Abstract
Introduction
The diagnosis and treatment of osteoporosis, a frequent age-related metabolic bone disorder, remain incomprehensive and challenging. The potential regulatory role of lncRNA XIST and sphingosine kinase 1 (SPHK1) pathway need experimental investigations.
Materials and methods
RAW264.7 cells and BMMs were obtained for in vitro studies and 30 ng/mL RANKL was implemented for induction of osteoclast differentiation. The suppressing of lncRNA XIST, SPHK1 and fused in sarcoma (FUS) was achieved using small hairpin RNA, while overexpression of XIST and FUS was constructed by pcDNA3.1 vector system. Tartrate-resistant acid phosphatase (TRAP) staining was used for observation of formation of osteoclasts. RNA-pulldown analysis and RNA binding protein immunoprecipitation (RIP) was implemented for measuring mRNA and protein interactions. RT-qPCR was conducted to determining mRNA expression, whereas ELISA and Western blotting assay was performed for monitoring protein expression.
Results
RANKL induced osteoclast differentiation and upregulated expression of osteoclastogenesis-related genes that included NFATc1, CTSK, TRAP and SPHK1 and the level of lncRNA XIST in both RAW264.7 cells and BMMs. However, knockdown of lncRNA XIST or suppressing SPHK1 significantly reserved the effects of RANKL. LncRNA XIST was further demonstrated to be interacted with FUS and increased the stability of SPHK1, indicating its ability in promoting osteoclast differentiation through SPHK1/S1P/ERK signaling pathway.
Conclusion
LncRNA XIST promoted osteoclast differentiation via interacting with FUS and upregulating SPHK1/S1P/ERK pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bone has been regarded as a complex tissue provided important biological function that included regulation of mineral homeostasis and protection of soft tissues [1]. Bone diseases such as osteoporosis is becoming a frequent age-related disorder, which is especially prevalent for the elderly with the aging of population [2]. Osteoporosis is a kind of metabolic bone disorder that could be characterized by impairment of bone mass and microstructure deterioration, leading to changes in bone volume and high risk of bone fragility fractures [3]. However, the diagnosis and treatment of osteoporosis still remain challenging.
Sphingosine kinase 1 (SPHK1) belongs to one of the isoforms of sphingosine kinase, which is a highly conserved lipid kinase that phosphorylate sphingosine to form sphingosine-1-phosphate (S1P) [4]. The SPHK1/S1P pathway has been reported to participate in various cellular processes including inflammation, obesity, cancer progression and hematopoietic immune system [5]. Osteoclasts and osteoblasts maintain the balance of bone resorption and formation, and the activation of osteoclast differentiation or osteoclast dysfunction might lead to the osteoporosis [6]. Previous studies reported that S1P regulated the movement of osteoclasts precursors (OCPs) between the blood and bone tissues and enhances expression of receptor activator of NF-κB ligand (RANKL), and further broke the balance of bone metabolism [7, 8]. A recent study reported that S1P stimulation increased MEK and ERK phosphorylation and promoted IL-6 expression [9]. Kim et al. demonstrated that S1P activatec ERK-1/-2 through transactivating epidermal growth factor receptor [10]. Therefore, S1P might be a potential target for the regulation of osteoclast differentiation and treatment of osteoporosis.
Long non-coding RNAs (lncRNAs) are nucleotides with higher than 200 transcriptional length, which could regulate various gene expressions and biological processes [11]. Gene ontology analysis in patients with postmenopausal osteoporosis revealed that upregulated lncRNA XIST has been identified as differentially expressed lncRNAs [12]. A previous study also indicated that lncRNA XIST could decrease the miR-124-3p expression, which further upregulated the expression of miR-124-3p targeted genes that included SPHK1 in ovarian cancer cells [13, 14]. Bioinformatics analysis using StarBase (http://starbase.sysu.edu.cn/) revealed that lncRNA XIST might interact with fused in sarcoma (FUS), while FUS was recruited to the promoters of microphthalmia-associated transcription factor-targeted genes including tartrate-resistant acid phosphatase (TRAP, Acp5) and CTSK [15]. Nevertheless, the interaction between lncRNA XIST and FUS and the regulatory role of lncRNA XIST through SPHK1/S1P pathway still need experimental confirmation and further investigations.
In the present study, we hypothesized that lncRNA XIST facilitated S1P-mediated osteoclastogenesis through interacting with FUS. Mice RAW264.7 cells and bone marrow-derived macrophages (BMMs) were obtained, and macrophage colony-stimulating factor (M-CSF) and RANKL was used for induction of osteoclast differentiation. LncRNA XIST, FUS and SPHK1 were inhibited or overexpressed to investigate their interactions and to elucidate the molecular mechanisms.
Materials and methods
Cell culture
RAW264.7 cells (CL-0190) and BMMs (CP-M141) were purchased from Procell Life Science (Wuhan, China). Cells were cultured in α- minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 30 ng/mL M-CSF, 100 U/mL penicillin and 100 U/mL streptomycin. The mixture was maintained at 37 °C in a humidified 5% CO2 atmosphere. For the induction of osteoclast differentiation, totally 30 ng/mL RANKL was added into the culture medium and then the cells were incubated at 37 °C for 0,1,3,5 days, respectively.
Cell transfection
To investigate the molecular mechanisms, XIST small hairpin RNA (sh-XIST), sh-SPHK1, sh-FUS and sh-NC was obtained from GeneChem, Shanghai, China using lentivirus expression system to achieve knockdown models. The construction of overexpression model of XIST and FUS was achieved through pcDNA3.1 vector. Subsequently, the cells were transfected with knockdown or overexpression vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s instructions. The expression of SPHK1 was also inhibited by addition of sphingosine kinase I-II (SKI-II, MedChemExpress, Monmouth Junction, NJ, cat.no. HY-13822), and the expression of S1PR1/3 was also regulated by addition of S1PR1/3 antagonist VPC23019 (MedChemExpress, cat.no. HY-108490). All the treated cells were incubated at 37 °C for 48 h and the harvested for further experiments.
Tartrate-resistant acid phosphatase staining
After the induction of osteoclastogenesis, the cells were washed with PBS for three times and fixed with 4% paraformaldehyde. TRAP staining was performed using TRAP-staining kit (Sigma-Aldrich, St Louis, USA) according to the protocol of manufacturer. The TRAP-positive multinucleated cells, which contained 3 or more nuclei, were observed and counted under a light microscope (IX71, Olympus, Tokyo, Japan).
RNA-pulldown analysis
The RNA-pulldown analysis was performed using Pierce™ Magnetic RNA–Protein Pull-Down Kit (Thermo Fischer Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, the transcribed mRNA was labeled using Biotin RNA Labeling Mix (Roche, Basel, Switzerland). The biotin-labeled RNA was mixed with DNase I, protease inhibitor and RNase inhibitor (Thermo Fischer Scientific, Waltham, MA, USA) and incubated at room temperature for 15 min and 50 μL of magnetic beads were added and incubated at 37 °C for 1 h to combine proteins. Complexes were isolated and washed with ice-cold buffer and detected by Western blotting analysis.
RNA binding protein immunoprecipitation
The RNA binding protein immunoprecipitation (RIP) assays were conducted with Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Sigma-Aldrich, St Louis, USA). After three PBS washes, the cells were treated with RIP lysis buffer and the supernatants were collected after centrifugation. Anti-FUS (ab245332, Abcam, Cambridge, UK) and Anti-IgG (ab150113, Abcam, Cambridge, UK) were immunoprecipitated with supernatants according to the manufacturer's protocol. Immunoprecipitated RNA was quantified by RT-qPCR.
Enzyme-linked immunosorbent assay
The supernatants of RAW264.7 cells and BMMs were collected and commercial enzyme-linked immunosorbent assay (ELISA, Solarbio, Beijing, China) kit was obtained for the detecting the concentration of S1P as per the manufacturer’s protocol. A microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) was obtained to detect the absorbance at 450 nm.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), and the single-stranded cDNA was synthesized by the reverse transcription using PrimeScript Reverse Transcription Master Mix Kit (TaKaRa Biotechnology, Otsu, Japan). The expression level of target gene was quantified by real-time RT-PCR using SYBR® Premix Ex Taq™ II (TaKaRa Biotechnology, Otsu, Japan) on a 7900HT Real-Time PCR System (Life Technologies, Carlsbad, CA, USA). The RT-qPCR was conducted as per the following program: 95 °C for 30 s; then 95 °C for 5 s, 60 °C for 15 s and 72 °C for 15 s, for 40 cycles. The relative gene expression level was calculated using the 2−ΔΔCt method via normalization to the GAPDH. The sequences of primers were listed in Table 1.
RNA stability assay
The stability of RNA was detected as previously described [16]. Briefly, the cells were seeded into 6-well plates and treated with 5 μg/mL actinomycin D (Sigma-Aldrich, St Louis, USA) for 0, 1, 2, 3, 4 h, respectively. Then RNA was isolated from cells and the quantity was determine according to RT-qPCR analysis.
Western blotting analysis
The cells were washed with PBS and lysed with radioimmunoprecipitation assay lysis buffer (RIPA, Beyotime, Shanghai, China) that containing protease inhibitor and phosphatase inhibitor. Cells were centrifugated at 12,000 g for 15 min at 4 °C and the protein concentration in the supernatants were determined by BCA protein assay kit (Beyotime, Shanghai, China). The samples were loaded and separated on a 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel, after which the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% bovine serum albumin (Solarbio, Beijing, China) for 1 h at 37 °C and incubated with primary antibodies at 4 °C overnight. The primary antibodies were list as follows: anti-SPHK1 antibody (ab109522, 1:1000), anti-NFATc1 antibody (ab25916, 1:1000), anti-CTSK antibody (ab37259, 1:1000), anti-TRAP antibody (ab191406, 1:1000), anti-S1PR1 antibody (ab77076, 1:1000), anti-S1PR3 antibody (ab108370, 1:1000), anti-p-ERK antibody (ab201015, 1:1000), anti-ERK antibody (ab17942, 1:1000) and anti-GAPDH (ab8245, 1:1000). Subsequently, the membranes were washed and incubated with a horseradish-peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The ImageQuant LAS 500 system was implemented for observing the immunoreaction and the results were analyzed using ImageJ software.
Statistical analysis
All the data were presented as mean ± standard deviation (SD). Student's t-test was performed for comparison between two groups, and one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was implemented for comparison among multiple groups. All data were analyzed using GraphPad Prism 7.0 software. P < 0.05 was consider to be statistically significant.
Results
RANKL induced osteoclast differentiation and upregulated lncRNA XIST expression
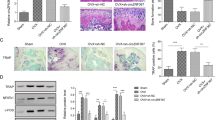
For inducing osteoclast differentiation, RAW264.7 cells and BMMs were treated with 30 ng/mL RANKL for 0,1,3,5 days. As presented in the graph of TRAP staining (Fig. 1A), the content of TRAP-positive cells was significantly increased under RANKL treatment for 1, 3, 5 days compared with 0 days. Moreover, with the increase of treatment duration, the ability of osteoclast differentiation was gradually increased. The relative mRNA expression of osteoclastogenesis-related genes that included NFATc1, CTSK, TRAP and SPHK1 and the level of lncRNA XIST was determined using RT-qPCR. The results showed that the level of XIST, SPHK1, and osteoclastogenesis-related genes were dramatically upregulated by RANKL induction, and most expressions were the highest after 5 days of treatment (Fig. 1B, C). Consistently, the level of S1P (detected by ELISA), as well as the protein expression of NFATc1, CTSK, TRAP and SPHK1 (detected by Western blotting, Fig. 1D, E) were all significantly upregulated in a time-dependent manner. Therefore, for the subsequent experiments, cells were treated with 30 ng/mL RANKL for 5 days.
The effects of RANKL treatment for 0,1,3,5-days on osteoclast differentiation in RAW264.7 cells and BMMs. A The formation of osteoclasts detected by TRAP staining. B, C The relative level of lncRNA XIST and mRNA expression of NFATc1, CTSK, TRAP, SPHK1 detected by RT-qPCR and calculated via normalization to the mRNA level of GAPDH. D The level of S1P determined using ELISA assay. E The protein expression of SPHK1, NFATc1, CTSK and TRAP measured by Western blotting and calculated via normalization to the protein level of GAPDH. *P < 0.05 vs RANKL 0-day group; **P < 0.01 vs RANKL 0-day group; ***P < 0.001 vs RANKL 0-day group
Knockdown lncRNA XIST inhibited osteoclast differentiation
To investigate the molecular mechanism of lncRNA XIST in osteoclast differentiation, sh-XIST was synthesized to knockdown lncRNA XIST. Compared with sh-NC group, the level of lncRNA XIST was dramatically decreased, indicating the successful construction of knockdown model (Fig. 2A). Inhibition of lncRNA XIST also significantly decreased the level of TRAP-positive cells (Fig. 2B). Furthermore, the Western blotting analysis and ELISA assay results demonstrated that the protein levels of NFATc1, CTSK, TRAP, SPHK1, S1P, S1PR1 and S1PR3, and the rate of p-ERK/total-ERK were all decreased in sh-XIST group compared with sh-NC group (Fig. 2C, D). The evidence indicated that lncRNA XIST might play a positive role in promoting osteoclastogenesis.
Knockdown of lncRNA XIST inhibited osteoclast differentiation in RAW264.7 cells and BMMs. A The relative level of lncRNA XIST detected by RT-qPCR and calculated via normalization to the mRNA level of GAPDH. B The formation of osteoclasts detected by TRAP staining. C The protein expression of SPHK1, S1PR1, S1PR3, p-ERK, ERK, NFATc1, CTSK and TRAP measured by Western blotting and calculated via normalization to the protein level of GAPDH. D The level of S1P determined using ELISA assay. *P < 0.05 vs sh-NC group; **P < 0.01 vs sh-NC group; ***P < 0.001 vs sh-NC group
Suppressing SPHK1 expression inhibited osteoclast differentiation
The suppression of SPHK1 was achieved through transfection of sh-SPHK1 or addition of SPHK1 inhibitor SKI-II. As presented in Fig. 3A, E, sh-SPHK1 could significantly decreased mRNA level of SPHK1, while SKI-II also significantly downregulated SPHK1 protein expression. Moreover, the transfection of sh-SPHK1 and treatment of SKI-II had no effects on the expression of lncRNA XIST (Fig. S1A, B). TRAP staining results showed that suppressing SPHK1 decreased the content of TRAP-positive cells and alleviated osteoclast differentiation (Fig. 3B, F). Western blotting analysis and ELISA results further elucidated that inhibition of SPHK1 dramatically decreased the protein levels of NFATc1, CTSK, TRAP, SPHK1, S1P, S1PR1 and S1PR3, and the rate of p-ERK/total-ERK in both RAW264.7 cells and BMMs (Fig. 3C, D, G, H). These data revealed that SPHK1 might also participate in the progression of osteoclastogenesis.
Suppressing SPHK1 inhibited osteoclast differentiation in RAW264.7 cells and BMMs. A, E The relative mRNA level of SPHK1 detected by RT-qPCR and calculated via normalization to the mRNA level of GAPDH. B, F The formation of osteoclasts detected by TRAP staining. C, G The protein expression of SPHK1, S1PR1, S1PR3, p-ERK, ERK, NFATc1, CTSK and TRAP measured by Western blotting and calculated via normalization to the protein level of GAPDH. D, H The level of S1P determined using ELISA assay. *P < 0.05 vs sh-NC or control group; **P < 0.01 vs sh-NC or control group; ***P < 0.001 vs sh-NC or control group
Interaction of lncRNA XIST with FUS increased the stability of SPHK1
Based on data from above experiments, we hypothesized that lncRNA XIST regulated SPHK1 expression. According to RT-qPCR analysis, knockdown of lncRNA XIST decreased SPHK1 expression (Fig. 4A). The distribution of lncRNA XIST was also investigated using RT-qPCR, which result suggested that most of lncRNA XIST and GAPDH was distributed in the cytoplasm of cells, while most of U1 was distributed in the nucleus in both RAW264.7 cells and BMMs (Fig. 4B). After actinomycin D treatment, the remaining of SPHK1 were also decreased in sh-XIST group compared with sh-NC group (Fig. 4C). Moreover, RNA-pull down assay revealed that sense-stranded lncRNA XIST and sense-stranded SPHK1 mRNA were interacted with FUS (Fig. 4D). Consistently, RIP analysis also proved that lncRNA XIST and SPHK1 mRNA were interacted with FUS (Fig. 4E). Knockdown of FUS downregulated the level of lncRNA XIST as well as the mRNA level of FUS and SPHK1 (Fig. 4F). The remaining of half-life period of lncRNA XIST and SPHK1 mRNA were both decreased in sh-FUS group compared with sh-NC group (Fig. 4G). Moreover, no direct interaction was observed between lncRNA XIST and SPHK1 (Fig. S1C). The above results suggested that lncRNA XIST might interact with FUS and maintain the stability of SPHK1.
LncRNA XIST interacted with FUS and maintained the stability of SPHK1 in RAW264.7 cells and BMMs. A The relative mRNA level of SPHK1 detected by RT-qPCR and calculated via normalization to the mRNA level of GAPDH. B The expression of lncRNA XIST in cytoplasm and nucleus detected by RT-qPCR. C The stability of SPHK1 mRNA determined by addition of actinomycin D and quantified by RT-qPCR. D The binding of SPHK1 mRNA and FUS, and lncRNA XIST and FUS measured by RNA-pulldown assay. E The binding abundance of FUS with SPHK1 mRNA or lncRNA XIST monitored using RIP analysis. F The relative level of lncRNA XIST and SPHK1 mRNA detected by RT-qPCR and calculated via normalization to the mRNA level of GAPDH after knockdown of FUS. G The stability of lncRNA XIST and SPHK1 mRNA determined by addition of actinomycin D and quantified by RT-qPCR after knockdown of FUS. *P < 0.05 vs sh-NC or IgG group; **P < 0.01 vs sh-NC or IgG group; ***P < 0.001 vs sh-NC or IgG group
lncRNA XIST upregulated S1PR1/S1PR3 expression
For elucidating the role of lncRNA XIST and S1PR1/S1PR3, an S1PR1/S1PR3 antagonist (VPC23019), was obtained for subsequent experiments. As presented in TRAP staining (Fig. S2A) revealed that osteoclast differentiation was inhibited, and ELISA assay (Fig. S2B) showed that the level of S1P was decreased by addition of VPC23019. However, overexpression of lncRNA XIST could significantly alleviate the effects of VPC23019 on osteoclast differentiation and S1P expression. Western blotting results (Fig. S2C) suggested that VPC23019 had no effects on SPHK1 expression, but decreased the level of S1PR1 and S1PR3, inactivated phosphorylation of ERK and inhibited expression of NFATc1, CTSK and TRAP. Overexpression of lncRNA XIST could reverse the VPC23019-induced protein expression changes. These data indicated that lncRNA XIST promoted osteoclast differentiation through upregulating S1PR1/S1PR3 expression.
Interaction of lncRNA XIST with FUS promoted osteoclast differentiation through SPHK1/S1P/ERK pathway
Overexpression models of lncRNA XIST and overexpression or knockdown of FUS models were established for further elucidating the molecular mechanisms. The successful construction of lncRNA XIST overexpression model was verified by RT-qPCR (Fig. S1D). As presented in Fig. 5A, B, the overexpression of lncRNA XIST and FUS both upregulated SPHK1 protein expression. Overexpression of lncRNA XIST increased the content of TRAP-positive cells, however, knockdown of FUS in NC group could decrease TRAP-positive rate and osteoclastogenesis induced by oe-XIST (Fig. 5C). Western blotting and ELISA results proved that knockdown of FUS reversed the positive effects of lncRNA XIST on the protein expression of NFATc1, CTSK, TRAP, SPHK1, S1P, S1PR1 and S1PR3, and the rate of p-ERK/total-ERK (Fig. 5D, E), indicating lncRNA XIST might interact with FUS and promoted osteoclast differentiation through SPHK1/S1P/ERK pathway.
Interaction between lncRNA XIST and FUS promoted osteoclast differentiation in RAW264.7 cells and BMMs. A, B The protein expression of SPHK1 measured by Western blotting and calculated via normalization to the protein level of GAPDH after overexpression of lncRNA XIST or FUS. C The formation of osteoclasts detected by TRAP staining after cotransfection with lncRNA XIST overexpression plasmids or/and shFUS. D The protein expression of SPHK1, S1PR1, S1PR3, p-ERK, ERK, NFATc1, CTSK and TRAP measured by Western blotting and calculated via normalization to the protein level of GAPDH after cotransfection with lncRNA XIST overexpression plasmids or/and shFUS. E The level of S1P determined using ELISA assay after cotransfection with lncRNA XIST overexpression plasmids or/and shFUS. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
Osteoporosis has been regarded the most common chronic metabolic bone disease in elderly patients, which could be characterized by increased bone fragility and reduction in bone mass [17]. It was reported that more than 6% of men and 21% of women has been suffered from osteoporosis [18]. During the dynamic remodeling of bone tissues, osteoblasts and osteoclasts were responsible for the formation and degradation of bone matrix, respectively [19]. Therefore, inadequate new bone formation or excessive bone resorption might both resulted in osteoporosis [20]. Previous studies reported that RANKL could initiate the cellular polarization of mature osteoblasts and enhance the activity of bone resorption, which finally resulted in the promoted osteoclastogenesis [21]. However, the comprehensive molecular mechanism has not been fully investigated. In the present study, RANKL was used for inducing osteoclast differentiation, and the role of lncRNA XIST, FUS and SPHK1 was explored to elucidate their relations in the progression of osteoclastogenesis.
A recent study found that the expression of NFATc1 and phosphorylation of MAPK, calcium oscillations and generation of reactive oxygen species (ROS) was upregulated in RANKL-treated mice-isolated BMMs [22]. Kim et al. also demonstrated that NFATc1, TRAP, CTSK and c-Fos signaling pathways was activated in RANKL-treated RAW 264.7 cells [23]. Consistently, our results indicated that the expression of NFATc1, CTSK, TRAP, SPHK1, S1P, p-ERK and lncRNA XIST was upregulated in both RANKL-treated RAW 264.7 cells and BMMs. Recent research also reported that several lncRNAs had ability on regulation progression of osteoporosis, for example, lncRNA Bmncr was demonstrated to alleviate RANKL-induced osteoclast differentiation [6]. Our further experiments showed that knockdown of lncRNA XIST or inhibition of SPHK1 reversed the effects of RANKL on osteoclastogenesis in vitro, indicating the important role of lncRNA XIST and SPHK1 in osteoclast differentiation. Moreover, the inhibition of S1PR1/S1PR3 using VPC23019 suppressed osteoclast differentiation, while overexpression of lncRNA XIST had the reserved effects, indicating that S1P receptors might be a potential target for in osteoporosis. Similarly, Chen et al. found that overexpression of lncRNA XIST significantly inhibited osteoblast differentiation, whereas interference of lncRNA XIST exhibited the reversed effects and promoted osteogenic differentiation [24].
The molecular mechanism of lncRNA XIST was further explored. A previous study elucidated that inhibiting lncRNA XIST reduced aging-induced bone loss and promoted osteogenic differentiation through sponging miR-19a-3p [25]. Knockdown of lncRNA XIST suppressed the progression of osteosarcoma through sponging miR-375-3p and regulating AKT/mTOR signaling pathway [26]. Barr et al. found that protein phosphatase 2A could inactive SPHK1 through interaction in human embryonic kidney cells and regulate cell necrosis and proliferation [27]. Yao et al. reported that miR-3677 could targeting SPHK1 and inhibit human osteosarcoma progression, which could be served as a potential therapeutic target [28]. In the present study, the relationship between lncRNA XIST, FUS and SPHK1 was verified by RNA-pulldown assay and RIP analysis. Results indicated that the interaction between lncRNA XIST and FUS increased the stability of SPHK1. FUS, a kind of RNA-binding proteins that contains an N‐terminal domain with a glutamine‐glycine‐serine‐tyrosine‐rich [29], was known as a nucleo‐cytoplasmic shuttling factor working during transcription [30]. It was reported that FUS could bind GluA1 mRNA in the vicinity of the 3' terminus and controls poly (A) tail maintenance, contributing to the stability of GluA1 mRNA, while depletion of FUS resulted in the downregulation of GluA1 [31]. Similarly, we found that FUS was bound with sense-stranded SPHK1 mRNA and inhibition of FUS decrease the stability of SPHK1 mRNA, indicating that FUS could regulate the stability of SPHK1 and finally resulted in the promoted osteoclast differentiation.
In conclusion, the present study newly demonstrated that lncRNA XIST facilitated osteoclast differentiation via interacting with FUS and upregulating SPHK1/S1P/ERK pathway, providing potential molecular mechanisms and therapeutic target for clinical treatment. However, further studies are needed and differentially expressed RNAs are worthy to be investigated for comprehensive understanding the progression mechanism of osteoclastogenesis.
References
Cohen MM Jr (2006) The new bone biology: pathologic, molecular, and clinical correlates. Am J Med Genet A 140:2646–2706 (Epub 2006/11/15)
Sözen T, Özışık L, Başaran N (2017) An overview and management of osteoporosis. Eur J Rheumatol 4:46–56 (Epub 2017/03/16)
Fonseca H, Moreira-Gonçalves D, Coriolano HJ, Duarte JA (2014) Bone quality: the determinants of bone strength and fragility. Sports Med 44:37–53 (Epub 2013/10/05)
Wang J, Feng W, Li F, Shi W, Zhai C, Li S, Zhu Y, Yan X, Wang Q, Liu L, Xie X (2019) SphK1/S1P mediates TGF-β1-induced proliferation of pulmonary artery smooth muscle cells and its potential mechanisms. Pulm Circ 9:2045894018816977 (Epub 2018/11/16)
Zhao Z, Ma J, Hu B, Zhang Y, Wang S (2018) SPHK1 promotes metastasis of thyroid carcinoma through activation of the S1P/S1PR3/Notch signaling pathway. Exp Ther Med 15:5007–5016 (Epub 2018/05/29)
Chen RS, Zhang XB, Zhu XT, Wang CS (2019) LncRNA Bmncr alleviates the progression of osteoporosis by inhibiting RANML-induced osteoclast differentiation. Eur Rev Med Pharmacol Sci 23:9199–9206 (Epub 2019/11/28)
Ishii M, Egen JG, Klauschen F, Meier-Schellersheim M, Saeki Y, Vacher J, Proia RL, Germain RN (2009) Sphingosine-1-phosphate mobilizes osteoclast precursors and regulates bone homeostasis. Nature 458:524–528 (Epub 2009/02/11)
Takeshita H, Kitano M, Iwasaki T, Kitano S, Tsunemi S, Sato C, Sekiguchi M, Azuma N, Miyazawa K, Hla T, Sano H (2012) Sphingosine 1-phosphate (S1P)/S1P receptor 1 signaling regulates receptor activator of NF-κB ligand (RANKL) expression in rheumatoid arthritis. Biochem Biophys Res Commun 419:154–159 (Epub 2012/02/14)
Hu SL, Huang CC, Tzeng TT, Liu SC, Tsai CH, Fong YC, Tang CH (2020) S1P promotes IL-6 expression in osteoblasts through the PI3K, MEK/ERK and NF-κB signaling pathways. Int J Med Sci 17:1207–1214 (Epub 2020/06/18)
Kim JH, Kim JH, Song WK, Kim JH, Chun JS (2000) Sphingosine 1-phosphate activates Erk-1/-2 by transactivating epidermal growth factor receptor in rat-2 cells. IUBMB Life 50:119–124 (Epub 2001/02/24)
Liu J, Yao L, Zhang M, Jiang J, Yang M, Wang Y (2019) Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging (Albany NY) 11:7830–7846 (Epub 2019/09/26)
Fei Q, Bai X, Lin J, Meng H, Yang Y, Guo A (2018) Identification of aberrantly expressed long non-coding RNAs in postmenopausal osteoporosis. Int J Mol Med 41:3537–3550 (Epub 2018/03/24)
Hai B, Pan X, Du C, Mao T, Jia F, Liu Y, Ma Y, Liu X, Zhu B (2020) LncRNA XIST promotes growth of human chordoma cells by regulating miR-124-3p/iASPP pathway. Onco Targets Ther 13:4755–4765 (Epub 2020/06/18)
Zhang H, Wang Q, Zhao Q, Di W (2013) MiR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J Ovarian Res 6:84 (Epub 2013/11/28)
Bronisz A, Carey HA, Godlewski J, Sif S, Ostrowski MC, Sharma SM (2014) The multifunctional protein fused in sarcoma (FUS) is a coactivator of microphthalmia-associated transcription factor (MITF). J Biol Chem 289:326–334 (Epub 2013/11/22)
Wawro M, Wawro K, Kochan J, Solecka A, Sowinska W, Lichawska-Cieslar A, Jura J, Kasza A (2019) ZC3H12B/MCPIP2, a new active member of the ZC3H12 family. RNA 25:840–856 (Epub 2019/04/17)
Cauley JA (2013) Public health impact of osteoporosis. J Gerontol A Biol Sci Med Sci 68:1243–1251 (Epub 2013/08/02)
Svedbom A, Hernlund E, Ivergård M, Compston J, Cooper C, Stenmark J, McCloskey EV, Jönsson B, Kanis JA (2013) Osteoporosis in the European Union: a compendium of country-specific reports. Arch Osteoporos 8:137 (Epub 2013/10/12)
Pietschmann P, Mechtcheriakova D, Meshcheryakova A, Föger-Samwald U, Ellinger I (2016) Immunology of osteoporosis: a mini-review. Gerontology 62:128–137 (Epub 2015/06/20)
Macías I, Alcorta-Sevillano N, Rodríguez CI, Infante A (2020) Osteoporosis and the potential of cell-based therapeutic strategies. Int J Mol Sci 21:1653 (Epub 2020/03/04)
Nagy V, Penninger JM (2015) The RANKL-RANK Story. Gerontology 61:534–542 (Epub 2015/02/28)
Liu Y, Wang C, Wang G, Sun Y, Deng Z, Chen L, Chen K, Tickner J, Kenny J, Song D, Zhang Q (2019) Loureirin B suppresses RANKL-induced osteoclastogenesis and ovariectomized osteoporosis via attenuating NFATc1 and ROS activities. Theranostics 9:4648–4662 (Epub 2019/08/02)
Kim M, Kim HS, Kim JH, Kim EY, Lee B, Lee SY, Jun JY, Kim MB, Sohn Y, Jung HS (2020) Chaenomelis fructus inhibits osteoclast differentiation by suppressing NFATc1 expression and prevents ovariectomy-induced osteoporosis. BMC Complement Med Ther 20:35 (Epub 2020/02/07)
Chen X, Yang L, Ge D, Wang W, Yin Z, Yan J, Cao X, Jiang C, Zheng S, Liang B (2019) Long non-coding RNA XIST promotes osteoporosis through inhibiting bone marrow mesenchymal stem cell differentiation. Exp Ther Med 17:803–811 (Epub 2019/01/18)
Chen S, Li Y, Zhi S, Ding Z, Huang Y, Wang W, Zheng R, Yu H, Wang J, Hu M, Miao J (2020) lncRNA Xist regulates osteoblast differentiation by sponging miR-19a-3p in aging-induced osteoporosis. Aging Dis 11:1058–1068 (Epub 2020/10/06)
Sun X, Wei B, Peng ZH, Fu QL, Wang CJ, Zheng JC, Sun JC (2019) Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR-375-3p. Int J Clin Exp Pathol 12:1507–1517 (Epub 2020/01/15)
Barr RK, Lynn HE, Moretti PA, Khew-Goodall Y, Pitson SM (2008) Deactivation of sphingosine kinase 1 by protein phosphatase 2A. J Biol Chem 283:34994–35002 (Epub 2008/10/15)
Yao C, Ruan JW, Zhu YR, Liu F, Wu HM, Zhang Y, Jiang Q (2020) The therapeutic value of the SphK1-targeting microRNA-3677 in human osteosarcoma cells. Aging (Albany NY) 12:5399–5410 (Epub 2020/03/2)
Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D’Agostino VG (2020) RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J Extracell Vesicles 10:e12043 (Epub 2021/01/05)
Loughlin FE, Wilce JA (2019) TDP-43 and FUS-structural insights into RNA recognition and self-association. Curr Opin Struct Biol 59:134–142 (Epub 2019/09/04)
Udagawa T, Fujioka Y, Tanaka M, Honda D, Yokoi S, Riku Y, Ibi D, Nagai T, Yamada K, Watanabe H, Katsuno M (2015) FUS regulates AMPA receptor function and FTLD/ALS-associated behaviour via GluA1 mRNA stabilization. Nat Commun 6:7098 (Epub 2015/05/15)
Acknowledgements
This work was supported by Guangdong Natural Science Foundation Committee, Doctoral Startup Fund (2018a03010282).
Author information
Authors and Affiliations
Contributions
(1) DWZ and LJY made substantial contributions to the conception. and design of the work; DWZ, HGW and KBZ are the acquisition, analysis, or interpretation of data; YQG is the creation of new software used in the work; (2) DWZ drafted the work or revised it critically for important intellectual content; (3) All authors approved the version to be published; (4) HL agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article did not involve clinical and animal experiments, so the ethical approval is not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
774_2021_1294_MOESM1_ESM.tif
Supplementary file1 Suppressing SPHK1 had no effect on the lncRNA XIST expression in RAW264.7 cells and BMMs. A, B The expression of XIST measured by RT-qPCR and calculated via normalization to the level of GAPDH after knockdown of SPHK1 or treatment with SPHK1 inhibitor SKI-II. C The binding of lncRNA XIST and SPHK1 protein measured by RNA-pulldown assay. D The expression of XIST measured by RT-qPCR and calculated via normalization to the level of GAPDH after overexpression of SPHK1. ***P<0.001 (TIF 979 KB)
774_2021_1294_MOESM2_ESM.tif
Supplementary file2 LncRNA XIST reversed VPC23019 inhibiting osteoclast differentiation in RAW264.7 cells and BMMs. A The formation of osteoclasts detected by TRAP staining. B The level of S1P determined using ELISA assay. C The protein expression of SPHK1, S1PR1, S1PR3, p-ERK, ERK, NFATc1, CTSK and TRAP measured by Western blotting and calculated via normalization to the protein level of GAPDH. *P<0.05; **P<0.01; ***P<0.001 (TIF 2987 KB)
About this article
Cite this article
Zhang, DW., Wang, HG., Zhang, KB. et al. LncRNA XIST facilitates S1P-mediated osteoclast differentiation via interacting with FUS. J Bone Miner Metab 40, 240–250 (2022). https://doi.org/10.1007/s00774-021-01294-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00774-021-01294-3