Abstract
Methyl-coenzyme M reductase (MCR) catalyses the reduction of methyl-coenzyme M (CH3-S-CoM) with coenzyme B (HS-CoB) to methane and CoM-S-S-CoB. It contains the nickel porphyrinoid F430 as prosthetic group which has to be in the Ni(I) oxidation state for the enzyme to be active. The active enzyme exhibits an axial Ni(I)-derived EPR signal MCR-red1. We report here on experiments with methyl-coenzyme M analogues showing how they affect the activity and the MCR-red1 signal of MCR from Methanothermobacter marburgensis. Ethyl-coenzyme M was the only methyl-coenzyme M analogue tested that was used by MCR as a substrate. Ethyl-coenzyme M was reduced to ethane (apparent K M=20 mM; apparent V max=0.1 U/mg) with a catalytic efficiency of less than 1% of that of methyl-coenzyme M reduction to methane (apparent K M=5 mM; apparent V max=30 U/mg). Propyl-coenzyme M (apparent K i=2 mM) and allyl-coenzyme M (apparent K i=0.1 mM) were reversible inhibitors. 2-Bromoethanesulfonate ([I]0.5 V=2 µM), cyano-coenzyme M ([I]0.5 V=0.2 mM), 3-bromopropionate ([I]0.5 V=3 mM), seleno-coenzyme M ([I]0.5 V=6 mM) and trifluoromethyl-coenzyme M ([I]0.5 V=6 mM) irreversibly inhibited the enzyme. In their presence the MRC-red1 signal was quenched, indicating the oxidation of Ni(I) to Ni(II). The rate of oxidation increased over 10-fold in the presence of coenzyme B, indicating that the Ni(I) reactivity was increased in the presence of coenzyme B. Enzyme inactivated in the presence of coenzyme B showed an isotropic signal characteristic of a radical that is spin coupled with one hydrogen nucleus. The coupling was also observed in D2O. The signal was abolished upon exposure of the enzyme to O2. 3-Bromopropanesulfonate ([I]0.5 V=0.1 µM), 3-iodopropanesulfonate ([I]0.5 V=1 µM), and 4-bromobutyrate also inactivated MCR. In their presence the EPR signal of MCR-red1 was converted into a Ni-based EPR signal MCR-BPS that resembles in line shape the MCR-ox1 signal. The signal was quenched by O2. 2-Bromoethanesulfonate and 3-bromopropanesulfonate, which both rapidly reacted with Ni(I) of MRC-red1, did not react with the Ni of MCR-ox1 and MCR-BPS. The Ni-based EPR spectra of both inactive forms were not affected in the presence of high concentrations of these two potent inhibitors.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Methyl-coenzyme M reductase (MCR) is a nickel enzyme found in all methanogenic archaea [1], in which it catalyses the methane forming step [2]. The nickel is bound within the prosthetic group F430, which is a nickel porphyrinoid. The active enzyme contains F430 in the Ni(I) oxidation state, as evidenced by EPR and UV/Vis spectroscopy [3, 4, 5]. It catalyses the reduction of methyl-coenzyme M (CH3-S-CoM) with coenzyme B (HS-CoB) to methane (CH4) and the heterodisulfide CoM-S-S-CoB (reaction 1):
Indirect evidence is available that MCR is also involved in the anaerobic oxidation of methane [6].
The crystal structure of the inactive MCR [with F430 in the Ni(II) oxidation state] from Methanothermobacter marburgensis has been resolved to 1.16 Å [7, 8, 9, 10]. The α2β2γ2 heterohexamer of molecular mass 300 kDa forms two separate active sites with F430 as prosthetic group deeply buried within the protein and accessible from the outside only via a 50-Å-long narrow channel, through which methyl-coenzyme M can diffuse to the bottom. The channel is sufficiently deep that the terminal thiol group of the heptanoyl arm of coenzyme B remains 8 Å from the Ni. Once coenzyme B is bound, methyl-coenzyme M can no longer enter the channel. Methyl-coenzyme M binding appears to facilitate that of coenzyme B, thus ensuring that methyl-coenzyme M enters first [9, 10].
Active MCR exhibits a characteristic axial Ni(I)-derived EPR signal designated MCR-red1 [3, 11, 12, 13]. In the absence of substrates this signal is referred to as MCR-red1a (a for absence) [5]. In this state the signal shows hyperfine splitting due to the interaction of the electron spin (S=1/2) with the nuclear spins (I=1) of the four nitrogens of the tetrapyrrole ring system. The active enzyme in the presence of the substrate methyl-coenzyme M is referred to as MCR-red1m (m for methyl-coenzyme M). The corresponding EPR signal shows improved resolution in comparison to MCR-red1a. Active MCR in the presence of coenzyme M, which inhibits the enzyme competitively to methyl-coenzyme M, is referred to as MCR-red1c (c for coenzyme M). Its EPR signal shows less resolved hyperfine splitting than MCR-red1m but is otherwise unchanged. When both coenzyme M and coenzyme B are present, the axial red1 signal is partially and reversibly converted into a strongly rhombic Ni(I)-derived EPR signal designated MCR-red2. The red2 signal but not the red1 signal broadens characteristically when H33S-CoM is employed, indicating coordination of the thiol group of coenzyme M to the Ni(I) center of F430 in the presence of coenzyme B [14, 15]. Apparently, binding of coenzyme B to active MCR induces a conformational change which is required for the Ni(I) to be able to interact with the thiol group of coenzyme M, and a corresponding change is assumed to be required for Ni(I)F430 to react with methyl-coenzyme M. Methyl-coenzyme M is not reduced to methane by active MCR in the absence of coenzyme B, as shown in a single turnover experiment [5, 16]. Also, free reduced F430 pentamethyl ester does not react with methyl-coenzyme M [17]. These findings indicate that either F430 or the substrate must be somehow activated for the reduction to occur. It was shown that methylsulfonium ions such as S-methyl methyl-coenzyme M [18] are readily reduced to methane by free Ni(I)F430 pentamethyl ester, and photolytically generated Ni(I)/thiyl radical pairs reacted with methyl thioether functions to give disulfides and methane [19, 20]. These reactions could be considered as models of substrate activation.
Presently, two catalytic mechanism for the enzymatic conversion of methyl-coenzyme M to methane and CoM-S-S-CoB are favored. Mechanism I assumes that the first step in the catalytic cycle is a nucleophilic attack by Ni(I) on the methyl group of methyl-coenzyme M, yielding methyl-Ni(III), which reacts with the coenzyme M thiolate to give methyl-Ni(II) and the coenzyme M thiyl radical. In this mechanism, methane is generated by protonolysis of methyl-Ni(II) in an electrophilic substitution reaction [7, 9, 10]. Mechanism II assumes that the first step in the catalytic cycle is the attack by Ni(I) on the thioether sulfur of methyl-coenzyme M, yielding a free methyl radical which reacts with the thiol group of coenzyme B to give methane and the coenzyme B thiyl radical [21, 22, 23]. In favor of mechanism I is that it can explain the observed inversion of stereoconfiguration in ethyl-coenzyme M reduction to ethane [24] and that it involves a methyl-Ni(II) intermediate, which has been shown to be an intermediate in the reduction of activated methyl thioethers to methane by free reduced F430 pentamethyl ester [18, 25]. Arguments against mechanism I are that, at least in solution, Ni(I) in F430 is not a strong enough nucleophile to attack the methyl group of unactivated methyl-coenzyme M in a nucleophilic substitution reaction [17], and that it is unknown whether methyl-Ni(III) is a strong enough oxidant to oxidize the coenzyme M thiolate to the thiyl radical. In favor of mechanism II is mainly that the calculated energy profile, especially that of the first step, appears to be much more favorable than that calculated for mechanism I [21, 22, 23]. An argument against mechanism II is that inversion of stereoconfiguration (as observed in the case of ethyl-coenzyme M) would require hydrogen abstraction by the intermediate methyl radical before it has time to rotate inside the active site. Although Ahn et al. [24] observed only ca. 60% of the maximal enantiomeric excess possible on the basis of the optical purity of their starting material, it is difficult to judge whether the enzyme reaction did indeed in part proceed under racemization or if the partial loss of enantiomeric purity was due to the very harsh conditions needed to convert the resulting ethane to acetic acid.
The oxidation state of Ni in the enzymatically inactive MCR-ox states, which like MCR-red1 exhibit a Ni-based EPR signal, is still an open question. The MCR-ox states are generated from MCR-red2 by the addition of polysulfide (MCR-ox1), sulfite (MCR-ox2) or O2 (MCR-ox3) [26]. EPR and ENDOR results have been interpreted as indicating a Ni(I) oxidation state for the MCR-ox states [27, 28] and Raman data led to the proposal that MCR-red1 and MCR-ox1 differ in the reduction state of the porphyrinoid ligand system rather than in the Ni oxidation state [27, 28, 29, 30]. However, this hypothesis is inconsistent with recently published redox titrations [31], optical/MCD spectroscopic data and density function calculations [32].
In this work we have probed the reactivity of Ni in MCR-red1 and MCR-ox1 with analogues of methyl-coenzyme M to further our understanding of the catalytic mechanism of MCR and of the oxidation state of Ni in the inactive MCR-ox states. The reaction of ethyl-coenzyme, allyl-coenzyme M, 2-bromoethanesulfonate (BES) and 3-bromopropanesulfonate (BPS) with MCR-red1 and MCR-ox1 in the absence and presence of coenzyme B are described in greater detail.
Material and methods
Methanothermobacter marburgensis (Methanobacterium thermoautotrophicum, strain Marburg [33]) is the strain deposited under DSM 2133 in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig). Coenzyme M (2-mercaptoethanesulfonate) was obtained from Merck (Darmstadt); methyl-coenzyme M was synthesized from coenzyme M by methylation with methyl iodide (Fluka) [5, 34]. According to the same protocol, ethyl-coenzyme M and propyl-coenzyme M were synthesized from their corresponding iodides. Coenzyme B [N-(7-mercaptoheptanoyl)threonine phosphate] was prepared from the symmetric disulfide CoB-S-S-CoB by reduction with NaBH4 as previously described [35, 36]. 2-Bromoethanesulfonic acid (BES) and 3-bromopropionic acid were obtained from Fluka and 4-bromobutyric acid from Aldrich. 3-Bromopropanesulfonate (BPS), 3-fluoropropanesulfonate and 3-iodopropanesulfonate were from the same preparations used by Rospert et al. [37].
Purification of active MCR
M. marburgensis was grown at 65 °C in a 13-L glass fermenter (New Brunswick) containing 10 L mineral medium stirred at 1200 rpm and gassed with 80% H2/20% CO2/0.1% H2S at a rate of 1200 mL/min [5]. When a ΔOD578 of 4.5 was reached, the gas supply was switched to 100% H2 for 30 min to induce the EPR signals MCR-red1 and -red2 in the cells. After 30 min the cells were cooled to 10 °C within 10 min under continuous gassing and harvested anaerobically by centrifugation using a flow-through centrifuge (Hettich, centrifuge 17 RS). Approximately 70 g of wet cells were obtained. From these cells, only the MCR isoenzyme I was purified [38, 39]. All steps of the purification were performed in the presence of 10 mM coenzyme M and in an anaerobic chamber (Coy Instruments) filled with 95% N2/5% H2 as described previously [5]. During purification the enzyme lost its MCR-red2 signal due to the removal of coenzyme B. In one purification, generally 150 mg active MCR in the red1c state (in 120 mL) were obtained. The spin concentration per mol F430 was higher than 0.8.
To obtain MCR-red1a the purified MCR-red1c was washed free of coenzyme M with 50 mM Tris/HCl (pH 7.6) by ultrafiltration with Amicon Ultra Centrifugal Filter Devices with a 100 kDa cut-off (Millipore, Bedford, Mass.). The spin concentration per mol F430 generally decreased to values between 0.5 and 0.8 during the washing procedure. MCR-red1a was converted into MCR-red1m by addition of methyl-coenzyme M to a final concentration of 10 mM.
The protein concentration was determined by using the method of Bradford [40] with bovine serum albumin (Serva) as standard or by measuring the absorbance difference of oxidized enzyme (MCR-silent) at 420 nm using ε=44,000 M−1 cm−1 for a molecular mass of 280 kDa. Both methods yielded almost the same results.
MCR activity determination
Methyl-coenzyme M reductase activity was determined by following methane formation at 65 °C gas chromatographically. The assays were performed in 8-mL serum bottles containing 0.4 mL assay solution and closed with a rubber stopper. Two different assay solutions were used. The DTT (dithiothreitol) assay solution was composed of 50 mM potassium phosphate (pH 7.6), 5 mM methyl-coenzyme M, 1 mM HS-CoB, 10 mM DTT and 10–200 µg of MCR. The Ti(III)/B12 assay solution was composed of 50 mM potassium phosphate (pH 7.6), 10 mM methyl-coenzyme M or ethyl-coenzyme M, 0.5 mM CoB-S-S-CoB, 10 mM Ti(III) citrate, 0.3 mM hydroxycobalamin and 10–200 µg of MCR. The gas phase was in both assays 95% N2/5% H2. The reaction was started by the addition of MCR or by raising the temperature from 4 °C to 65 °C. At intervals of 0.5 to 3 min, 0.2 mL gas samples were withdrawn and analyzed for methane and ethane by gas chromatography [3, 39].
EPR spectroscopy
Samples (0.35 mL) were analyzed for EPR spectra at 77 K in 0.3-cm (inner diameter) quartz tubes with 95% N2/5% H2 as the gas phase and sealed with a closed-off rubber tube. The samples contained at least 1–4.7 mg MCR (3.6–17 nmol) in 10 or 50 mM Tris/HCl (pH 7.6). EPR spectra at X-band (9.4 GHz) were obtained with a Bruker EMX-6/1 EPR spectrometer comprising the EMX 1/3 console, ER 041 X6 bridge with built-in ER-0410-116 microwave frequency counter, ER-070 magnet and ER-4102st standard universal rectangular cavity. All spectra were recorded with a field modulation frequency of 100 kHz. Cooling of the sample was performed with liquid nitrogen in a finger Dewar at 77 K.
EPR spin quantitations were carried out under non-saturating conditions using 10 mM copper perchlorate as the standard (10 mM CuSO4, 2 M NaClO4, 10 mM HCl). All signal intensities are expressed as spin per mol F430. The programs of S.P.J. Albracht were used for computer simulations of the EPR signals [41].
Syntheses of allyl-coenzyme M, trifluoromethyl-coenzyme M, cyano-coenzyme M and seleno-coenzyme M
Sodium 2-(allysulfanyl)ethanesulfonate (allyl-coenzyme M)
Sodium 2-mercaptoethanesulfonate (1.64 g, 10 mmol) was dissolved under N2 at 0 °C in a solution of sodium methoxide [from 10 mL of dry methanol and 252 mg (11 mmol) of sodium metal]. Under vigorous stirring, allyl iodide (1.68 g, 10 mmol) was added dropwise at 0 °C and the resulting precipitate was resuspended by adding another 5 mL of methanol. The reaction mixture was allowed to warm to room temperature, stirred overnight and evaporated to dryness. The product was isolated by repeated precipitation from 1–2 mL H2O with acetone (3×, until a test for iodide with AgNO3 was negative) to give 450 mg (23%, mp >200 °C) of sodium 2-(allylsulfanyl)ethanesulfonate. 1H NMR (DMSO-d 6, 400 MHz): δ 5.67 (ddt, J=7.1, 9.9, 17.0 Hz, 1H), 5.05 (dd, J=1.4, 17.0 Hz, 1H), 4.98 (d, J=9.9 Hz, 1H), 3.06 (d, J=7.1 Hz, 2H), 2.57 (s, 4H). 13C NMR (DMSO-d 6, 125 MHz): δ 135.0, 117.5, 51.9, 34.1, 26.2. MS (ESI-Q1, m/z): monoisotopic mass calc. for [C5H9O3S2]−: 181.00; found: 180.7 (M−, 100%), 181.7 (M−, 8), 384.8 (M2Na−, 52), 385.8 (M2 −Na, 10), 792.8 (M4Na3 −, 10), 793.8 (M4Na3 −, 4), 996.6 (M5Na4 −, 6), 997.6 (M5Na4 −, 4). Elemental analysis (C, H, S): calc. for C5H9NaO3S2 (204.25 g mol−1): C 29.40, H 4.44, S 31.40; found: C 29.24, 29.32, H 4.45, 4.67, S 31.
Ammonium 2-(trifluoromethylsulfanyl)ethanesulfonate (trifluoromethyl-coenzyme M)
Sodium 2-mercaptoethanesulfonate (4.1 g, 25 mmol) was placed in a jacket-cooled photolysis apparatus under N2, and ca. 90 mL of liquid NH3 was condensed in at −45 °C. Trifluoroiodomethane was condensed into a Schlenk flask at −75 °C and 3.1 mL (1.6 equiv, 40 mmol) was rapidly transferred into the photolysis apparatus with a precooled (−80 °C) pipette. The resulting homogeneous mixture was irradiated with a central high-pressure mercury lamp for 4.5 h (negative Ellman test). The ammonia was allowed to evaporate overnight, the residue was dissolved in 5 mL H2O and repeatedly precipitated from acetone until a test for iodide was negative (4.71 g). The mixed Na+/NH4 + salt (3.0 g) was converted into the ammonium form by acidification of a solution in 20 mL H2O with 7 mL of amberlite IR-120 (H+ form) to pH 1, filtration, addition of conc. aq. NH3 to reach pH 12, and lyophilization (2.71 g, 64% overall yield). 1H NMR (DMSO-d 6, 400 MHz)*: δ 7.07 (s, 4NH), 3.15 (2H), 2.77 (2H); AA′BB′ system: J AA=J BB=−13.0 Hz, J AB1=J AB3=6.1 Hz, J AB2=J AB4=11.25 Hz. 13C NMR (DMSO-d 6, 100 MHz): 133.6 (q, J=3.0 Hz), 53.6 (s), 26.8 (s). 19F NMR (CD3OD, PhCF3, 282 MHz): −41.8 (s). MS (ESI-Q1, m/z): monoisotopic mass calc. [C3F3H4O3S2]−: 208.96; found: 208.8 (M−, 100%), 209.9 (M−, 4), 210.8 (M−, 10), 440.8 (M2 −, 8), 442.8 (M2 −, 1). Elemental analysis (C, H, N, F, S: calc. for C3H8F3NO3S2 (227.23 g mol−1): C 15.99, H 3.55, F 25.08, N 6.16, O 21.12, S 28.22; found: C 15.99, H 3.59, F 25.23, N 5.96, O 21.12, S 28.41.
Sodium 2-thiocyanatoethanesulfonate (cyano-coenzyme M)
Sodium 2-bromoethanesulfonate (614.1 mg, 2.91 mmol) and potassium thiocyanate (291.5 mg, 3.0 mmol) were suspended in dry DMF (12 mL) and stirred at 120 °C for 4 h. The solvent was evaporated under vacuum and the residue dissolved in 10 mL H2O. The product was converted into the ammonium form by acidification with amberlite IR-120 (H+ form) to pH 1, filtration, addition of conc. aq. NH3 to reach pH 12, and lyophilization. The resulting solid was suspended in 100 mL of diethyl ether, stirred for 24 h and filtered. Recrystallization from methanol gave 160 mg (29%) of pure product. 1H NMR (CD3OD, 400 MHz): δ 3.38 (2H), 3.21 (AA′BB′ system, J AA=−16.3, J BB=−15.4, J AB1=5.3, J AB2=11.3, J AB3=11.1, J AB4=5.1 Hz, 2H). 1H NMR (D2O, 300 MHz): δ 3.23 (2H), 3.18 (2H, AA′BB′ system, J AA=−10.3, J BB=−11.9, J AB1=7.5, J AB2=J AB3=4.7, J AB4=8.1 Hz). 13C NMR (CD3OD, 125 MHz): δ 113.4, 52.4, 30.3. 13C NMR (D2O, 75 MHz): δ 113.6, 50.0, 27.7. MS (ESI-Q1, m/z): monoisotopic mass calc. for [C3H4NO3S2]−: 165.96; found: 165.7 (M−, 100%), 167.5 (M−, 10).
Disodium 2-selenolatoethanesulfonate (seleno-coenzyme M)
All operations were carried out under N2 with solvents that had been degassed by three freeze–thaw cycles. To a solution of NaBH4 (606 mg, 16 mmol) in 5 mL H2O kept at 4 °C, selenium metal (3×300 mg, 3×3.8 mmol) was carefully added in three portions. After 10 min, a solution of sodium 2-bromoethanesulfonate (1.60 g, 7.6 mmol) in H2O (5 mL) was added dropwise and the resulting solution was stirred overnight at 25 °C. The solvent was removed by lyophilization, the residue was ground into a fine powder, washed with ethanol (200 mL) and dried under vacuum, giving 1.43 g (45%) of product. 1H NMR (D2O/K2DPO4/NaOD, pH 7.1, 500 MHz): δ 3.26 (2H), 2.93 (2H), AA′BB′ system. 13C NMR (D2O/K2DPO4/NaOD, pH 7.1, 75 MHz): δ 54.7, 19.1 (t, J CSe=0.26 Hz). 77Se NMR (D2O/K2DPO4/NaOD, pH 7.1, 93 MHz): δ 201.9 (s). MS (ESI-Q1, m/z): monoisotopic mass calc. for [C2H5O3SSe]−: 188.91; found: 185.2 (M−, 20%), 186.1 (M−, 52), 187.0 (M−, 93), 188.0 (M−, 100), 189.0 (M−, 46).
Results
M. marburgensis grows at 65 °C on 80% H2/20% CO2 with a doubling time of approximately 2 h to a cell concentration of 10 g (wet mass) per L [42] and contains approximately 7 mg MCR per g cells (wet mass) after growth under these conditions [36]. The MCR fraction obtained by ammonium sulfate precipitation of between 70% and 100% consists of two MCR isoenzymes designated MCR I (Mcr) and MCR II (Mrt) [38, 39]. Isoenzyme I predominates (70%) in cells grown under conditions of H2 limitation, which were used for mass cultivation of the organism in 13-L fermenters [43, 44]. The high cell yield and MCR concentration in the cells is the reason why almost all biochemical studies with purified active MCR have been performed with MCR I from M. marburgensis.
The following investigations were performed with isoenzyme I from M. marburgensis, which was purified and separated from isoenzyme II to apparent homogeneity in the active MCR-red1c state [5]. This state was induced in the cells by incubating them after growth under 100% H2 at 65 °C for 30 min before harvest of the cells. During purification, all solutions contained 10 mM coenzyme M since in its absence MCR-red1 is very labile and almost impossible to purify. On the other hand, coenzyme M is a competitive inhibitor to methyl-coenzyme M. Coenzyme M therefore had to be removed before assay of the enzyme. Complete removal was very difficult to achieve without considerable loss in activity. Thus in most of the MCR-red1 preparations used there was still some coenzyme M left.
The presence of some coenzyme M in all of the MCR preparations is the reason why in Table 1 the apparent K M and V max values are given for the substrates and apparent K i values for the reversible inhibitors. Another reason is that it was not always possible to obtain initial velocities since the activity of the enzyme sometimes increased in the first minutes, as shown for the kinetics of ethane formation from ethyl-coenzyme M and coenzyme B in Fig. 1A. In these cases the highest rate reached was used for the graphic determination of K M and V max from Lineweaver–Burk plots and of K i from Dixon plots.
Kinetics of ethyl-coenzyme M reduction to ethane catalyzed by methyl-coenzyme M reductase (MCR) from M. marburgensis. A Kinetics of ethane formation in the presence of (squares) 40 mM, 20 mM, (circles) 10 mM, 6.7 mM, 4 mM and (triangles) 3 mM ethyl-coenzyme M. B Reciprocal plots of the rates versus the ethyl-coenzyme M concentration. The 0.4-mL assay mixture contained 50 mM potassium phosphate (pH 7.6), 0.5 mM CoB-S-S-CoB, 10 mM Ti(III) citrate, 0.3 mM hydroxycobalamin, 0.1 mg of purified MCR-red1c (<0.1 mM coenzyme M) and ethyl-coenzyme M in the concentrations indicated
For the irreversible inhibitors in Table 1, [I]0.5 V values are given, which indicate the concentration of the inhibitor at which 50% of the activity in the absence of the inhibitor was observed under the experimental conditions. These inhibitors were found to inactivate MCR by reacting with the active site Ni(I), as revealed by EPR spectroscopy. Inhibition of methane formation from methyl-coenzyme M by the irreversible inhibitors is the result of both competitive binding and inactivation. Therefore, there are no K i values for these inhibitors.
The specific activity of the preparations was generally near 30 U per mg protein under the standard assay conditions and the spin concentration determined from the EPR signal was above 80%. The standard assay mixture contained hydroxycobalamin and Ti(III) citrate for the continuous reduction of the product CoM-S-S-CoB [45], which is a very potent inhibitor of MCR activity, and for the reductive activation of the enzyme before start of the reaction. In some of the assays, hydroxycobalamin and Ti(III) had to be left out since cobalamins very effectively catalyze the reductive dehalogenation of compounds such as 2-bromoethanesulfonate and 3-bromopropanesulfonate with Ti(III). In the absence of hydroxycobalamin and Ti(III), product formation relatively rapidly leveled off and there was no reductive activation of the enzyme before start of the reaction. Therefore the specific activities were much lower. In Table 1 the footnotes a and b indicate which assays contained cobalamin and Ti(III) and which did not.
Substrate specificity
It is long known that MCR can catalyze the reduction of ethyl-coenzyme M with coenzyme B [46, 47, 48]. We found that ethyl-coenzyme M reduction proceeds with less than 0.1% of the catalytic efficiency of methyl-coenzyme M reduction (Table 1). The apparent K M and V max for methyl-coenzyme M and ethyl-coenzyme M were obtained from kinetic analyses, as shown for ethyl-coenzyme M in Fig. 1A. They were determined with the same enzyme preparation and under the same assay conditions. The only difference was that the enzyme concentration was much higher in the assays for ethyl-coenzyme M reduction. In the kinetic analysis the coenzyme B concentration (1 mM) was not varied since MCR has been shown to have a ternary complex catalytic mechanism [39]; reciprocal plots of the rates versus the methyl-coenzyme M concentration at different constant coenzyme B concentrations were intersecting, with the intersecting point on the abscissa to the right of the ordinate. The K M values obtained are thus independent of the coenzyme B concentration.
The apparent K M for methyl-coenzyme M was found to be 5 mM and the apparent V max to be 30 U/mg (Table 1). In previous studies, a lower K M value for methyl-coenzyme M of 1 mM was obtained [39]. An explanation most probably is that the apparent K M given in Table 1 was determined with MCR-red1 purified in the presence of 10 mM coenzyme M, whereas the lower K M of 1 mM was determined with enzyme preparations partially purified in the presence of 12 mM methyl-coenzyme M. We observed that MCR-red1 purified in the presence of coenzyme M is much more stable than MCR-red1 purified in the presence of methyl-coenzyme M or generated from MCR-ox1 by reduction in the presence of methyl-coenzyme M. At equal spin concentrations, however, the specific activity of MCR-red1c is only half that of MCR-red1m, and this difference in specific activity was not abolished when most of the coenzyme M was removed from the MCR-red1c sample by ultrafiltration and washing [5]. This is still not understood.
We also tested propyl-coenzyme M and allyl-coenzyme M as possible substrates. If they were reduced to propane and propene, respectively, this was at rates below the detection limit of the gas chromatographic method employed. Propyl-coenzyme M (apparent K i=2 mM) and allyl-coenzyme M (apparent K i=0.2 mM) reversibly inhibited methyl-coenzyme M reduction with coenzyme B (Table 1), indicating that they competed with methyl-coenzyme in binding to the active site. Binding to the active site was also evidenced by the finding that when allyl-coenzyme M or propyl-coenzyme M were added to MCR-red1c, the hyperfine splitting became much more pronounced, just as was the case when methyl-coenzyme M or ethyl-coenzyme M were added. This is shown for methyl-coenzyme M and allyl-coenzyme M in Fig. 2A.
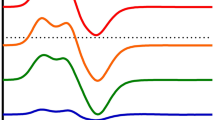
EPR spectra of purified MCR-red1a after addition of coenzyme M, methyl-coenzyme M or allyl-coenzyme M (A) in the absence and (B) presence of coenzyme B and (C) after exposure to air. A (1) MCR-red1c=active methyl-coenzyme M reductase in the presence of 10 mM coenzyme M (g z =2.250; g x,y =2.066); (2) MCR-red1m=active MCR in the presence of 10 mM methyl-coenzyme M (g z =2.252; g x,y =2.068); (3) MCR-red1c after the addition of 10 mM allyl-coenzyme M. B Samples as in A but supplemented with 5 mM coenzyme B. In the case of (1) the rhombic MCR-red2 signal was induced (g z =2.288; g y =2.235; g x =2.179). C Samples as in B but after exposure to air. In the case of (1) the MCR-ox3 signal was induced (g z =2.217; g x,y =2.137). The samples in (2) and (3) contained small amounts of MCR-ox1 signal, which has been subtracted for better comparison. The concentration of purified enzyme in all samples was 4.7 mg (17 nmol) in 0.35 mL 50 mM Tris/HCl (pH 7.6). Where indicated, substrate and/or substrate analogues were added to the samples at room temperature and the samples were incubated at the same temperature for 5 min before freezing in liquid nitrogen. Spectra were recorded under the following conditions: microwave frequency, 9434 MHz; microwave power incident to the cavity, 2.05 mW; temperature, 77 K; modulation amplitude, 0.6 mT
In line 1 of Fig. 2A the EPR spectrum of MCR-red1 in the presence of coenzyme M is given, and in lines 2 and 3 the EPR spectra when additionally methyl-coenzyme M or allyl-coenzyme M were present. Figure 2B shows the spectra of the enzyme supplemented subsequently with coenzyme B, and Fig. 2C shows what happened when the solutions were finally exposed to air. When MCR-red1c was supplemented with coenzyme B, the rhombic MCR-red2 signal was induced (Fig. 2B, line 1), which was converted to the MCR-ox3 signal upon exposure of MCR-red2 to air (Fig. 2C, line 1). In the presence of methyl-coenzyme M (Fig. 2, line 2) or allyl-coenzyme M (Fig. 2, line 3), neither the red2 signal nor the ox3 signal were induced. Upon subsequent exposure of the enzyme solutions to air the MCR-red1 signal was quenched, leaving behind only a small radical signal (Fig. 2C, lines 2 and 3).
The results obtained for allyl-coenzyme M are of special interest. Hydroxycobalamin and Ti(III) were found to chemically reduce allyl-coenzyme to an unidentified gaseous product, which was observed gas chromatographically. It thus could have been that allyl-coenzyme M is reduced by the active site Ni(I). Quenching of the EPR signal was, however, not observed when active MCR was incubated in the presence of allyl-coenzyme M either in the absence or presence of coenzyme B. Allyl-coenzyme M thus behaved quite differently from 2-bromoethanesulfonate and 3-bromopropanesulfonate, which both were also chemically reduced by hydroxycobalamin and Ti(III), but which inactivated MCR as indicated by the quenching of the MCR-red1 signal or conversion of the MCR-red1 signal to the MCR-BPS signal (see below).
Inhibitors that quench the MCR-red 1 signal
2-Bromoethanesulfonate, cyano-coenzyme M, seleno-coenzyme M and trifluoromethyl-coenzyme M belong in this category (Table 1). When added to MCR-red1 in 10 mM concentrations, 2-bromoethanesulfonate quenched the MCR-red1 signal both in the absence and presence of coenzyme B (Fig. 3A and B, lines 1). The three other inhibitors oxidized the active site Ni(I) to Ni(II) only at significant rates in the presence of coenzyme B (Fig. 3B). The rate of inactivation was highest with 2-bromoethanesulfonate and lowest with trifluoromethyl-coenzyme M. The differences in inactivation rates reflected the differences in the [I]0.5 V values (Table 1), but there was no linear correlation.
Inactivation of MCR-red1a by 2-bromoethanesulfonate (BES), cyano-coenzyme M, seleno-coenzyme M and trifluoromethyl-coenzyme M (A) in the absence and (B) in the presence of coenzyme B and (C) after exposure to air. A EPR spectra of MCR-red1a after 5 min incubation in the presence of 10 mM analogues: (1) 2-bromoethanesulfonate; (2) cyano-coenzyme M; (3) seleno-coenzyme M; and (4) trifluoromethyl-coenzyme M. B Samples as in A but supplemented with 5 mM coenzyme B; the radical signal at g=2.008 had a signal intensity of approximately 0.1 spin per mol F430 and showed a resolved hyperfine structure with I=1/2. C Samples as in B but after exposure to air; the radical signal at g=2.008 had a signal intensity of less than 0.1 spin per mol F430. The concentration of purified enzyme in all samples was 4.7 mg (17 nmol) in 0.35 mL 10 mM Tris/HCl (pH 7.6). Sample handling was performed at room temperature. Samples were frozen after 5 min. For EPR conditions see Fig. 2; microwave frequency, 9434 MHz
The finding that the inactivation of MCR-red1 by cyano-coenzyme M, seleno-coenzyme M and trifluoromethyl-coenzyme M was dependent on coenzyme B prompted us to investigate the effect of coenzyme B on the inactivation of MCR-red1 by 2-bromoethanesulfonate at low concentrations of the inhibitor. Indeed, at concentrations of 2-bromoethanesulfonate of 25 µM (Fig. 4A) and 250 µM (Fig. 4B), MCR-red1 inactivation was stimulated by coenzyme B (Fig. 4). The rate of inactivation by 2-bromoethanesulfonate at 25 µM was 2- to 3-fold lower than that at 250 µM, which is consistent with the observed [I]0.5 V of 2 µM (Table 1). In the absence of coenzyme M, which competes with 2-bromoethanesulfonate for binding in the active site, the rate of inactivation was much higher than in its presence (Fig. 4A). Coenzyme M thus protected the enzyme from inactivation by 2-bromoethanesulfonate.
Kinetics of MCR-red1c inactivation by BES in the (open circles) absence or (filled circles) presence of coenzyme B as followed by EPR spectroscopy. A (open triangles) MCR-red1c in the absence of BES; (open circles) MCR-red1c in the presence of 25 µM BES; (filled circles) MCR-red1c in the presence of 5 mM coenzyme B and 25 µM BES; (open squares) MCR-red1a (active MCR in the absence of coenzyme M) in the presence of 25 µM BES. B (open triangles) MCR-red1c in the absence of BES; (open circles) MCR-red1c in the presence of 250 µM BES; (filled circles) MCR-red1c in the presence of 5 mM coenzyme B and 250 µM BES. The experimental conditions were as described in Fig. 2. The concentration of purified enzyme in all samples was 2.5 mg (8.8 nmol) in 0.35 mL. The samples were incubated at room temperature for the times indicated, then frozen in liquid nitrogen and analyzed for EPR signal intensities. The relative signal intensities of the red1 signal were determined by double integration of the EPR signals. The intensity of the MCR-red1 signal in the absence of coenzyme B and BES was taken as 1. The kinetic results labeled with open squares were obtained by following the decrease of absorbance at 385 nm spectrophotometrically
When MCR-red1 was inactivated by 2-bromoethanesulfonate, cyano-coenzyme M, seleno-coenzyme M or trifluoromethyl-coenzyme M in the presence of coenzyme B, a novel radical signal with doublet hyperfine structure was induced (Fig. 3B). Upon exposure of the inactivated enzyme to air, the doublet was replaced by a new radical signal without resolved hyperfine structure (Fig. 3C). The hyperfine splitting of the radical signal is characteristic of coupling to a single hydrogen nucleus (I=1/2). We therefore repeated the experiment with 2-bromoethanesulfonate in D2O. In Fig. 5A, line 1, the EPR spectrum of MCR in H2O after the addition of 2-bromoethanesulfonate is shown and in Fig. 5B, line 1, the respective spectrum of MCR in D2O. The spectra in H2O and D2O before (not shown) and after inactivation by 2-bromoethanesulfonate were almost identical, indicating that, if the hyperfine splitting of the radical signal was derived from spin coupling to a 1H nucleus, this nucleus was not exchangeable by deuterons of D2O.
EPR spectra of MCR-red1c in the presence of coenzyme B after inactivation by BES in (A) H2O or (B) D2O. A (1) MCR-red1c in the presence of 5 mM coenzyme B after the addition of 10 mM BES; (2) the same sample after exposure to air. B (1) MCR-red1c in the presence of 5 mM coenzyme B after the addition of 10 mM BES; (2) the same sample after exposure to air. The concentration of purified enzyme in all samples was 4.7 mg (17 nmol) in 0.35 mL 10 mM Tris/HCl (pH 7.6). Sample handling was performed at room temperature. For EPR conditions, see Fig. 2; microwave frequency, 9434 MHz
3-Bromopropionate was found to also quench the MCR-red1 signal (result not shown). We did not determine whether inactivation by this 2-bromoethanesulfonate analogue was coenzyme B dependent.
Inhibitors that convert the MCR-red1 signal to the MCR-BPS signal
3-Bromopropanesulfonate, 3-iodopropanesulfonate and 4-bromobutyrate belong in this category. All three compounds were found to irreversibly inhibit MCR, most of them at relatively low concentrations (Table 1). 3-Bromopropanesulfonate with an [I]0.5 V<0.1 µM is the most potent inhibitor of MCR known to date [49]. Via titration we found that MCR-red1 is completely inactivated when only twice the molar amount of the inhibitor is added. The EPR spectrum of the enzyme inactivated by 3-bromopropanesulfonate in the absence of coenzyme B is given in Fig. 6A, line 1. The signal, which was first described by Rospert et al. [37], has been designated the MCR-BPS signal. The EPR spectrum after inactivation in the presence of coenzyme B is shown in Fig. 6A, line 2. It differs from the MCR-BPS signal only very slightly in the g values. The MCR signal generated by 3-bromopropanesulfonate in the absence or presence of coenzyme B was quenched upon exposure of the enzyme to O2 (Fig. 6A), but not by 10 mM 2-bromoethanesulfonate (not shown), which had absolutely no effect on the EPR signal. When the enzyme had been inactivated in the presence of coenzyme B, exposure to O2 led to the induction of a novel radical signal (Fig. 6A, line 3). Almost identical spectra were obtained when the enzyme was inactivated by 3-iodopropanesulfonate (not shown) and 4-bromobutyrate (Fig. 6B).
Inactivation of MCR-red1c by (A) 3-bromopropanesulfonate (BPS) and (B) 4-bromobutyrate in the absence and presence of coenzyme B as followed by EPR spectroscopy. A (1) MCR-red1c after the addition of 10 mM BPS; the MCR-BPS signal was induced (g z =2.219; g x,y =2.116); (2) sample as in (1) but supplemented with 5 mM coenzyme B (g z =2.229; g x,y =2.111); (3) sample as in (2) but after exposure to air; a new signal appeared at g z =2.035 and g x,y =2.007. The signal intensity was approximately 0.1 spin per mol F430. B (1) MCR-red1c after the addition of 10 mM 4-bromobutyrate; (2) sample as in (1) but supplemented with 5 mM coenzyme B; (3) sample as in (2) but after exposure to air. The concentration of purified enzyme in all samples was 3.2 mg (11.4 nmol) in 0.35 mL. Sample handling was performed at room temperature. Samples were frozen after 5 min. For EPR conditions, see Fig. 2; microwave frequency: (A) 9272 MHz, (B) 9434 MHz
3-Bromopropanesulfonate has been reported to be a reversible inhibitor [49]. Reversible inhibition was concluded from investigations with enzyme preparations that contained low amounts of enzymatically inactive MCR-ox1, which is not affected by the inhibitor (see below). After removal of the excess 3-bromopropanesulfonate by gel filtration, the enzyme regained some activity. We know now that MCR-ox1 was converted into active enzyme under the reducing assay conditions [26]. That 3-bromopropanesulfonate irreversibly inhibits MCR was already mentioned by Rospert et al. [37].
3-Fluoropropanesulfonate ([I]0.5 V=50 µM) has been reported to also inhibit MCR and to induce the MCR-BPS signal. We found that, for full signal induction, 50 mM 3-fluoropropanesulfonate had to be present. At 10 mM 3-fluoropropanesulfonate, only approximately 30% of MCR-red1 was converted into MCR-BPS and this percentage did not increase during incubation at room temperature for many hours (not shown). The easiest interpretation of these findings is that the 3-fluoropropanesulfonate used was contaminated with minute amounts of 3-bromopropanesulfonate. It was synthesized starting from 3-bromo-1-fluoropropane via sulfitolysis [37].
Effect of the inhibitors on MCR-ox1
MCR-ox1 is an enzymatically inactive form of MCR, which exhibits a nickel-based EPR spectrum. It can be either generated in vivo by “oxidizing” the cells of M. marburgensis with CO2 after growth on 80% H2/20% CO2 [50, 51] or in vitro by “oxidizing” MCR-red1 in the presence of coenzyme M and coenzyme B with polysulfide [26]. MCR-ox1 can also be generated in vivo by incubation of M. marburgensis cells in the presence of high concentrations of sulfide or dithionite [4]. MCR-ox1 is relatively stable under oxic conditions and less stable under reducing conditions, in which it is slowly converted into MCR-red1. It also reacts over 1000-fold slower with chloroform than MCR-red1 [26, 30]. We report here that MCR-ox1 is also relatively stable in the presence of 2-bromoethanesulfonate (Fig. 7A) or 3-bromopropanesulfonate (Fig. 7B, [30]). At 10 mM concentrations of the two inhibitors the intensity of the MCR-ox1 signal only decreased slowly. The slow decrease observed can be explained by a slow conversion of MCR-ox1 into MCR-red1 under the reducing assay conditions (10 mM coenzyme M). This interpretation is supported by the finding that in the presence of 3-bromopropanesulfonate some MCR-BPS was formed after 50 h incubation, as indicated from the EPR spectrum.
EPR spectra of MCR-ox1 before and after the addition of (A) BES and (B) BPS. In black: MCR-ox1 (g z =2.231; g x,y =2.160)=MCR-red1c supplemented with 5 mM coenzyme B and subsequently with 1 mM polysulfide. In gray: MCR-ox1, 2 h after the addition of (A) 10 mM BES or (B) 10 mM BPS. In light gray: the same samples 50 h after the addition of BES or BPS. The concentration of purified enzyme in all samples was 1 mg (3.6 nmol) in 0.35 mL. For EPR conditions, see Fig. 2; microwave frequency, 9434 MHz
It was previously reported that MCR-ox1 is slowly converted into MCR-BPS in the presence of 3-bromopropanesulfonate [30]. This result, which differs from ours, can be explained by the presence of Ti(III) in the assays of [30]. Ti(III) is known to reduce MCR-ox1 to MCR-red1 [3], which we have shown to react with 3-bromopropanesulfonate.
Discussion
In the Results section it was shown that there are three types of methyl-coenzyme M analogues: (1) analogues such as ethyl-coenzyme M and allyl-coenzyme M that are alternate substrates or reversible inhibitors; (2) analogues such as 2-bromoethanesulfonate that irreversibly inhibit the enzyme by oxidation of its active site Ni(I) to Ni(II); and (3) analogues such as 3-bromopropanesulfonate that irreversibly inhibit the enzyme by reacting with the active site Ni(I) to give a state that exhibits a Ni-derived EPR signal similar in line shape to that of the inactive MCR-ox states.
Alternate substrates
MCR was shown to catalyze the reduction of ethyl-coenzyme M to ethane with a catalytic efficiency less than 0.1% of that of methyl-coenzyme M reduction to methane. The large difference in catalytic efficiency was mainly due to differences in V max (0.1 U/mg versus 30 U/mg) and only partly due to differences in K M (20 mM versus 5 mM). In 1978 it was reported that, at a concentration of 10 mM, ethyl-coenzyme M was reduced to ethane at only 20% of the rate of methyl-coenzyme M reduction to methane (2 mU/mg versus 10 mU/mg) [34]. Wackett et al. [47] published an apparent K M for ethyl-coenzyme M of 1.3 mM and a V max of 7.4 mU/mg, to be compared with an apparent K M for methyl-coenzyme M of 0.1 mM and V max of 11 mU/mg. Both results were obtained with cell extract, which was inactive (note the low specific activities) but which was slowly activated under the assay conditions in an ATP- and H2-dependent process. Thus the results cannot really be compared.
In the Introduction it was indicated that presently two catalytic mechanisms are proposed which differ mainly in how methyl-coenzyme M is attacked by the active site’s Ni(I). Mechanism I assumes that the methyl thioether bond is heterolytically cleaved in a nucleophilic substitution reaction at the methyl carbon [9, 10] and mechanism II by a homolytic substitution at sulfur [21, 22, 23]. For maximal reactivity, in mechanism I the methyl group of methyl-coenzyme M has to be positioned above the Ni(I), as shown in Fig. 8A, and in mechanism II the thioether sulfur has to be positioned as shown in Fig. 8B. From Fig. 8 it can be deduced that when R in Fig. 8 is a methyl group as in ethyl-coenzyme M, rather than a hydrogen as in methyl-coenzyme M, the reacting carbon of ethyl-coenzyme M (R=CH3) cannot be positioned above the Ni at the same distance as the reacting carbon of methyl-coenzyme M (R=H), whereas in the case of mechanism II the positioning of the sulfur is not affected by the size of R. When modeled into the crystal structure [7, 9, 10] in this position, there was enough space for the ethyl group. Therefore, mechanism I can easily explain that MCR is less efficient in ethyl-coenzyme M reduction than in methyl-coenzyme M reduction, whereas mechanism II cannot. This interpretation is naturally based on the assumption that the first step in the catalytic cycle is rate determining, which is supported by the calculated energy profiles [22, 23].
Optimal position of methyl-coenzyme M in the active site of MCR assuming (A) catalytic mechanism I [9, 10] and (B) catalytic mechanism II [21, 22, 23]. The long aliphatic arm of coenzyme B can reach into the channel to the extent where its terminal thiol group is still at a distance of 8 Å from the Ni [7]
MCR has been reported to catalyze the reduction of methyl-selenocoenzyme M (K M=0.3 mM; V max=35 mU/mg), difluoromethyl-coenzyme M (K M=2.5 mM; V max=20 mU/mg) and 3-(methylthio)propionate (K M=1.3 mM; V max=1.3 mU/mg) [47]. As seen from the extremely low V max, these results have been obtained with cell extracts containing mainly inactive MCR, which was slowly reactivated under the assay conditions. The data are mentioned here only because they qualitatively show that these substrate analogues can be used as alternative substrates. Interestingly, difluoromethyl-coenzyme M was found to inactivate the enzyme at higher concentrations, and was thus not only an alternative substrate but also an irreversible inhibitor [47]. It could therefore be that MCR also catalyzes the reduction of trifluoromethyl-coenzyme M but is inactivated by the substrate analogue so rapidly that the reduction is overlooked.
Reversible inhibitors
Allyl-coenzyme M was found to be a reversibe inhibitor of MCR (apparent K i=0.2 mM) and not a substrate. The previously reported formation in very low amounts of a gaseous product in cell extracts [52] can be explained by chemical reduction (see Results), since the extracts contain cobalamins. The finding that allyl-coenzyme M is not reduced to propene is in favor of catalytic mechanism I, which predicts that allyl-coenzyme M reduction should be sterically hindered (R too bulky; see Fig. 8A). In the case of mechanism II, allyl-coenzyme M should have been reduced to propene or inactivated the enzyme due to the facile generation of the allyl radical. The same arguments also hold true for the finding that propyl-coenzyme M was not a substrate but a reversible inhibitor.
2-Methoxyethanesulfonate has been reported to be an inhibitor of MCR (apparent K i=8.3 mM) [47]. The reason this analogue is not reduced to methane is probably an energetic rather than a steric one: The C–O bond is much stronger than the C–S bond. This makes the first step more difficult in both the discussed catalytic mechanisms.
Irreversible inhibitors of the 2-bromoethanesulfonate type
2-Bromoethanesulfonate is a well-known inhibitor of MCR and of methanogenesis [34]. In the Results section it was shown that this inhibitor ([I]0.5 V=2 µM) quenched the MCR-red1 signal of active MCR already at very low concentration. The results indicate that the Ni(I) in the active site of MCR-red1 was oxidized to Ni(II) by 2-bromoethanesulfonate.
In solution, 2-bromoethanesulfonate has been shown to react with free Ni(I)F430 to bromide, ethene and sulfite (reaction 2) [53]:
Probably the following reaction sequence takes place:
An analogous fragmentation giving ethene was also observed as a side reaction when Ni(I)F430 pentamethyl ester was reacted with the sulfonium ion S-methyl methyl-coenzyme M [54].
Reaction of F430 in the active site of MCR with 2-bromoethanesulfonate cannot proceed via the same sequence (reactions 3, 4 and 5), since only one Ni(I)F430 is present per active site. Namely, the last step in which the sulfur trioxide anion radical is reduced must be different. The sulfur trioxide anion radical is a strong oxidant that can probably generate other radicals within the active site. Indeed, when MCR-red1 was oxidized by 2-bromoethanesulfonate in the presence of coenzyme B, a radical signal was induced. Its hyperfine structure was characteristic of the electron spin being coupled to the nuclear spin of one hydrogen atom. In D2O the hyperfine splitting remained, indicating that the hydrogen did not exchange with protons of water. When the enzyme was exposed to O2, the split signal was replaced by a new signal without resolved hyperfine structure. Within the active site there is a thioglycine [7, 55] and two tyrosine residues [9, 10] that could be oxidized by the sulfur trioxide anion radical to a thioglycyl radical or a tyrosyl radical [56], respectively. The split EPR signal is most similar to that reported for the glycyl radical in proteins [57, 58]. More detailed studies are needed to make an assignment.
Interestingly, also 3-bromopropionate, cyano-coenzyme M, seleno-coenzyme M and trifluoromethyl-coenzyme quenched the red1 signal and induced the split radical EPR signal when coenzyme B was present, indicating that they in principle all reacted like 2-bromoethanesulfonate.
Quenching of the red1 signal of active MCR by 2-bromoethanesulfonate was more than 10-fold enhanced in the presence of coenzyme B. This effect was most pronounced in the case of trifluoromethyl-coenzyme M, which quenched the EPR signal noticeably only in the presence of coenzyme B (Fig. 3). The coenzyme B dependence is taken as further evidence that coenzyme B has to bind to MCR before the nickel in MCR-red1 becomes reactive enough to react with methyl-coenzyme M or its substrate analogues.
2-Chloroethanesulfonate (apparent K i=70 µM) [34] and 2-azidoethanesulfonate (apparent K i=1 µM) [49] have been reported to be inhibitors of MCR. From their structure they are predicted to belong to the group of irreversible MCR inhibitors headed by 2-bromoethanesulfonate. Bromomethanesulfonate (apparent K i=1.5 µM) [59] and chloromethanesulfonate ([I]0.5 V=0.25 mM) [49] are also predicted to quench the red1 signal of active MCR, but their reduction products will be different from those generated by 2-bromoethanesulfonate reduction.
Irreversible inhibitors of the 3-bromopropanesulfonate type
3-Bromopropanesulfonate is the MCR inhibitor with the by far the lowest [I]0.5 V of <0.1 µM. When this compound reacts with MCR-red1, the enzyme is almost immediately inactivated and its EPR signal converted into the Ni-based EPR signal MCR-BPS (Fig. 6A), which is similar in line shape to that of the MCR-ox signals (Fig. 7). The induction of the signal was not dependent on the presence of coenzyme B. In its presence, almost the same signal was induced, differing, however, only very slightly in the g values. The MCR-BPS signal was quenched when the inactive enzyme was exposed to air, but it was stable when 10 mM 2-bromoethanesulfonate was added, which is quite remarkable considering that 25 µM 2-bromoethanesulfonate are sufficient to very rapidly quench the MCR-red1 signal. Since the stable MCR-BPS signal was induced in the absence of coenzyme B, it cannot be argued that the entrance to the active site was blocked by coenzyme B and thus the active site was not accessible to the 2-bromoethanesulfonate. The results thus indicate that the EPR signal of MCR-BPS must be derived from Ni that can no longer be oxidized by 2-bromoethanesulfonate. This theoretically could be a Ni(III) or a high-spin Ni(II) axially coordinated to a radical [60], which are both resonating structures. Four-coordinated or axially weakly six-coordinated Ni(III) is most certainly excluded since the EPR signal of MCR-BPS by no means resembles that of Ni(III)F430 [61].
High-spin Ni(II)F430 axially coordinated to a radical as shown in Fig. 9 is predicted to exhibit an EPR signal that shows hyperfine splitting due to the coupling with the four nitrogens of the tetrapyrrolic ring system. Probably owing to line broadening, the expected hyperfine splitting is not apparent (Fig. 6). The hyperfine splitting can, however, readily be seen in the MCR-ox signals [26], which are thought to be derived from a high-spin Ni(II) axially coordinated to a thiyl radical [62, 63]. It is therefore proposed that MCR-BPS is generated from MCR-red1 as shown in Fig. 9. Consistent with this interpretation is the finding that 3-iodopropanesulfonate and 4-bromobutyrate also induced the MCR-BPS signal. The three compounds have in common that the radical formed by reduction cannot decompose in the manner described for 2-bromoethanesulfonate. 3-Azidopropanesulfonate ([I]0.5 V=40 µM) and 4-bromobutanesulfonate ([I]0.5 V=6 µM), which have been reported to be strong inhibitors of MCR [49], are predicted to react similarly to 3-bromopropanesulfonate.
When MCR-BPS formed in the presence of coenzyme B was exposed to O2, a radical signal was induced, which differed from that induced by 2-bromoethanesulfonate in the presence of coenzyme B. The O2 induced signal had some structure, which has to be analyzed in detail by high-field EPR and ENDOR spectroscopy before an interpretation is possible.
Conclusions
Most of the results are consistent with catalytic mechanism I, in which the first step in the catalytic cycle is a nucleophilic substitution, yielding methyl-Ni(III). The low catalytic efficiency of ethyl-coenzyme M reduction, the inertness of allyl-coenzyme M as a substrate and the observed inversion of stereoconfiguration are much less easy to explain with the alternative mechanism II, in which methyl-coenzyme M is reduced to a free methyl radical as the first step in the catalytic cycle. However, none of the findings completely exclude mechanism II. The finding that the oxidation of Ni(I) to Ni(II) in the active site of MCR-red1 by 2-bromoethanesulfonate is dependent on coenzyme B shows that, upon coenzyme B binding, the reactivity of the Ni(I) species is increased. This had previously already become evident from the finding that the thiol group of coenzyme M interacts with Ni(I) of MCR-red1 only in the presence of coenzyme B [14]. Because the structural consequences of coenzyme B binding, which lead to the enhanced reactivity, are still unknown, they cannot be taken into consideration in density functional calculations which, so far, constitute the main argument in support of mechanism II [21, 22, 23]. The formation of MCR-BPS from MCR-red1 and 3-bromopropanesulfonate was interpreted by us to proceed in a reaction yielding an alkylated Ni, most probably a high-spin Ni(II) with the alkyl radical as axial ligand (Fig. 9). The reaction can be formulated as nucleophilic substitution or as oxidative addition and could serve as model reaction for the first step in methyl-coenzyme M reduction.
Abbreviations
- BES:
-
2-bromoethanesulfonate
- BPS:
-
3-bromopropanesulfonate
- CH3-S-CoM:
-
methyl-coenzyme M
- HS-CoB:
-
coenzyme B
- HS-CoM:
-
coenzyme M
- MCR:
-
methyl-coenzyme M reductase
- MCR-ox:
-
MCR exhibiting the EPR signals ox1, ox2 or ox3
- MCR-red1:
-
MCR exhibiting the EPR signals red1a, red1c or red1m
- MCR-red1a:
-
MCR-red1c or MCR-red1m after extensive washing by ultrafiltration in the absence of coenzyme M and methyl-coenzyme M
- MCR-red1c:
-
MCR-red1 in the presence of coenzyme M
- MCR-red1m:
-
MCR-red1 in the presence of methyl-coenzyme M
- MCR-red2:
-
MCR exhibiting both the red1 and red2 EPR signals
References
Wolfe RS (2004) ASM News 70:15–18
Thauer RK (1998) Microbiology 144:2377–2406
Goubeaud M, Schreiner G, Thauer RK (1997) Eur J Biochem 243:110–114
Becker DF, Ragsdale SW (1998) Biochemistry 37:2639–2647
Mahlert F, Grabarse W, Kahnt J, Thauer RK, Duin EC (2002a) J Biol Inorg Chem 7:101–112
Krüger M, Meyerdierks A, Glockner FO, Amann R, Widdel F, Kube M, Reinhardt R, Kahnt J, Bocher R, Thauer RK, Shima S (2003) Nature 426:878–881
Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK (1997) Science 278:1457–1462
Grabarse W, Mahlert F, Shima S, Thauer RK, Ermler U (2000) J Mol Biol 303:329–344
Grabarse W, Mahlert F, Duin EC, Goubeaud M, Shima S, Thauer RK, Lamzin V, Ermler U (2001) J Mol Biol 309:315–330
Grabarse W, Shima S, Mahlert F, Duin EC, Thauer RK, Ermler U (2001) Methyl-coenzyme M reductase. In: Messerschmidt A, Huber R, Poulos T, Wieghardt K (eds) Handbook of metalloproteins. Wiley, Chichester, pp 897–914
Jaun B, Pfaltz A (1986) J Chem Soc Chem Commun 1327–1329
Rospert S, Böcher R, Albracht SPJ, Thauer RK (1991) FEBS Lett 291:371–375
Holliger C, Pierik AJ, Reijerse EJ, Hagen WR (1993) J Am Chem Soc 115:5651–5656
Finazzo C, Harmer J, Bauer C, Jaun B, Duin EC, Mahlert F, Goenrich M, Thauer RK, Van Doorslaer S, Schweiger A (2003) J Am Chem Soc 125:4988–4989
Finazzo C, Harmer J, Jaun B, Duin EC, Mahlert F, Thauer RK, Van Doorslaer S, Schweiger A (2003) J Biol Inorg Chem 8:586–593
Horng YC, Becker DF, Ragsdale SW (2001) Biochemistry 40:12875–12885
Jaun B (1993) Methane formation by methanogenic bacteria: redox chemistry of coenzyme F430. In: Sigel H, Sigel A (eds) Metal ions in biological systems, vol XXXX. Dekker, New York, pp 287–337
Lin S-K, Jaun B (1992) Helv Chim Acta 75:1478–1490
Signor L, Knuppe C, Hug R, Schweizer B, Pfaltz A, Jaun B (2000) Chem Eur J 6:3508–3516
Tada M, Masuzawa Y (1997) Chem Commun 2161–2162
Ghosh A, Wondimagegn T, Ryeng H (2001) Curr Opin Chem Biol 5:744–750
Pelmenschikov V, Blomberg MR, Siegbahn PE, Crabtree RH (2002) J Am Chem Soc 124:4039–4049
Pelmenschikov V, Siegbahn PE (2003) J Biol Inorg Chem 8:653–662
Ahn Y, Krzycki JA, Floss HG (1991) J Am Chem Soc 113:4700–4701
Lin S-K, Jaun B (1991) Helv Chim Acta 74:1725–1738
Mahlert F, Bauer C, Jaun B, Thauer RK, Duin EC (2002b) J Biol Inorg Chem 7:500–513
Telser J, Horng YC, Becker DF, Hoffman BM, Ragsdale SW (2000) J Am Chem Soc 122:182–183
Telser J, Davydov R, Horng YC, Ragsdale SW, Hoffman BM (2001) J Am Chem Soc 123:5853–5860
Tang Q, Carrington PE, Horng YC, Maroney MJ, Ragsdale SW, Bocian DF (2002) J Am Chem Soc 124:13242–13256
Singh K, Horng YC, Ragsdale SW (2003) J Am Chem Soc 125:2436–2443
Piskorski R, Jaun B (2003) J Am Chem Soc 125:13120–13125
Craft JL, Horng YC, Ragsdale SW, Brunold TC (2004) J Biol Inorg Chem 9:77–89
Wasserfallen A, Nölling J, Pfister P, Reeve J, de Macario EC (2000) Int J Syst Evol Microbiol 50:43–53
Gunsalus RP, Romesser JA, Wolfe RS (1978) Biochemistry 17:2374–2377
Kobelt A, Pfaltz A, Ankel-Fuchs D, Thauer RK (1987) FEBS Lett 214:265–268
Ellermann J, Hedderich R, Böcher R, Thauer RK (1988) Eur J Biochem 172:669–677
Rospert S, Voges M, Berkessel A, Albracht SPJ, Thauer RK (1992) Eur J Biochem 210:101–107
Rospert S, Linder D, Ellermann J, Thauer RK (1990) Eur J Biochem 194:871–877
Bonacker LG, Baudner S, Mörschel E, Böcher R, Thauer RK (1993) Eur J Biochem 217:587–595
Bradford MM (1976) Anal Biochem 72:248–254
Beinert H, Albracht SPJ (1982) Biochim Biophys Acta 683:245–277
Schönheit P, Moll J, Thauer RK (1980) Arch Microbiol 127:59–65
Bonacker LG, Baudner S, Thauer RK (1992) Eur J Biochem 206:87–92
Reeve JN, Nölling J, Morgan RM, Smith DR (1997) J Bacteriol 179:5975–5986
Hedderich R, Thauer RK (1988) FEBS Lett 234:223–227
Gunsalus RP, Wolfe RS (1978) FEMS Lett 3:191–193
Wackett LP, Honek JF, Begley TP, Wallace V, Orme-Johnson WH, Walsh CT (1987) Biochemistry 26:6012–6018
Belay N, Daniels L (1988) Antonie van Leeuwenhoek J Microbiol Serol 54:113–125
Ellermann J, Rospert S, Thauer RK, Bokranz M, Klein A, Voges M, Berkessel A (1989) Eur J Biochem 184:63–68
Albracht SPJ, Ankel-Fuchs D, van der Zwaan JW, Fontijn RD, Thauer RK (1986) Biochim Biophys Acta 870:50–57
Albracht SPJ, Ankel-Fuchs D, Böcher R, Ellermann J, Moll J, van der Zwaan JW, Thauer RK (1988) Biochim Biophys Acta 955:86–102
Wackett LP, Honek JF, Begley TP, Shames SL, Niederhoffer EC, Hausinger RP, Orme-Johnson WH, Walsh C (1988) Methyl-S-coenzyme-M reductase: a nickel-dependent enzyme catalyzing the terminal redox step in methane biogenesis. In: Lancaster J Jr (ed) The bioinorganic chemistry of nickel. VCH, Weinheim, pp 249–274
Holliger C, Kengen SW, Schraa G, Stams AJ, Zehnder AJ (1992) J Bacteriol 174:4435–4443
Lin S-K (1992) Doctoral thesis, ETH, Zürich
Selmer T, Kahnt J, Goubeaud M, Shima S, Grabarse W, Ermler U, Thauer RK (2000) J Biol Chem 275:3755–3760
Stubbe JA, van der Donk WA (1998) Chem Rev 98:705–762
Wagner AF, Frey M, Neugebauer FA, Schafer W, Knappe J (1992) Proc Natl Acad Sci USA 89:996–1000
Knappe J, Wagner AF (2001) Adv Protein Chem 58:277–315
Olson KD, Chmurkowska-Cichowlas L, McMahon CW, Wolfe RS (1992) J Bacteriol 174:1007–1012
Wondimagegn T, Ghosh A (2001) J Am Chem Soc 123:1543–1544
Jaun B (1990) Helv Chim Acta 73:2209–2217
Craft JL, Horng YC, Ragsdale SW, Brunold TC (2004) J Am Chem Soc 126:4068–4069
Duin EC, Signor L, Piskorski R, Mahlert F, Clay MD, Goenrich M, Thauer RK, Jaun B, Johnson MK (2004) J Biol Inorg Chem (in press)
Acknowledgements
This work was supported by the Max Planck Society, the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie and by the Swiss National Science Foundation. We thank Reinhard Böcher for technical assistance. M.G. is grateful to the Claussen-Simon-Foundation for a fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Goenrich, M., Mahlert, F., Duin, E.C. et al. Probing the reactivity of Ni in the active site of methyl-coenzyme M reductase with substrate analogues. J Biol Inorg Chem 9, 691–705 (2004). https://doi.org/10.1007/s00775-004-0552-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-004-0552-1