004) J Biol Chem 279:43336–43344
Xu X, Wang Y, Lauer-Fields JL, Fields GB, Steffensen B (2004) Matrix Biol 23:171–181
Gioia M, Monaco S, Fasciglione GF, Coletti A, Modesti A, Marini S, Coletta M (2007) J Mol Biol 368:1101–1113
Ottani V, Marini D, Franchi M, Ruggeri A, Raspanti M (2002) Micron 33:587–596
Orgel JPRO, Irving TC, Miller A, Wess TJ (2006) Proc Natl Acad Sci USA 103:9001–9005
Prockop DJ, Kivirikko KI (1995) Annu Rev Biochem 64:403–434
Murphy G, Allan JA, Willenbrock F, Cockett MI, O’Connell JP, Docherty AJP (1992) J Biol Chem 267:9612–9618
Hirose T, Patterson C, Pourmotabbed T, Mainardi CL, Hasty KA (1993) Proc Natl Acad Sci USA 90:2569–2573
Aimes RT, Quigley JP (1995) J Biol Chem 270:5872–5876
Monaco S, Sparano V, Gioia M, Sbardella D, Di Pierro D, Marini S, Coletta M (2006) Protein Sci 15:2805–2815
Wilson CL, Matrisian LM (1996) Int J Biochem Cell Biol 28:123–136
Marchenko GN, Ratnikov BI, Rozanov DV, Godzik A, Deryugina EI, Strongin AY (2001) Biochem J 356:705–718
Ohuchi I, Imai K, Fujii Y, Sato H, Seiki M, Okada Y (1997) J Biol Chem 272:2446–2451
Marini S, Fasciglione GF, De Sanctis G, D’Alessio S, Politi V, Coletta M (2000) J Biol Chem 275:18657–18663
Wike-Hooley JL, Haveman J, Reinhold RS (1984) Radiother Oncol 2:343–366
Jebens EH, Monk-Jones ME (1959) J Bone Joint Surg 41B:388–400
Stack MS, Gray RD (1989) J Biol Chem 264:4277–4281
Stack MS, Gray RD (1990) Arch Biochem Biophys 281:257–263
Stein RL, Izquierdo-Martin M (1994) Arch Biochem Biophys 308:274–277
Holman CM, Kan CC, Gehring MR, van Wart HE (1999) Biochemistry 38:677–681
Fasciglione GF, Marini S, D’Alessio S, Politi V, Coletta M (2000) Biophys J 79:2138–2149
Leikina E, Mertts MV, Kuznetsova N, Leikin S (2002) Proc Natl Acad Sci USA 99:1314–1318
Laemmli UK (1970) Nature 227:680–683
Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H (2004) EMBO J 23:3020–3030
Chandrakasan G, Torchia DA, Piez KA (1976) J Biol Chem 251:6062–6067
Wallon UM, Overall CM (1997) J Biol Chem 272:7473–7481
Bradford M (1976) Anal Biochem 72:248–254
Knight CG, Willenbrock F, Murphy G (1992) FEBS Lett 296:263–266
Lauer-Fields JL, Tuzinski KA, Shimokawa K, Nagase H, Fields GB (2000) J Biol Chem 275:13282–13290
Tsukada H, Pourmotabbed T (2002) J Biol Chem 277:27378–27384
Bertini I, Fragai M, Luchinat C, Melikian M, Mylonas E, Sarti N, Svergun DI (2009) J Biol Chem 284(19):12821–12828
Rosenblum G, Van den Steen PE, Cohen SR, Grossmann JG, Frenkel J, Sertchook R, Slack N, Strange RW, Opdenakker G, Sagi I (2007) Structure 15:1227–1236
Acknowledgments
This work was financially supported by the Italian Space Agency (ASI 2005 OSMA to U.T. and M.C.). The authors are grateful to Hideaki Nagase and Tayebeh Pourmotabbed for very fruitful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Gioia and G. F. Fasciglione contributed equally to the work.
Rights and permissions
About this article
Cite this article
Gioia, M., Fasciglione, G.F., Monaco, S. et al. pH dependence of the enzymatic processing of collagen I by MMP-1 (fibroblast collagenase), MMP-2 (gelatinase A), and MMP-14 ectodomain. J Biol Inorg Chem 15, 1219–1232 (2010). https://doi.org/10.1007/s00775-010-0680-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-010-0680-8