Abstract
This review intends not only to discuss the current possibilities to gain 100% sequence coverage for proteins, but also to point out the critical limits to such an attempt. The aim of 100% sequence coverage, as the review title already implies, seems to be rather surreal if the complexity and dynamic range of a proteome is taken into consideration. Nevertheless, established bottom-up shotgun approaches are able to roughly identify a complete proteome as exemplary shown by yeast. However, this proceeding ignores more or less the fact that a protein is present as various protein species. The unambiguous identification of protein species requires 100% sequence coverage. Furthermore, the separation of the proteome must be performed on the protein species and not on the peptide level. Therefore, top-down is a good strategy for protein species analysis. Classical 2D-electrophoresis followed by an enzymatic or chemical cleavage, which is a combination of top-down and bottom-up, is another interesting approach. Moreover, the review summarizes further top-down and bottom-up combinations and to which extent middle-down improves the identification of protein species. The attention is also focused on cleavage strategies other than trypsin, as 100% sequence coverage in bottom-up experiments is only obtainable with a combination of cleavage reagents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Prologue
Initially, it is necessary not only to present the importance of a high or 100% protein sequence coverage, but also the constraints that hinder the realization. It could even be said that the idea for 100% sequence coverage is comparable to the way of thinking in surrealism. Nevertheless, proteomics pioneer M. R. Wilkins already pointed out in the beginning of the proteomics era that 100% protein identification is worthwhile (Wilkins et al. 1996). The question is why every protein should be identified with 100% sequence coverage and why all this should be “beyond the realism”, in other words, “surrealism”.
In Anno Domini 1994, Wilkins defined the proteomics concept (Wasinger et al. 1995). According to him, the proteome can be described as the complete protein map of the genome of an organism, a cell or a cellular compartment. More than a decade later, the complete proteome analysis seems to be feasible. In 2006, Mann et al. were able to show that half of the approximately 4,500 predicted proteins of the popular model system, yeast, can be identified using a high-throughput bottom-up approach, termed GeLCMS (de Godoy et al. 2006). The same research group later published a quantitative proteomics study of haploid and diploid yeast cells that covered the whole proteome without discriminating difficult to detect proteins, such as membrane proteins, or low abundant ones (de Godoy et al. 2008). Furthermore, the identified peptides represented average protein sequence coverage of 38%. It would seem that all doubts concerning the enormous challenge of the dynamic range of every proteome and the analysis of difficult to detect protein groups, such as the above-mentioned membrane proteins, are overcome. The “amour impossible” to membrane proteins, as phrased nicely by T. Rabilloud, would no longer be valid (Santoni et al. 2000). Even the satirist Quintus Horatius Flaccus, who was mentioned by P. G. Righetti in his review “The art of observing rare protein species in proteomes with peptide ligand libraries”, would be incorrect with his famous statement “Est modus in rebus, sunt certi denique fines” (“There is moderation in all things; there are, in short, fixed limits”; Boschetti and Righetti 2009). In reality, the term “protein species” is abandoned a bit too early, but this issue will be addressed later on (Jungblut et al. 1996, 2008). For the moment, the topic should remain on complete proteome analysis, membrane proteins and dynamic range, and to put all this into perspective.
Certainly, T. Rabilloud changed his dictum about membrane proteins to a “possible, but difficult love” (Rabilloud 2009). Earlier, P. G. Righetti compared the challenges in proteomics, such as the dynamic range, to the situation of the main character in Rudyard Kipling’s story “The Strange Ride of Morrowbie Jukes” (Righetti et al. 2005a). Owing to the irresistible technological progress, proteomics can overcome a number of difficulties just like rescue came to poor Morrowbie from nowhere. The technical advances as the essential driving force to the impressive results in the complete yeast proteome analysis were, therefore, appreciated by the responsible protagonists of the aforementioned study. Current limitations were also addressed, but all answers were linked to the prospective development of the “rescuer”, or, in other words, progress. Indeed, progress may be fast, as in the case of the yeast proteome, but not ensured (de Godoy et al. 2006; de Godoy et al. 2008). Or to say it in the words of the famous Swiss author Friedrich Dürrenmatt:”Je planmäßiger die Menschen vorgehen, desto wirksamer vermag sie der Zufall zu treffen.” (The more methodically humans proceed, the more effective they might be hit by coincidence). Therefore, it is fair to be encouraged that the state of the yeast proteome analysis continues to advance. However, the detection of the two- to threefold amount of expressed gene products in mammalian cells will depend on some coincidence: The positive one that moves the situation in the right direction, and the negative one that has a vice versa effect. Hence, even if, the complete proteome of a mammalian cell could finally be analyzed by a high-throughput bottom-up approach, only one representative protein for any expressed gene would be identified in most cases (de Godoy et al. 2008). Therefore, a different strategy is needed, besides refined versions of high-throughput bottom-up (Cox and Mann 2007), to analyze a proteome with all its modifications and isoforms, because a gene encodes several protein species (Jungblut et al. 1996, 2008). An analysis for modifications or isoforms can certainly be done using a high-throughput bottom-up approach on defined subproteomes (de Godoy et al. 2006), but there is a need for a more original approach to face the problem of protein species analysis. The purification of subproteomes is nevertheless important. For the isolation of subproteomes, it is generally accepted that the purification is required to take place under well-defined conditions to guarantee high levels of purity and reproducibility. However, this is nearly impossible to achieve (Jungblut et al. 2008).
The previously mentioned term, protein species, was introduced roughly at the same time as proteomics (Jungblut et al. 1996). An exact definition and a clear nomenclature, however, were recently conceived (Jungblut et al. 2008; Schlüter et al. 2008). In short, the main aspect of the protein species concept could be summarized with the following citation: “The term protein refers to its coding gene and, therefore, is the umbrella term for all of the developing protein species” (Jungblut et al. 2008). Any of these primary translation products, which could be named initial protein species, has various possibilities of transformation. For instance, it could be processed by an enzyme or environmental influences. Single nucleotide polymorphisms also result in new protein species. All this can certainly lead to different functions at the various locations in a cell, a tissue or organism. The 170 identified protein species of Histon 3.2 are a concrete example (Garcia et al., 2007). The full complexity behind the protein species concept has extensively been addressed in the work of Schlüter et al. (2008) and Jungblut et al. (1996). In summary, a protein species represents the smallest unit of the proteome that can be correlated with a function (Schlüter et al. 2008). As a consequence, it should be obvious that an absolute bottom-up strategy is very fast and sensitive, but out of a complex mixture of protein species from a selected compartment (cell, tissue or organism), in most cases, only one representative protein per expressed gene is identified. The peptides are necessarily distributed over the whole LC run so that a subsequent assignment of the peptides to the corresponding protein species is more or less impossible. The same applies to the quantification of a protein species as the same peptide can originate from different protein species. The separation of a protein species mixture on the protein level, as practiced in the classical 2D-electrophoresis (2-DE; first dimension: IEF, isoelectric focusing; second dimension: SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis) combined with a mass spectrometric bottom-up analysis or the top-down approach (Kelleher et al. 1999), allows for protein species detection. Both strategies, however, are not as efficient as a bottom-up strategy on the peptide level (Cox and Mann 2007; de Godoy et al. 2008; Jungblut et al. 1996; Schlüter et al. 2008). This in turn leads to unrealistic analysis times (Hoehenwarter et al. 2006).
However, there are almost insuperable hurdles for all available proteomic strategies. For instance, all protein species from a eukaryotic cell should be detected. First of all, cell lines actually constitute bad models for cells in their physiological context, i.e. the tissue (Godovac-Zimmermann et al. 2005; Dumont et al. 2002). A relevant analysis of all present protein species would consequently be based on a selection of a few cells out of a defined area of the tissue and such a sample preparation would contain protein species in a quantity range of low zeptomol to the high femtomol. Except for the highly abundant protein species, this dynamic range is beyond the detection capabilities of available techniques. Every pre-fractionation technique or subsequent separation strategy on the protein level (PAGE or liquid chromatography, LC) requires more starting material. Indeed, the dynamic range could be reduced by the use of “peptide ligand libraries” (Boschetti and Righetti 2009), although a high sample amount would be necessary, which in turn could only be obtained using cell lines. Furthermore, this technique cannot be applied to all protein classes, such as membrane proteins. Quantification experiments are, however, after a standardization of the protein amounts, no longer reasonable. The importance to comparatively quantify protein species in two cell states or to perform an absolute quantification for the functional characterization is quite obvious.
Real protein species function characterization, particularly with regard to the recently suggested protein code (Sims and Reinberg 2008) requires very high sequence coverage together with the identification of all post-translational modifications. The combination of all modified and unmodified amino acids of a protein species would finally construct the protein code. Such a protein surface of an isolated protein species provides a variety of targets for other protein species which can cause various reactions. Nevertheless, even for a given protein species, there are many possibilities. Therefore, the meaning of the modification pattern must be evaluated on every occasion in the context of the compartment of the organism.
Owing to this enormous complexity, it seems logical that a more detailed look is necessary. Without a good overview of a proteome, in-depth research makes no sense. Before a search for protein species can be initiated, it is important to determine which proteins are present. Therefore, high-throughput bottom-up strategies, as defined by Mann et al. (Cox and Mann 2007) constitute an absolute necessity. There is, without a doubt, a need for many further developments concerning this strategy to avoid “years of misdirected work” in the case of in-depth analysis (Mann and Kelleher 2008). Moreover, the question is if the time has already come to think about a more detailed look or if the danger is still too high to get misdirected. Owing to the complex situation, which has been adequately discussed, it is rather unrealistic, even for a well-defined problem, to hope for a perfect 100% sequence coverage of every protein species. Although 100% sequence coverage is quite surreal, it is negligent to describe cell events on the protein level without 100% sequence coverage. The situation is comparable to the study of a living cell using conventional confocal microscopy. Due to the diffraction, the resolving power is limited to approximately 200 nm. Abbe’s resolution limit was accepted as an insuperable law until the group of Stefan Hell circumvented it (Hell 2007). The STED (Stimulated Emission Depletion) microscope improved the resolving power by a factor of ten so that events in living cells can be clearly displayed, e.g. synaptic vesicles during exocytose (Willig et al. 2006). In analogy, in order to see cell events on the protein level, a proteomics platform is needed which provides the potential for 100% sequence coverage for every protein species.
Mass spectrometrists in proteomics—servant of two masters
One can compare the search for a proteomic strategy that achieves 100% sequence coverage for every protein species with the famous play of the Commedia dell’arte “Servant of two masters” from Carlo Goldoni. Truffaldino has the idea to serve two masters, Beatrice and Florindo, concurrently and without them being aware of the situation. His main goal was to satisfy his lust for food, his exorbitant hunger. The situation becomes even more complicated because the two masters end up falling in love and searching for each other. Likewise, in a haste to get 100% sequence coverage for a protein species, two trends are served in proteomics; the bottom-up and top-down approach. Towards the end of the play, Beatrice and Florindo finally meet each other. Truffaldino is not only forgiven, but also allowed to marry his beloved. In much the same manner, just as Truffaldino gained from the masters’ bonding, the daily user could potentially gain from the combination of bottom-up and top-down to middle-down (Forbes et al. 2001). In middle-down, proteins are initially enzymatically or chemically cleaved into bigger fragments as compared to the bottom-up approach (3–20 kDa). Nevertheless, drastic changes and twists in proteomics workflows, resulting in bottom-up and top-down approaches, do not solve the 100% sequence coverage problem, but only lead to slight and steady improvements. The real question is to which extent the daily user’s hunger for sequence coverage is already satisfied. One should not forget that, even though Truffaldino gained a wife, his hunger was not satisfied.
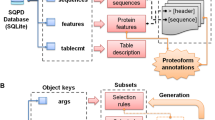
That being said, the following bottom-up chapter summarizes which strategies on the peptide level are available to gain a high or even 100% sequence coverage in protein identification. Specific, less specific and unspecific cleavage strategies are presented. In addition, an outlook is given for the rarely applied chemical cleavage agents. Furthermore, the influence on the sequence coverage of in-gel proteolysis, matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) are carefully reviewed. Most of the proteomic workflows, referred in the bottom-up chapter, have been established for high-throughput approaches which prevent the detection of protein species as it has already been pointed out in the prologue. Nevertheless, these studies can contribute to improve the identification of protein species if applied to a combination of top-down and bottom-up strategies. Although top-down strategies can already identify protein species attaining high sequence coverage, there are several limitations which are discussed later on (see top-down chapter). Some could perhaps be solved using the discussed strategies of the bottom-up chapter in a middle-down or a combined top-down/bottom-up approach. Only the second strategy, however, makes unambiguous protein species identification possible as middle-down strategies still separate proteomes on the peptide level. In our point of view, a combined top-down/bottom-up approach is consequently the best strategy for a protein species analysis. Figure 1 provides a summary of all mentioned strategies.
Bottom-up
The saying “Quod latet, ignotum est, ignoti nulla cupido (What is hidden is unknown: for what is unknown there is no desire)” from Ovid could come in one’s mind of Washburn’s shotgun approach “multidimensional protein identification technology” (MudPIT) and every other bottom-up shotgun workflow (Washburn et al. 2001). In other words, many proteins remain undetected with no interest to search them out. Meanwhile, protein identification is so efficient that ~7,000 proteins are identifiable on the basis of peptides in a single mass spectrometry (MS) experiment (Wisniewski et al. 2009; Garcia, 2010). Nevertheless, a trend-setting study from the group of Aebersold demonstrates that a serious problem is still present in the background. The basic message shall be summarized here: “Although such analyses typically assume that a protein’s peptide fragments are observed with equal likelihood, only a few so-called ‘proteotypic’ peptides are repeatedly and consistently identified for any given protein present in a mixture.” (Mallick et al. 2007). It could also be shown that “proteotypic peptides” of low abundant proteins in a complex mixture are reproducibly detectable. Naturally, “proteotypic peptides” differ from one proteomic workflow to another. SDS-PAGE followed by MALDI-MS results in different “proteotypic peptides” than SDS-PAGE coupled with LC–ESI or MudPIT-ESI. The aforementioned indication, however, was only verified using trypsin, the most popular protease for MS-based protein identification. In addition, a prediction program was developed for the evaluated proteomic workflows and tested on real datasets. This program enables the calculation of “proteotypic peptides” resulting from tryptic digestions of any protein sequence. One advantage of “proteotypic peptides” is obviously the extension of the dynamic range for protein identification leading to the above-mentioned high numbers of protein identification (Mann and Kelleher 2008). The consequential disadvantage is the occurrence of “non-proteotypic peptides”, which are irreproducibly detectable or absent. Therefore, the frequent absence of “non-proteotypic peptides” may serve as the main obstacle for 100% sequence coverage. The sequence ranges of proteins, which are covered by “non-proteotypic peptides”, remain unrevealed in proteome analysis but, in most cases, the need to close the sequence gaps is not essential. If protein identification of one single representative protein species per expressed gen is sufficient, high sequence coverage is certainly not necessary (de Godoy et al. 2008). Finally, this confirms Ovid’s expression that hidden things remain unknown and the interest for unknown things is low as long as a significant identification is possible.
Nevertheless, the variety of enzymatic and chemical cleavage strategies should provide enough possibilities to approach 100% sequence coverage for every protein, when sufficient protein amounts are available. All listed cleavage strategies in Table 1 should generate reproducibly different “proteotypic peptides” for a given proteomic workflow. For a determined workflow, even less specific enzymes, such as elastase, continually provide the same abundant peptides in the same region of the analyzed proteins (Rietschel et al. 2009a). Perhaps, this is questionable for the unspecific candidates such as proteinase K, subtilisin and thermolysin, but some hints for repeated peptide detection in the same sequence region of proteins are reported for proteinase K (Speers and Wu 2007; Bendz et al. 2008). “Proteotypic peptides”, however, prevent 100% sequence coverage. Particularly the studies of MacCoss et al. (2002) using trypsin, elastase and subtilisin together with the relevant work of Coon’s group using conventional specific enzymes (Swaney et al. 2010) prove that the answer to the 100% sequence coverage challenge lie in the combination of enzymes. Additional optimizations in a proteomic workflow such as the complementary use of MALDI and ESI, more efficient MALDI matrices, strategies to improve the peptide fragmentation and a better peptide recovery from gel-separated proteins, all contribute to higher numbers of detected peptides for an identified protein and, therefore, result in a higher sequence coverage (Fig. 2). Indeed, many points are left out of this article, for instance, the rarely used protease thermolysin (Schlosser et al. 2005; Chen et al. 2009). Apart from the selected examples of enzymatic and chemical cleavage strategies in the following three subchapters, further studies prove the possibility to gain high sequence coverage for proteins (Distler et al. 2006; John et al. 2006; Chmelik et al. 2009; Chen et al. 2010; Zvonok et al. 2010). Microwave-assisted protein cleavage is only described for acids. Microwave-assisted digestions using enzymes are not mentioned, although a positive effect for the protein sequence coverage should be certainly gained. Microwave-assisted protein cleavage and many other strategies, such as ultrasonic energy or high pressure, however, were recently reviewed (Capelo et al. 2009). The same is true for immobilized enzymes (Ma et al. 2009; Krenkova and Svec 2009; Spross and Sinz 2009, 2010). Furthermore, the challenges of data interpretation and the software solutions are not presented in detail although both points are important for a maximum possible and correct peptide assignment to the protein sequence and an in-depth analysis of tandem mass (MS/MS) spectra. This list could certainly be continued. Nevertheless, the discussed studies in the following subchapters should be seen as an attempt to summarize knowledge and literature references of bottom-up approaches which could give direction to improved bottom-up/top-down combinations. Until now, most of such combinations more or less ignore this know-how (see chapter “Top-down and bottom-up”). We strongly believe that the insights of the selected bottom-up studies can contribute to increase the sequence coverage in combined top-down/bottom-up workflows, but also to provide more variability to approach the different challenges in the protein species analysis, for instance, membrane proteomes or the detection of phosphorylation sites. At the end of the day, this should make a more sophisticated analysis of protein species possible. Indeed, most of the bottom-up studies are alone unsuitable for the identification of protein species.
Specific enzymatic cleavage strategies
A recently introduced study by the research group of J. J. Coon evaluates all popular specific enzymes, which can be used as an alternative specific proteolysis strategy to trypsin in a proteomic workflow (Swaney et al. 2010). Besides trypsin, the yeast proteome was digested using endoproteinase Lys-C, Arg-C, Asp-N or Glu-C and fractionated by strong cation exchange (SCX). Every fraction was then separated via nano-LC and analyzed using an ESI-linear ion trap (LIT)-orbitrap. Theoretic calculations, however, showed that the combination of these five proteases can cover 95% of all amino acids in the yeast proteome with at least one peptide consisting of 7–35 residues, suitable for mass spectrometric sequence analysis. Therefore, the average sequence coverage of every protein should be significantly improved. Owing to the existence of the aforementioned “proteotypic peptides” in a proteomics workflow, 95% are certainly not obtainable. The following discussion in this chapter, however, will consistently show that every step in the workflow influences the peptide population. The more variants used in every step and the more optimization done, respectively, the less the information gets lost in the process. The optimal peptide length has already been mentioned, but every peptide also needs distinct fragmentation conditions. In the case of the selected study, the situation is even more complex due to the use of different proteases, other than trypsin, which generate peptides with internal basic amino acids. The researchers of this study, nevertheless, had previously developed the “decision tree”, a concept to gain an optimal fragmentation using collision-induced dissociation (CID) or electron transfer dissociation (ETD) (Swaney et al. 2008). The charge state and the mass-to-charge ratio (m/z ratio) constitute critical factors for a successful CID- and ETD experiment, respectively, e.g. all twofold charged peptides, including threefold charged over m/z = 600, fragment optimally using CID, whereas for threefold charged below m/z = 600, ETD is the optimal fragmentation method. Therefore, different “decision trees” were initially tested for all proteases even though, finally, no differences were noticed. As expected, for tryptic digestions, more peptides were fragmented via CID, although for all other proteases, more peptides were sequenced by ETD. The two main reasons for a more frequent ETD selection are the above-mentioned internal basic amino acids yielding high charge states and an extended average peptide length. Finally, the average sequence coverage for every identified protein could be improved from 20 to 40% by the use of five different enzymes. The number of identified proteins was marginally enhanced between 2,700 and 3,300 (depends on the used enzyme) to 3,900, only when the 92,000 peptides from the digests of the five enzymes were pooled. Furthermore, it could be proven that low abundant proteins (<100 copies/cell) were considerably better detectable when this combination of five enzymes is used. The sequence coverage was improved by a factor of three, but the yield was only 7%. In contrast, high abundant proteins (100,000 copies/cell) provided sequence coverage of ~75%. Indeed, the application of five enzymes is pretty laborious, not to mention that two different proteases already made a significant difference in this study. Moreover, three digestions using three different enzymes provided more information than three replicates with the same protease. Finally, the study performed theoretical calculations for the analysis of phosphorylation sites. Therefore, several datasets of published yeast proteome analyses, based on tryptic digestions, were investigated concerning the coverage of serines and threonines in the complete proteome by tryptic peptides. Only a combination of their dataset and a previously published Lys-C dataset from the group of M. Mann (de Godoy et al. 2006) with the tryptic datasets resulted in an improvement of 65%. Beside this study, earlier applications of Lys-C, Arg-C, Asp-N and Glu-C, including combinations of these enzymes, are available (Biringer et al. 2006; Choudhary et al. 2003). Furthermore, earlier studies included less specific or unspecific enzymes such as chymotrypsin and proteinase K (see “Less specific and unspecific cleavage strategies”).
In addition to these five specific proteases, the group of A. J. Heck recently introduced an additional enzyme of significant interest. It is a metalloendopeptidase which cuts specifically at the N-terminal of lysine (Lys-N; Nonaka et al. 1995). Therefore, the generated peptides provide simple ETD-/ETcaD- and MALDI-TOF/TOF-spectra containing a clear c- and b-ion-series, respectively (Taouatas et al. 2008; Boersema et al. 2009). ETcaD is a further development of ETD which considerably improves the ETD fragmentation of doubly charged peptides by additional CID (Swaney et al. 2007). Labile posttranslational modifications (PTM), however, still remain at the peptide backbone. Moreover, it could be shown that peptides of a Lys-N digest can be fractionated by SCX into N-terminal acetylated peptides (I), monophosphorylated peptides containing a single lysine (II), peptides containing one lysine (III) and peptides with more than one basic amino acid (IV). Fractions I and IV can be optimally sequenced via CID or ETcaD, whereas fractions II and III using ETcaD. Indeed, all MS/MS spectra of peptides of fraction IV contain z-fragments depending on the position(s) of the internal basic amino acid(s). Owing to the blocked N-terminus, MS/MS spectra fraction I peptides yield only z-fragments. Recently, a special DeNovo-algorithm for ETD spectra of Lys-N digested samples was also published (van Breukelen et al. 2010). Furthermore, it was investigated if, when using Lys-N and trypsin digested samples, modifications at lysine and the N-terminus of a peptide improve the quality of ETD spectra, thus resulting in directed and complete sequence ladders. Guanidinated, dimethylated, and imidazolinylated peptides of Lys-N digests yielded significant progress whereas only guanidinated and imidazolinylated peptides of tryptic digests, containing a single lysine at the C-terminus, provided simplified MS/MS spectra (Hennrich et al. 2009). The tested nicotinylation suppresses peptide backbone fragmentation in ETD. In summary, MS/MS spectra with directed sequence ladders, such as obtained from peptides of Lys-N digests are easy to interpret. Therefore, the peptide identification rate increases and, along with it the average protein sequence coverage. Furthermore, data of Lys-N digested samples are complementary to tryptic ones (Gauci et al. 2009). During the analysis of a purified phosphoproteome, the number of identified phosphorylated peptides was ~70% higher using a combination of Lys-N and trypsin. In contrast, replicates of tryptic digests yielded only 25% improvement. Therefore, the data are consistent with experimental and theoretical results of Swaney et al. (2010).
Less specific and unspecific cleavage strategies
For the analysis of membrane proteins, the available specific enzymes are insufficient due to the low number of cleavage sites. Beside the specific proteases, some alternative less specific or unspecific enzymatic or chemical cleavage strategies have been consequently established for this problematic protein group in the last years, and in most cases for shotgun approaches. Many protocols, however, are based on old ideas. Chymotrypsin and elastase were introduced by Lucas et al. (1969) and Morris et al. (1974), respectively, for the mass spectrometric determination of the primary structure of purified proteins. Morris stated that they found elastase to be an ideal proteolytic enzyme for combination with the mass spectrometric study of dehydrofolate reductase. Nevertheless, Wu et al. (2003) considered elastase and subtilisin to be unsuitable for the analysis of complex proteomes. For instance, in the case of the vesicle proteome they found elastase and subtilisin activities to be substantially diminished when applied to complex membrane-containing samples. However, they established a protocol based on the unspecific protease proteinase K in an alkaline milieu. In an earlier study, MacCoss et al. (2002) showed that digestions of a standard protein mix, purified protein complexes and lens tissue with the enzyme combination elastase, subtilisin and trypsin provide high sequence coverage and a better detection of PTMs and such samples, though, are less complex compared to the vesicle proteome of Wu et al. (2003). Using the three enzyme proteolysis strategy, the analyzed standard proteins yielded sequence coverages of approx. 90%. Abundant proteins in the purified protein complexes or lens tissue revealed significantly higher sequence coverages. Nevertheless, the various overlapping peptides for abundant proteins, which build up clusters in certain sequence ranges, are the most striking point in this study. Before McCoss, however, Schlosser et al. (2001, 2002, 2005) demonstrated in several studies that elastase is an efficient enzyme for phosphoprotein analysis. Later studies with elastase using membrane proteomes (purple membranes, Corynebacterium membranes) or a phosphoproteome could finally disprove Wu et al. (2003) that elastase is inapplicable for the analysis of subproteomes (Rietschel et al. 2009a; Wang et al. 2008).
An alternative proteolysis strategy for membrane proteins was introduced by Fischer et al. (2006). Initially, the theoretical digestion of membrane proteomes using different cleavage reagents demonstrated that a combination of trypsin and chymotrypsin could be beneficial for the membrane protein analysis in terms of sequence coverage and number of identified proteins (Fischer and Poetsch 2006). The authors summarized that a combination of a cleavage at hydrophobic and hydrophilic amino acids is advantageous for the membrane proteome analysis. Therefore, the combination of chymotrypsin and Glu-C would also be well suited. In practice, however, the results were not convincing when using the model membrane protein bacteriorhodopsin. A following study based on the analysis of the membrane proteome of Corynebacterium glutamicum, after trypsin and chymotrypsin digests, proved the expected effect of this enzyme combination (Fischer et al. 2006).
Many of the previously mentioned alternative cleavage protocols are summarized in an actual review by Speers and Wu (2007). In addition, detailed tables and references for chaotropic reagents, detergents and organic solvent systems, including threshold amounts for different cleavage strategies, are provided. These additives are obligatory for membrane protein solubilization, denaturation and accessibility into the membrane; such additives certainly also improve the accessibility to soluble proteins. Another main focus of this and another review is the recovery of hydrophobic peptides using LC separation at enhanced temperature (60°C; Blackler et al. 2008a). Apart from the already mentioned protocols, a pepsin method was introduced by Han and Schey (2004). LC–ESI-MS/MS analysis of aquaporin digested by pepsin showed that 100% sequence coverage is possible in this case. A pepsin proteolysis protocol for MALDI has recently been evaluated and tested using purple membranes (Rietschel et al. 2009b). The most abundant membrane protein bacteriorhodopsin (~90–99%) yielded sequence coverage of nearly 60%. A number of 40 peptides could be assigned to the sequence of bacteriorhodopsin after direct MALDI measurement and approximately 70 using a LC–MALDI approach. High peptide numbers were also obtained for lower abundant proteins, e.g. 80 peptides using LC–MALDI for the S-layer cell surface glycoprotein. Indeed, this bacteriorhodopsin sequence coverage is not as impressive as the 100% sequence coverage observed after a tryptic digestion in 60% methanol (MeOH) in combination with LC–ESI (Blonder et al. 2004). For the excellent coverage, however, CID fragment spectra of low quality were also taken into consideration, especially from large tryptic fragments. The straightforward idea to use the MS compatible MeOH as a solubilization and denaturation agent, nevertheless, led to following studies based on digestions with chymotrypsin, elastase and pepsin to apply the same buffer system which significantly improved the quality of the digestions (Fischer et al. 2006; Rietschel et al. 2009a, b). The denaturing and solubility-improving properties of MeOH were also emphasized in the review by Speers and Wu (2007). One last protocol from this review worth mentioning combines a proteinase K digest under basic conditions with a cyanogen bromide (CNBr) cleavage in formic acid (Blackler et al. 2008b). First of all, the hydrophilic protein parts are removed using proteinase K. Then, the remaining residues in the membrane are delipidated by the formic acid and cleaved by CNBr at methionine into smaller fragments. Thereafter, the hydrophilic and hydrophobic peptide fraction can be separately analyzed by, for instance, MudPIT.
Chemical cleavage strategies
The application of CNBr for methionine cleavage has been extensively used throughout the years. Washburn et al. (2001) has evaluated the MudPIT approach using CNBr and trypsin as cleavage reagents. Van Montfort (2002a, b) established an in-gel proteolysis protocol for the CNBr/trypsin combination. Using solely CNBr cleavage, 100% sequence coverage was immediately gained for the model membrane protein bovine rhodopsin (Ablonczy et al. 2005). Recently, a strategy concerning 2-nitro-5-thiocyanobenzoic acid (NTCB) cleaving at cysteine, combined with a tryptic digestion, resurfaced for membrane proteome analysis (Iwasaki et al. 2009). Besides cysteine and methionine, further possible amino acid positions, which can be cut by chemical reagents, are tryptophan, aspartic acid and the amino acid combination asparagine–glycine. Chemical reagents which are generally used for protein digestion include: cyanogen bromide (CNBr) cleaving at methionine (Met) residues, BNPS-skatole or iodosobenzoic acid cleaving at tryptophan (Trp) residues, formic acid cleaving at aspartic acid (Asp) peptide bonds, hydroxylamine cleaving at asparagine–glycine (Asn-Gly) peptide bonds, and 2-nitro-5-thiocyanobenzoic acid (NTCB) cleaving at cysteine (Cys) residues. Except for iodosobenzoic acid, Crimmins et al. (2001, 2005) provides an overview of the basic protocols, including links to the original literature, for the in-solution cleavage and the cleavage on PVDF membranes, respectively. An older review of Han et al. (1982) also contains general information about the cleavage protocol with iodosobenzoic acid. Additional variants of the chemical cleavage strategies are supplied by a compilation of protocols from Smith (2003). In general, chemical cleavage strategies generate high mass peptides due to the infrequency of the specifically cleaved peptide bonds in protein sequences. Therefore, they are interesting cleavage tools for the middle-down approach or the strategy which combines top-down and bottom-up (see corresponding chapters). Besides CNBr and acidic cleavage, however, only very few actual studies are available which apply other chemical reagents. Nevertheless, it can be suggested from the sparse literature that the neglected chemical cleavages with NTCB, BNPS-skatol, iodosobenzoic acid or hydroxylamine, especially in combination with other enzymatic or chemical cleavage strategies, yield additional peptides which improve the sequence coverage of the protein of interest (Freemont et al. 1988; Vestling et al. 1994; Rahali and Gueguen 1999; Wu et al. 1996; Yamagami and Ishiguro 1998).
In contrast, there are some interesting new applications of the acidic cleavage at Asp which is why this method is exemplarily summarized as follows. Diluted acid (pH ~2) cleaves at 108°C in 2 h C-terminal at Asp (Schultz et al. 1962; Inglis 1983). N-terminal cleavage occurs only rarely. Frequently used acids are formic acid (Li et al. 2001), diluted hydrochloric acid (Zubarev et al. 1994; Vorm and Roepstorff 1994) or trifluoroacetic acid (TFA; Tsugita et al. 1992). The use of a microwave reduces the reaction time from 2 h to 1 and 10 min (Zhong et al. 2005; Swatkoski et al. 2006, 2007a, b). During the investigation of the protein components of a virus, Swatkoski et al. (2007b) were able to gain very high sequence coverage via microwave-assisted acidic cleavage followed by MALDI-time-of-flight (TOF)-MS. The same group tested the influence of acidic cleavage on some common modifications using model proteins and peptides (Swatkoski et al. 2008). For example, methionine and cysteine were not oxidized, but phosphate groups were partially cleaved. Furthermore, the ribosomal proteome of yeast was investigated using an LC–ESI–LIT-Orbitrap or LC–MALDI-TOF/TOF (Swatkoski et al. 2007a). All above-mentioned acidic cleavages, however, were conducted offline. Meanwhile Hauser and Basile (2008) developed a platform which performs the reduction in the proteins (optional) as well as the microwave-assisted acidic cleavage, followed by the peptide separation via LC and an ESI-MS/MS acquisition. The cleavage at Asp, nevertheless, doubles the peptide length on average when compared with tryptic digests if miss cleavage sites are not taken into consideration (16 residues vs. 9). Owing to internal basic amino acids, the resulting peptides are higher charged as compared to tryptic ones. Therefore, ETD and electron capture dissociation (ECD) are better suited for fragmentation experiments than CID, i.e. both fragmentation techniques result in significantly extended sequence ladders (Hauser et al. 2008). Besides the microwave-assisted acidic cleavage in diluted acids, a variant was evaluated using concentrated acids. Zhong et al. (2005) recovered nearly 100% sequence coverage when using the model membrane protein bacteriorhodopsin and 25% TFA as cleavage reagent. 25% TFA cleaved quite unspecifically, but cleavages at glycine were detected more frequently. Moreover, C-terminal and N-terminal sequence ladders were visible in the MS spectra, especially when low sample concentrations were used. As a real sample, the membrane fraction of a human breast cancer cell was analyzed using LC–MALDI. A further variant is the recently published specific acidic cleavage at Asp using MALDI matrices (Remily-Wood et al. 2009). Protocols for in-solution and in-gel cleavage were evaluated using standard proteins.
In-gel proteolysis
In-gel digestions remain unsatisfactory till the present day. Hydrophobic and larger peptides are hardly extractable which is understandable, as the acrylamide network provides an attractive adhesion surface (Speers and Wu 2007; Bornemann et al. 2010). In general, the loss from in-gel digest protocols is 15–50% (Hellman et al. 1995; Speicher et al. 2000; Stewart et al. 2001). The size of the protease is also of certain importance, i.e. proteases beyond the mass of 25 kDa, for example pepsin and Glu-C, can hardly penetrate into the gel matrix, which hinders the effective protein cleavage (Rabilloud et al. 2009). Furthermore, proteins are fixated during standard staining procedures and SDS is removed whereby considerable amounts of protein might be not accessible to an enzyme due to aggregation (Speers and Wu 2007). High accessibility of the protease to the protein and an effective extraction would consequently result in higher sequence coverages. Many strategies, including the usage of detergents, organic solvents, microwave excitation, ultrasonic treatment and immobilized enzymes, are only partially successful to solve the both mentioned problems (Lazarev et al. 2009). A current publication shows that after protein separation, the controlled extension of the acrylamide network using the cleavable crosslinker ethylene-glycol-diacrylate (EDA) improves enzyme penetration (Bornemann et al. 2010). The sequence coverage and number of assigned peptides increased for every analyzed protein using a tryptic digestion and MALDI-peptide mass fingerprinting (PMF). Whether the gel system is practicable for real proteomic studies will be shown in the future.
Another main problem of enzymatic in-gel digestions is missing robust standard protocols, except for tryptic in-gel protocols. A frequently applied trypsin in-gel digestion procedure was developed, for instance, by Shevchenko et al. (2006). Indeed, a number of other enzymes, such as chymotrypsin (Galkin et al. 2008; Chmelik et al. 2009; Papasotiriou et al., 2010), elastase (Galkin et al. 2008; Schlosser et al. 2002, 2005; Papasotiriou et al. 2010), proteinase K (Schlosser et al. 2005; Jansson et al. 2008; Bendz et al. 2008; Papasotiriou et al. 2010) and pepsin (Jansson et al. 2008; Chen et al. 2010), have also been used. But, as far as we know, protocols as well-evaluated as the tryptic one have not been established. In fact, the same is true for in-solution digestion protocols of other enzymes than trypsin. We discussed many protocols in the previous chapters but they all do not have the robustness of tryptic proteolysis protocols. The importance of well-evaluated proteolysis protocols shall be exemplary demonstrated by an in-gel digest using proteinase K. The adjustment of a pH value of 11 using sodium carbonate buffer is critical, because carbon dioxide from the air would change it over time, resulting in too many short peptides (Jansson et al. 2008). A pH of 12 circumvents this problem but also causes more background in the mass spectrometric analysis as the gel gradually starts to hydrolyze. Another gel system based on N-acryloyl-aminopropanol is more stable against hydrolysis but is costly (Simo-Alfonso et al. 1996a, b; Bendz et al. 2008).
MALDI- and ESI-MS
Even without detection losses during a gel extraction or LC separation, all peptides ought to ionize equally efficiently. In reality, this is not the case. ESI particularly ionizes analytes which concentrate in the charged droplet surface phase (Cech and Enke 2000; Speers and Wu 2007). Therefore, hydrophobic peptides are detected more frequently than hydrophilic. This could also be proved by derivatizing peptides with alkyl tags (Frahm et al. 2007). In contrast, MALDI detects basic, polar and aromatic residues quite efficiently (Krause et al. 1999; Speers and Wu 2007). Particularly, arginine-containing peptides induce intense signals in MALDI-spectra. Due to the various peptides of an elastatic digestion as compared to a tryptic one, in terms of physicochemical properties, the systematic investigation of elastatic digests from Rietschel et al. (2009a) constitute a good example for the above-mentioned differences between ESI and MALDI. Elastatic digestions generate, for example, many peptides with a pI of 6 which are prevalently detected by ESI. Due to their frequently small peptide size below 700 Da, which correspond to the MALDI matrix region, the comparison is somehow unfair using this peptide group. Nevertheless, small and hydrophobic peptides are generally better detectable using ESI. On the contrary, MALDI is the preferred method for basic peptides. Acidic peptides show no performance differences. Due to the above, MALDI and ESI are considered complementarities as noted by several studies (Yang et al. 2007; Irungu et al. 2008; Zhang et al. 2008; Molle et al. 2009). In the study of Rietschel et al. (2009a), the authors were able to identified more peptides using ESI, not only as a result of more efficient detection of small peptides, but also because of the more precise precursor mass (5 ppm ESI; 50 ppm MALDI) of the acquired MS/MS spectra and the considerably better established LC–ESI approach compared to the relatively new LC–MALDI-Workflow. Following studies demonstrated the enormous positive influence of a precise precursor mass between 3 to 5 ppm for the significant identification of MALDI-MS/MS spectra (Rietschel et al. 2009c). These spectra were generally of higher quality compared to ESI-MS/MS spectra, i.e. more extended sequence ladders, but were ambiguous in consequence of the insufficient precursor mass accuracy. Due to the broad specificity of elastase, since it cuts with a frequency of 80% at the C-terminal of the amino acids Ala, Val, Leu, Ile, Ser, Thr, database search was carried out without a defined enzyme specificity. Thus, a high mass accuracy was extremely important. A further problem in significance issues may be the general abundant presence of internal fragments in MALDI-MS/MS spectra. This, however, has only been investigated for tryptic digests (Khatun et al. 2007). In general, a directed fragmentation, as provided by the previously discussed Lys-N digests, would be desirable. Various modification strategies also try to achieve this aim such as N-terminal sulfating, guanidination, dimethylation, imidazolinylation, nicotinylation or itraq (Hennrich et al. 2009; Ross et al. 2004). Except for sulfating reagents, all others have a positive effect on the detection of peptides. A study from Ernoult et al. (2008) using itraq labeled tryptic sample and a proteomic workflow based on nicotinylation combined with proteinase K and pepsin digests (Jansson et al. 2008), are selected as representative examples.
Good detectability of an analyte in MALDI usually depends on the used matrix. Phosphorylated peptides, for instance, can be optimally analyzed by using 2,5-dihydroxybenzoic acid (DHB)/1% phosphoric acid (Kjellström and Jensen 2004). A current research paper concerning the rational design of MALDI matrices, however, has shown that, even though alpha-cyano-4-hydroxycinnamic acid (CHCA) is the most widely used matrix for MALDI analysis of peptides, there is still room for improvement by the development of new matrices (Jaskolla et al. 2008). A newly designed matrix, 4-chloro-alpha-cyanocinnamic acid (Cl-CCA), not only enhances the ionization efficiency of acidic and neutral peptides, but also provides the same excellent performance for basic peptides as CHCA (Jaskolla et al. 2009). In addition, a higher number of phosphorylated peptides is detected. Therefore, sensitivity, number of detected peptides and sequence coverage is already improved for tryptic in-solution and in-gel digests which could be proven with the standard protein bovine serum albumin BSA (Jaskolla et al. 2008). Using the three proteases trypsin, chymotrypsin and pepsin, a following study investigated the advantages of Cl-CCA for in-solution digestions of diverse proteins (Jaskolla et al. 2009). The efficiency of Cl-CCA is especially obvious in the case of more complex peptide mixtures of the less specific enzyme chymotrypsin and the relatively unspecific enzyme pepsin. Figure 3 displays sequence coverage recovery from trypsin and pepsin digestions of beta-casein using CHCA and Cl-CCA. Independent of the enzyme, the number of assigned peptides and sequence coverage is always higher for the Cl-CCA preparation. The higher complexity of a pepsin digest, i.e. the number of possible cleavage positions, peptide hydrophobicity and pI distribution of peptides, can simply be more efficiently detected using Cl-CCA. Indeed, some gaps remain in the sequence using a final on-target peptide amount of 10 fmol. Nevertheless, the dilution effect of unspecific or less specific enzymes due to the various cleavage possibilities compared to trypsin is less apparent when using Cl-CCA as matrix. As a result, nearly doubled sequence coverage and an enhanced detection of clustered peptides are achieved. In addition, detection of phosphorylation sites in the tryptic digest was only possible in the case of Cl-CCA. This outlines once again the importance to apply different enzymes.
Sequence coverage of 10 fmol beta-casein (P02666) after in-solution digestions using the following enzymes and MALDI matrices for measurement: a using trypsin as enzyme and CHCA as matrix, b using trypsin as enzyme and Cl-CCA as matrix, c using pepsin as enzyme and CHCA as matrix, and d using pepsin as enzyme and Cl-CCA as matrix. Gray boxes represent the position of identified peptides in the protein sequence. Several gray boxes at the same position indicate the detection of the corresponding methionine oxidations. Black colored arrow reveals the serine phosphorylation detected only in the case of trypsin as enzyme and Cl-CCA as matrix (data source Jaskolla et al. 2009)
Top-down
In an actual publication, one of the top-down pioneers N. L. Kelleher sums up the newly established protocol in the following way: “this is sufficient for top-down experimentation across a wide range of masses and should extend the number of laboratories able to perform top-down proteomics in a routine fashion” (Vellaichamy et al. 2010). Nevertheless, certain cautiousness is generally advisable, especially in the case of the complicated analysis of intact proteins. Positive aspects for the fast development of top-down in the future include the impressive progress of fourier transform ion cyclotron resonance (FT-ICR) instruments in the last years (Schaub et al. 2008), the Orbitrap (Makarov 2000; Macek et al. 2006; Bondarenko et al. 2009), the fragmentation techniques ECD (Zubarev et al. 1998) and ETD (Syka et al. 2004) and the considerably improved strategies for data analysis (Garcia 2010; Zamdborg et al. 2007). Both above-mentioned fragmentation techniques are advantageous over others as only the backbone bonds of a protein are cleaved. Labile PTMs such as phosphorylations or glycosylations are preserved (Breuker et al. 2008). Particularly, however, the increasing knowledge concerning the handling of tertiary protein structures in the gas phase has in the past and will in the future extend the mass range for the detection and fragmentation in top-down proteomics (Breuker et al. 2008). ECD, for instance, only reduces stepwise the charge state of a protein larger than 20 kDa. On the contrary, “activated ion” (AI) ECD can generate sequence information in the range of 20–50 kDa, but the efficiency drops with increasing protein mass (Horn et al. 2000). “Prefolding dissociation” (PFD) makes top-down MS of proteins larger than 200 kDa possible (Han et al. 2006; Karabacak et al. 2009).
Nevertheless, there are still some difficult hurdles to overcome, some of which are exemplarily cited below out of a review from Garcia (2010). First, there is the charge distribution of a protein in ESI which dilutes the protein amount to different charge states. This reduces the sensitivity, especially for large proteins. Furthermore, different charge states of the same protein can result in different MS/MS spectra, which complicate the interpretation via a software algorithm. Due to the fact that high charge states are advantageous for ECD and ETD, every reduction of the charge distribution and shift to higher charge states would improve the sensitivity of the protein detection and the fragmentation performance in top-down approaches. Secondly, instrumental developments are needed to extend the dynamic range, accelerate the duty cycle and gain sensitivity. Particulary, the 30 kDa limit should be negotiated (Garcia 2010). Up to this limit, top-down analyses of diverse protein types, even of hard candidates such as integral membrane proteins, yield high or 100% sequence coverage if a sufficient protein amount is available and a proper time-scale for the measurement is given (Kelleher et al. 1999; Horn et al. 2000; Sze et al. 2002; Whitelegge et al. 2006; Zabrouskov and Whitelegge 2007; Breuker et al. 2008; Ayaz-Guner et al. 2009; Ryan et al. 2010).
Nevertheless, the most striking gap between top-down and high-throughput bottom-up approaches is the performance difference in the separation of intact proteins compared to peptides (Garcia 2010; Vellaichamy et al. 2010). Indeed, the 2-DE is not a high-throughput technique but it provides an excellent resolving power on the protein level (Görg et al. 2009). Surely, there are well-known limitations, such as extremely basic, membrane and very large proteins, which are all difficult, if not impossible, to separate (Görg et al. 2009). Owing to the challenging extraction of intact proteins from gels, 2-DE is not used as a separation strategy in top-down approaches. Different strategies, such as electroblotting, direct analysis of thin gel slices via MALDI, extraction using passive diffusion or a variety of electroelution approaches, have been tested (Razunguzwa et al. 2009). Low protein recovery, low sensitivity and time consumption, however, constitute disadvantages found in all the aforementioned techniques. A similar situation is summarized by an actual publication which introduces a microfluidics-based electroelution strategy (Razunguzwa et al. 2009). The new system perhaps performs better in terms of the previously mentioned critical points. The future, however, will reveal if the new technique is sufficient to solve real problems. Nevertheless, a top-down strategy based on the pre-fractionation performing solely SDS-PAGE or IEF followed by SDS-PAGE has already been evaluated with real samples (yeast and human cell lines; Vellaichamy et al. 2010). The advantage of the established pre-fractionation approach is the fact that commercial systems are available, such as the off-gel electrophoresis for IEF and the GELFrEE system (gel-eluted liquid fraction entrapment electrophoresis) for PAGE (Ros et al. 2002; Tran and Doucette 2008). In the study by Vellaichamy et al. (2010), all collected fractions were subsequently separated by liquid chromatography using a polymeric stationary phase. In terms of sensitivity, the instrumentation setup performed better than C4 material for proteins up to 80 kDa. Finally, 10–60 proteins per fraction could be identified by an online coupled FT-ICR mass spectrometer (MS) using nonselective nozzle skimmer dissociation (NSD; Loo et al. 1988), whereas the number of identified proteins decreased from the low-mass to the high mass fractions. Nonselective means that a precursor selection is not possible. This reveals one existing problem in top-down experiments. A charge state selection of a large protein is very difficult when the mass spectrometer is coupled online to a LC. The second problem is the low sequence coverage for high mass proteins. The evaluation of the top-down method using three standard proteins, resulted in sequence coverages of 40–50% for cytochrome c (12 kDa) and carbonic anhydrase (29 kDa), but only approximately 6% in the case of BSA (66 kDa). However, this result is not considered remarkable if the increase in complexity of the tertiary gas phase structure from a small to a large protein is taken into consideration (Breuker et al. 2008). When compared with NSD, ECD would certainly perform better and preserve labile PTMs, but until now it is incompatible with online LC–MS/MS experiments (Garcia 2010). On the contrary, ETD can be used in online experiments. The group of Burlingame obtained sequence coverages ranging from 30 to 70% after analyzing histones (10–15 kDa) from embryonic murine stem cells and even characterized a number of PTMs (Eliuk et al. 2010). However, it was also indicated that the identification of larger proteins is still possible but an analysis of PTMs is then limited to the protein termini. For the protein separation in this study, a classical reversed-phase LC (C18) was used online coupled to a LIT-Orbitrap.
Apart from the few given examples, several other pre-fractionation and separation strategies based, for example, on ion exchange- (IEX), reversed-phase- (RP) and hydrophilic interaction chromatography (HILIC) or free-flow electrophoresis (FFE), are frequently used. A more detailed overview about protein pre-fractionation and separation is provided by the reviews of Garcia (2010) and Righetti et al. (2005b). In summary, however, it must be pointed out once more that competitive separation techniques of intact proteins as opposed to peptides are still missing. In conclusion, it should be apparent that without an effective protein separation, high sequence coverage for a protein is quite unrealistic.
Middle-down
Due to the discussed difficulties in protein separation and fragmentation, an old concept has recently been reanimated, and termed middle molecule MS or middle-down (Yergey et al. 1984; Forbes et al. 2001; Wu et al. 2005; Garcia 2010). In 2001, Kelleher’s group introduced one of the first elaborated concepts based on the FT-ICR-MS analysis of peptide fragments between 10 and 40 kDa produced by limited proteolysis (Forbes et al. 2001). First of all, a purified 159 kDa protein was digested using Lys-C. Large peptide fragments in the mass range of 5–48 kDa were then assigned to the protein sequence with a mass accuracy of 50 ppm. Finally, these assigned peptides covered 100% of the protein sequence. Another model protein with a mass of 199 kDa, however, attained only 15% sequence coverage due to failure of limited proteolysis using Lys-C, as too many small peptides were generated. MS/MS experiments were not conducted during this study. Based on the preliminary data, the following theoretical concept was developed: pre-fractionation of the proteome followed by limited proteolysis, identification of proteins by MS/MS experiments of peptides in the mass range of 10–40 kDa using high-resolution MS, e.g. FT-ICR MS, and, finally, closing of sequence gaps with targeted MS/MS experiments. In the summary of the study, however, it was pointed out that more robust cleavage methods are necessary, which mainly generate fragments in the mass range of 10–40 kDa, as Lys-C was unsuitable for one chosen model protein. The chemical cleavage reagent CNBr was suggested as an alternative (Kelleher et al. 1999). Furthermore, it was stated that instrumentation development is not sufficient enough for their suggested concept, particularly the fast acquisition of MS/MS spectra in the required mass range.
The group of Karger revived the concept of Kelleher and termed it with the abbreviation extended range proteomic analysis (ERPA). ERPA analyses peptides in the mass range 0.5–10 kDa (Wu et al. 2005, 2007; Zhang et al. 2007). The group refined ERPA to such an extent that the sample throughput, sensitivity and performance of the chromatography are nearly comparable to bottom-up approaches, whereas the sequence coverage and detection of PTMs, such as phosphorylations and glycosylations, is significantly improved. It should be mentioned, however, that the method was only evaluated with single proteins. The potential of middle down is obvious. The possibility to sequence larger peptides facilitates the detection of PTM patterns and small changes in the primary sequence, which are not detected in the bottom-up approach (Garcia 2010). The identification of 170 histone species via middle-down, which was already given as a straightforward example for the identification of protein species in the prologue, proves this (Garcia et al. 2007). Garcia et al. fractionated the purified N-terminus of histone 3.2 (5–6 kDa) using HILIC. The protein species of every fraction were then detected offline via FT-ICR and sequenced by ECD. Due to the strict focus on the N-terminus, the study provides an insight into the variability of the posttranslational modification pattern, but much information remains hidden, as the histone has not completely been characterized after all. In the same way, the above-mentioned top-down study of Burlingame’s group used not only strictly this approach, but also a middle-down strategy to analyze the highly N-terminal modified region of histone 4 from embryonic murine stem cells (Eliuk et al. 2010). A digest with endoproteinase Asp-N generated an N-terminal 23-mer of Histone 4 which could be detected in several variants. Recently, the pioneers of middle down demonstrated for a subproteome (human nuclei) that some information of unexpected or multiple modifications are provided and the middle-down approach can compete with the classical bottom-up approach in terms of identified proteins (Boyne et al. 2009). Nevertheless, the middle-down approach is still suboptimal because the direct link between peptides and their original source, the protein, is lacking. The probability, however, to assign larger fragments correctly to a protein species is certainly higher.
Top-down and bottom-up
The main advantage of classical 2D-PAGE is certainly the link between the original protein and the enzymatically or chemically generated peptides which is not lost. Owing to the separation by protein pI and size, protein species, such as phosphorylated or glycosylated variants can be mapped (Görg et al. 2009). The critical factors hindering 100% sequence coverage as well as the improvement possibilities have already been discussed in the bottom-up chapter. The same is true for the extraction of intact proteins and the subsequent top-down analysis which was referred to in the top-down chapter. Therefore 2D-PAGE could be termed as the oldest combination of top-down and bottom-up even though the protein mass is only determined quite inaccurately by marker proteins of known molecular weight.
Therefore, Lubman’s group developed an alternative technique to 2D-SDS-PAGE which has been further refined over the years and tested on different samples such as “breast cancer cells” (Wall et al. 2000, 2001; Kachman et al. 2002; Yan et al. 2003; Zhu et al. 2003). Initially, proteins were pre-fractionated according to their pI value and then separated via RP-LC coupled to a fraction collector. Finally, all collected RP-LC fractions were tryptically digested and analyzed by MALDI-TOF in the workflow prototype. A mass determination of the proteins from all collected RP-LC fractions was subsequently performed using MALDI-TOF MS. To acquire more accurate protein masses of the different RP-LC fractions, the fractions of the IEF pre-fractionation were separated using the same RP-LC setup coupled online to an ESI-TOF. Owing to the reproducible elution of the proteins in the used RP-LC setup, the MALDI-PMF data could be easily correlated to the protein masses of the ESI-TOF data. Knowledge of the pI and mass of every detected protein allowed for the calculation of a virtual 2D-gel which additionally contained the organic solvent content of every detected protein at the elution time point, i.e. a value for the hydrophobicity. The assignment of the protein mass of the ESI-TOF to the MALDI-PMF-data is finally based on this value. Besides a MALDI-TOF analysis, in later studies, the peptide mixtures of every RP-LC fraction were also analyzed by ESI-TOF or sequenced after further separation via capillary electrophoresis using an ESI-quadrupol ion-trap (QIT)-TOF. Because the link between protein mass and tryptic peptides is never lost during the analysis, this coupling of bottom-up with top-down is certainly suited for the analysis of protein species. This is confirmed for all analyzed samples by the excellent sequence coverage gained for proteins up to 70 kDa and the identification of various modifications based on the accurate protein mass and the bottom-up data.
Indeed, there are various combinations of bottom-up and top-down approaches which independently analyze the peptide and protein level of less complex protein mixtures but also complex proteomes after a pre-fractionation on the protein level (VerBerkmoes et al. 2002; Strader et al. 2004; Simpson et al. 2006; Sharma et al. 2007). Owing to the missing direct link between protein and its corresponding peptides, these strategies are unusable for protein species analysis and are therefore neglected. Contrary, Millea et al. (2006) introduced a platform which preserves this link. The cytosol of E. coli was fractionated on the protein level via strong anion exchange (SAX) and subsequently separated using RP-LC. The flow was split; 10% were used for the online coupling to an ESI-TOF for the protein mass determination and 90% were collected into fractions. The single fractions were then analyzed after a tryptic digestion using a MALDI-QIT-TOF. Using a more improved instrumentation for protein mass determination, such as a 12 T FT-ICR mass spectrometer, a similar concept was recently evaluated using standard proteins (phosphorylated/unphosphorylated) and the yeast proteasome or a complex mixture of purified yeast phosphoproteins (Wu et al. 2009a, b). Owing to the lower complexity of the samples, only a 1D-LC separation was performed with a flow rate of 5.5 µl/min. For the analysis of phosphorylated proteins, a special metal-free LC platform was developed to ensure the best possible recovery of phosphorylated proteins. From the flow of 5.5 µl/min, 300 nl were online acquired with the FT-ICR using a nanoESI chip for ionization (Advion BioSciences, Inc., Ithaca, NY). The remaining 5.2 µl were fractionated into 96-well plates. One part of every fraction was digested with trypsin and analyzed using a LIT after LC separation. The other part was used for offline top-down experiments using the FT-ICR MS. Another example, using once more an ESI-TOF for the protein mass determination, is the analysis of ribosomal proteins of Bacillus subtilis, which were two-dimensionally separated by SCX and RP-LC (Lauber et al. 2009). In this case, the analysis of the digested fractions was conducted by an ESI-LIT-FT-ICR MS online coupled to an LC. This study is significant, as two different enzymes, trypsin and Glu-C, were applied resulting in high sequence coverage for all identified proteins.
Corresponding MALDI platforms for protein separation, tryptic digest and MALDI TOF/TOF analysis have also been established (Yoo et al. 2006; Zheng et al. 2006). Pre-fractionated protein mixtures, such as from tumor tissue, were separated using capillary LC and spotted onto MALDI plates, which had been pre-coated with trypsin (Harris and Reilly 2002). The digest was finished in approx. 10 min on the target. Thereafter, the samples were co-crystallized with matrix followed by an MALDI-TOF/TOF analysis of every spot. Although a protein mass could not be determined, the link between protein and corresponding peptides is preserved during the workflow, wherefore protein species are identifiable when the sequence coverage is high enough. A slightly modified variant of such a LC–MALDI approach was evaluated by Getie-Kebtie et al. (2008). In addition, in a recent study, highly charged MALDI-spectra of standard proteins have been fragmented using ETD (Trimpin et al. 2010a, b). Therefore, one could start to speculate about the numerous interesting possibilities for LC–MALDI platforms on the protein level, provided that the technique is evaluated for realistic samples and biological problems in the future.
Summary
All reviewed approaches, which combine bottom-up and top-down, should have the potential to provide 100% sequence coverage along with protein species identification and characterization. The know-how of established procedures in bottom-up proteomics should be more widely used by these approaches, such as the various possibilities of enzymatic and chemical cleavage strategies or the application of superior matrices in MALDI such as Cl-CCA. This could significantly improve the sequence coverage, but it should be obvious that the obstacles are vast. In addition, two other main challenges still remain unsolved. Protein separation must obtain the performance of peptide separation and a general strategy is needed that partitions the extremely complex proteome conglomerate into analyzable sub-problems.
Epilogue
Certainly, we were not able to provide a patent remedy for 100% sequence coverage. Therefore, a pessimist would assess the situation as hopeless. An optimist, however, would see the prospects available from the discussed approaches. A critic, nevertheless, could object to the fact that a number of approaches were neglected. For reasons of precaution, two wise men are cited; Quintus Horatius Flaccus and Marcus Valerius Martialis. The first stated: “nes scire fas est omnia (It is impossible to know everything)”, while the latter “bonus vir semper tiro (A good man is always learning)”. In other words, anyone who is interested in 100% sequence coverage is open for new and overlooked ideas of the past to approach the goal.
Nevertheless, after studying the scientific literature, we decided to undertake a quest for “100% sequence coverage”, comparable to the protagonist in Miguel de Cervantes’ famous novel “El ingenioso hidalgo Don Quixote de la Mancha” (The Ingenious Hidalgo Don Quixote of La Mancha). After reading numerous knight tales, Don Quixote dons a rusty suit of armour and a paper hat to go out as a knight-errant in search of adventure. Indeed, it has been obvious from the beginning that 100% sequence coverage for every protein, having in mind the enormous complexity of a proteome, might be far away from reality. Regardless of the consequences, we have tried to tilt at windmills in memory of Don Quixote. As it is in most of Don Quixote’s adventures, he suffered damages. Some less famous adventures of him, however, had a happy ending. Such rays of hope in Don Quixote’s adventures could be compared with the progress concerning the 100% sequence coverage problem. Nevertheless, our knowledge and techniques appear to be “the rusty suit of armour and paper hat of Don Quixote” wherewith we face this enormous challenge. Without any doubt, however, persistence, time and money improve the rusty suite of armour. Therefore, the answer to the following question should be easy to give: is someone, who is interested to solve the 100% sequence coverage problem, more “a fool” or “an idealistic realist”? We won’t provide an answer even though both could be true for Don Quixote.
References
Ablonczy Z, Crouch RK, Knapp DR (2005) Mass spectrometric analysis of integral membrane proteins at the subpicomolar level: application to rhodopsin. J Chromatogr B Analyt Technol Biomed Life Sci 825:169–175
Ayaz-Guner S, Zhang J, Li L, Walker JW, Ge Y (2009) In vivo phosphorylation site mapping in mouse cardiac troponin I by high resolution top-down electron capture dissociation mass spectrometry: Ser22/23 are the only sites basally phosphorylated. Biochemistry 48:8161–8170
Bendz M, Möller MC, Arrigoni G, Wahlander A, Stella R, Cappadona S, Levander F, Hederstedt L, James P (2008) Quantification of membrane proteins using nonspecific protease digestions. J Proteome Res 8:5666–5673
Biringer RG, Amato H, Harrington MG, Fonteh AN, Riggins JN, Hühmer AF (2006) Enhanced sequence coverage of proteins in human cerebrospinal fluid using multiple enzymatic digestion and linear ion trap LC–MS/MS. Brief Funct Genomic Proteomic 5:144–153
Blackler AR, Speers AE, Wu CC (2008a) Chromatographic benefits of elevated temperature for the proteomic analysis of membrane proteins. Proteomics 8:3956–3964
Blackler AR, Speers AE, Ladinsky MS, Wu CC (2008b) A shotgun proteomic method for the identification of membrane-embedded proteins and peptides. J Proteome Res 7:3028–3034
Blonder J, Conrads TP, Yu LR, Terunuma A, Janini GM, Issaq HJ, Vogel JC, Veenstra TD (2004) A detergent- and cyanogen bromide-free method for integral membrane proteomics: application to Halobacterium purple membranes and the human epidermal membrane proteome. Proteomics 4:31–45
Boersema PJ, Taouatas N, Altelaar AF, Gouw JW, Ross PL, Pappin DJ, Heck AJ, Mohammed S (2009) Straightforward and de novo peptide sequencing by MALDI-MS/MS using a Lys-N metalloendopeptidase. Mol Cell Proteomics 8:650–660
Bondarenko PV, Second TP, Zabrouskov V, Makarov A, Zhang Z (2009) Mass measurement and top-down HPLC/MS analysis of intact monoclonal antibodies on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. J Am Soc Mass Spectrom 20:1415–1424
Bornemann S, Rietschel B, Baltruschat S, Karas M, Meyer B (2010) A novel polyacrylamide gel system for proteomic use offering controllable pore expansion by crosslinker cleavage. Electrophoresis 31:585–592
Boschetti E, Righetti PG (2009) The art of observing rare protein species in proteomes with peptide ligand libraries. Proteomics 9:1492–1510
Boyne MT, Garcia BA, Li M, Zamdborg L, Wenger CD, Babai S, Kelleher NL (2009) Tandem mass spectrometry with ultrahigh mass accuracy clarifies peptide identification by database retrieval. J Proteome Res 8:374–379
Breuker K, Jin M, Han X, Jiang H, McLafferty FW (2008) Top-down identification and characterization of biomolecules by mass spectrometry. J Am Soc Mass Spectrom 19:1049–1053
Capelo JL, Carreira R, Diniz M, Fernandes L, Galesio M, Lodeiro C, Santos HM, Vale G (2009) Overview on modern approaches to speed up protein identification workflows relying on enzymatic cleavage and mass spectrometry-based techniques. Anal Chim Acta 650:151–159
Cech NB, Enke CG (2000) Relating electrospray ionization response to nonpolar character of small peptides. Anal Chem 72:2717–2723
Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H (2009) Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res 8:651–661
Chen WQ, Priewalder H, John JP, Lubec G (2010) Silk cocoon of Bombyx mori: proteins and posttranslational modifications-heavy phosphorylation and evidence for lysine-mediated cross links. Proteomics 10:369–379
Chmelik J, Zidkova J, Rehulka P, Petry-Podgorska I, Bobalova J (2009) Influence of different proteomic protocols on degree of high-coverage identification of nonspecific lipid transfer protein 1 modified during malting. Electrophoresis 30:560–567
Choudhary G, Wu SL, Shieh P, Hancock WS (2003) Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J Proteome Res 2:59–67
Cox J, Mann M (2007) Is proteomics the new genomics? Cell 130:395–398
Crimmins DL, Mische SM, Denslow ND (2001) Chemical cleavage of proteins on membranes, Chap 11. Curr Protoc Protein Sci Unit 11.5
Crimmins DL, Mische SM, Denslow ND (2005) Chemical cleavage of proteins in solution, Chap 11. Curr Protoc Protein Sci Unit 11.4
de Godoy LMF, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M (2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol 7:R50
de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455:1251–1254
Distler AM, Kerner J, Peterman SM, Hoppel CL (2006) A targeted proteomic approach for the analysis of rat liver mitochondrial outer membrane proteins with extensive sequence coverage. Anal Biochem 356:18–29
Dumont JE, Dremier S, Pirson I, Maenhaut C (2002) Cross signaling, cell specificity, and physiology. Am J Physiol Cell Physiol 283:C2–C28
Eliuk SM, Maltby D, Panning B, Burlingame AL (2010) High resolution electron transfer dissociation (ETD) studies of unfractionated intact histones from murine embryonic stem cells using online capillary LC separation: determination of abundant histone isoforms and post-translational modifications. Mol Cell Proteomics 9:824–837
Ernoult E, Gamelin E, Guette C (2008) Improved proteome coverage by using iTRAQ labelling and peptide OFFGEL fractionation. Proteome Sci 6:27
Fischer F, Poetsch A (2006) Protein cleavage strategies for an improved analysis of the membrane proteome. Proteome Sci 4:2
Fischer F, Wolters D, Rögner M, Poetsch A (2006) Toward the complete membrane proteome: high coverage of integral membrane proteins through transmembrane peptide detection. Mol Cell Proteomics 5:444–453
Forbes AJ, Mazur MT, Patel HM, Walsh CT, Kelleher NL (2001) Toward efficient analysis of >70 kDa proteins with 100% sequence coverage. Proteomics 1:927–933
Frahm JL, Bori ID, Comins DL, Hawkridge AM, Muddiman DC (2007) Achieving augmented limits of detection for peptides with hydrophobic alkyl tags. Anal Chem 79:3989–3995
Freemont PS, Dunbar B, Fothergill-Gilmore LA (1988) The complete amino acid sequence of human skeletal-muscle fructose-bisphosphate aldolase. Biochem J 249:779–788
Galkin A, Meyer B, Wittig I, Karas M, Schägger H, Vinogradov A, Brandt U (2008) Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem 283:20907–20913
Garcia BA (2010) What does the future hold for top down mass spectrometry? J Am Soc Mass Spectrom 21:193–202
Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL (2007) Pervasive combinatorial modification of histone H3 in human cells. Nat Methods 4:487–489
Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem 81:4493–4501
Getie-Kebtie M, Franke P, Aksamit R, Alterman MA (2008) Experimental evaluation of protein identification by an LC/MALDI/on-target digestion approach. J Proteome Res 7:3697–3707
Godovac-Zimmermann J, Kleiner O, Brown LR, Drukier AK (2005) Perspectives in spicing up proteomics with splicing. Proteomics 5:699–709
Görg A, Drews O, Lück C, Weiland F, Weiss W (2009) 2-DE with IPGs. Electrophoresis 30:S122–S132
Han J, Schey KL (2004) Proteolysis and mass spectrometric analysis of an integral membrane: aquaporin 0. J Proteome Res 3:807–812
Han KK, Richard C, Biserte G (1982) Current developments of chemical cleavage of proteins. Int J Biochem 15:875–884
Han X, Jin M, Breuker K, McLafferty FW (2006) Extending Top-Down Mass Spectrometry to Proteins with Masses Greater than 200 Kilodaltons. Science 314:109–112
Harris WA, Reilly JP (2002) On-probe digestion of bacterial proteins for MALDI-MS. Anal Chem 74:4410–4416
Hauser NJ, Basile F (2008) Online microwave D-cleavage LC–ESI-MS/MS of intact proteins: site-specific cleavages at aspartic acid residues and disulfide bonds. J Proteome Res 7:1012–1026
Hauser NJ, Han H, McLuckey SA, Basile F (2008) Electron transfer dissociation of peptides generated by microwave D-cleavage digestion of proteins. J Proteome Res 7:1867–1872
Hell SW (2007) Far-field optical nanoscopy. Science 316:1153–1158
Hellman U, Wernstedt C, Gonez J, Heldin CH (1995) Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem 224:451–455
Hennrich ML, Boersema PJ, van den Toorn H, Mischerikow N, Heck AJ, Mohammed S (2009) Effect of chemical modifications on peptide fragmentation behavior upon electron transfer induced dissociation. Anal Chem 81:7814–7822
Hoehenwarter W, Ackermann R, Zimny-Arndt U, Kumar NM, Jungblut PR (2006) The necessity of functional proteomics: protein species and molecular function elucidation exemplified by in vivo alpha A crystallin N-terminal truncation. Amino Acids 31:317–323
Horn DM, Ge Y, McLafferty FW (2000) Activated ion electron capture dissociation for mass spectral sequencing of larger (42 kDa) proteins. Anal Chem 72:4778–4784
Inglis AS (1983) Cleavage at aspartic acid. Methods Enzymol 91:324–332
Irungu J, Go EP, Zhang Y, Dalpathado DS, Liao HX, Haynes BF, Desaire H (2008) Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J Am Soc Mass Spectrom 19:1209–1220
Iwasaki M, Masuda T, Tomita M, Ishihama Y (2009) Chemical cleavage-assisted tryptic digestion for membrane proteome analysis. J Proteome Res 8:3169–3175
Jansson M, Warell K, Levander F, James P (2008) Membrane protein identification: N-terminal labeling of nontryptic membrane protein peptides facilitates database searching. J Proteome Res 7:659–665
Jaskolla TW, Lehmann WD, Karas M (2008) 4-Chloro-alpha-cyanocinnamic acid is an advanced, rationally designed MALDI matrix. Proc Natl Acad Sci USA 105:12200–12205
Jaskolla TW, Papasotiriou DG, Karas M (2009) Comparison between the matrices alpha-cyano-4-hydroxycinnamic acid and 4-chloro-alpha-cyanocinnamic acid for trypsin, chymotrypsin, and pepsin digestions by MALDI-TOF mass spectrometry. J Proteome Res 8:3588–3597
John JP, Anrather D, Pollak A, Lubec G (2006) Mass spectrometrical verification of stomatin-like protein 2 (SLP-2) primary structure. Proteins 64:543–551
Jungblut PR, Thiede B, Zimny-Arndt U, Muller EC, Scheler C, Wittmann-Liebold B, Otto A (1996) Resolution power of two-dimensional electrophoresis and identification of proteins from gels. Electrophoresis 17:839–847
Jungblut PR, Holzhütter HG, Apweiler R, Schlüter H (2008) The speciation of the proteome. Chem Cent J 2:16
Kachman MT, Wang H, Schwartz DR, Cho KR, Lubman DM (2002) A 2-D liquid separations/mass mapping method for interlysate comparison of ovarian cancers. Anal Chem 74:1779–1791
Karabacak NM, Li L, Tiwari A, Hayward LJ, Hong P, Easterling ML, Agar JN (2009) Sensitive and specific identification of wild type and variant proteins from 8 to 669 kDa using top down mass spectrometry. Mol Cell Proteomics 8:846–856
Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW (1999) Top-down versus bottom-up protein characterization by tandem high resolution mass spectrometry. J Am Chem Soc 121:806–812
Khatun J, Ramkissoon K, Giddings MC (2007) Fragmentation characteristics of collision-induced dissociation in MALDI TOF/TOF mass spectrometry. Anal Chem 79:3032–3040
Kjellström S, Jensen ON (2004) Phosphoric acid as a matrix additive for MALDI MS analysis of phosphopeptides and phosphoproteins. Anal Chem 76:5109–5117
Krause E, Wenschuh H, Jungblut PR (1999) The dominance of arginine-containing peptides in MALDI-derived tryptic mass fingerprints of proteins. Anal Chem 71:4160–4165
Krenkova J, Svec F (2009) Less common applications of monoliths: IV. Recent developments in immobilized enzyme reactors for proteomics and biotechnology. J Sep Sci 32:706–718
Lauber MA, Running WE, Reilly JP (2009) B. subtilis ribosomal proteins: structural homology and post-translational modifications. J Proteome Res 8:4193–4206
Lazarev AV, Rejtar T, Dai S, Karger BL (2009) Centrifugal methods and devices for rapid in-gel digestion of proteins. Electrophoresis 30:966–973
Li A, Sowder RC, Henderson LE, Moore SP, Garfinkel DJ, Fisher RJ (2001) Chemical cleavage at aspartyl residues for protein identification. Anal Chem 73:5395–5402
Loo JA, Udseth HR, Smith RD (1988) Collisional effects on the charge distribution of ions from large molecules, formed by electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom 2:207–210
Lucas F, Barber M, Wolstenholme WA, Geddes AJ, Graham GN, Morris HR (1969) Mass-spectrometric determination of the amino acid sequences in peptides isolated from the protein silk fibroin of Bombyx mori. Biochem J 114:695–702
Ma J, Zhang L, Liang Z, Zhang W, Zhang Y (2009) Recent advances in immobilized enzymatic reactors and their applications in proteome analysis. Anal Chim Acta 632:1–8
MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR 3rd (2002) Shotgun identification of protein modifications from protein complexes and lens tissue. Proc Natl Acad Sci USA 99:7900–7905
Macek B, Waanders LF, Olsen JV, Mann M (2006) Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol Cell Proteomics 5:949–958
Makarov A (2000) Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal Chem 72:1156–1162
Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25:125–131
Mann M, Kelleher NL (2008) Precision proteomics: the case for high resolution and high mass accuracy. Proc Natl Acad Sci USA 105:18132–18138
Millea KM, Krull IS, Cohen SA, Gebler JC, Berger SJ (2006) Integration of multidimensional chromatographic protein separations with a combined “top-down” and “bottom-up” proteomic strategy. J Proteome Res 5:135–146
Molle D, Jardin J, Piot M, Pasco M, Leonil J, Gagnaire V (2009) Comparison of electrospray and matrix-assisted laser desorption ionization on the same hybrid quadrupole time-of-flight tandem mass spectrometer: application to bidimensional liquid chromatography of proteins from bovine milk fraction. J Chromatogr A 1216:2424–2432
Morris HR, Batley KE, Harding NC, Bjur RA, Dann JG, King RW (1974) Dihydrofolate reductase: low-resolution mass-spectrometric analysis of an elastase digest as a sequencing tool. Biochem J 137:409–411
Nonaka T, Ishikawa H, Tsumuraya Y, Hashimoto Y, Dohmae N (1995) Characterization of a thermostable lysine-specific metalloendopeptidase from the fruiting bodies of a basidiomycete, Grifola frondosa. J Biochem 118:1014–1020
Papasotiriou DG, Jaskolla TW, Markoutsa S, Baeumlisberger D, Karas M, Meyer B (2010) Peptide mass fingerprinting after less specific in-gel proteolysis using MALDI-LTQ-Orbitrap and 4-chloro-alpha-cyanocinnamic acid. J Proteome Res 9:2619–2629
Rabilloud T (2009) Membrane proteins and proteomics: love is possible, but so difficult. Electrophoresis 30:S174–S180
Rabilloud T, Vaezzadeh AR, Potier N, Lelong C, Leize-Wagner E, Chevallet M (2009) Power and limitations of electrophoretic separations in proteomics strategies. Mass Spectrom Rev 28:816–843
Rahali V, Gueguen J (1999) Chemical cleavage of bovine B-lactoglobulin by BNPSSkatole for preparative purposes: comparative study of hydrolytic procedures and peptide characterization. J Protein Chem 18:1–12
Razunguzwa TT, Biddle A, Anderson H, Zhan D, Powell M (2009) Development of a microfluidic-based gel protein recovery system. Electrophoresis 30:4020–4028
Remily-Wood E, Dirscherl H, Koomen JM (2009) Acid hydrolysis of proteins in matrix assisted laser desorption ionization matrices. J Am Soc Mass Spectrom 20:2106–2115
Rietschel B, Arrey TN, Meyer B, Bornemann S, Schuerken M, Karas M, Poetsch A (2009a) Elastase digests: new ammunition for shotgun membrane proteomics. Mol Cell Proteomics 8:1029–1043
Rietschel B, Bornemann S, Arrey TN, Baeumlisberger D, Karas M, Meyer B (2009b) Membrane protein analysis using an improved peptic in-solution digestion protocol. Proteomics 9:5553–5557
Rietschel B, Baeumlisberger D, Arrey TN, Bornemann S, Rohmer M, Schuerken M, Karas M, Meyer B (2009c) The benefit of combining nLC–MALDI-Orbitrap MS data with nLC–MALDI-TOF/TOF data for proteomic analyses employing elastase. J Proteome Res 8:5317–5324
Righetti PG, Castagna A, Antonucci F, Piubelli C, Cecconi D, Campostrini N, Rustichelli C, Antonioli P, Zanusso G, Monaco S, Lomas L, Boschetti E (2005a) Proteome analysis in the clinical chemistry laboratory: myth or reality? Clin Chim Acta 357:123–139
Righetti PG, Castagna A, Antonioli P, Boschetti E (2005b) Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis 26:297–319
Ros A, Faupel M, Mees H, van Oostrum J, Ferrigno R, Reymond F, Michel P, Rossier JS, Girault HH (2002) Protein purification by off-gel electrophoresis. Proteomics 2:151–156
Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3:1154–1169
Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan S, Baniulis D, Cramer WA, Faull KF, Whitelegge JP (2010) Post-translational modifications of integral membrane proteins resolved by top-down Fourier-transform mass spectrometry with collisionally activated dissociation. Mol Cell Proteomics 9:791–803
Santoni V, Molloy M, Rabilloud T (2000) Membrane proteins and proteomics: un amour impossible? Electrophoresis 21:1054–1070
Schaub TM, Hendrickson CL, Horning S, Quinn JP, Senko MW, Marshall AG (2008) High-performance mass spectrometry: Fourier transform ion cyclotron resonance at 14.5 Tesla. Anal Chem 80:3985–3990
Schlosser A, Pipkorn R, Bossemeyer D, Lehmann WD (2001) Analysis of protein phosphorylation by a combination of elastase digestion and neutral loss tandem mass spectrometry. Anal Chem 73:170–176
Schlosser A, Bodem J, Bossemeyer D, Grummt I, Lehmann WD (2002) Identification of protein phosphorylation sites by combination of elastase digestion, immobilized metal affinity chromatography, and quadrupole-time of flight tandem mass spectrometry. Proteomics 2:911–918
Schlosser A, Vanselow JT, Kramer A (2005) Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal Chem 77:5243–5250
Schlüter H, Apweiler R, Holzhütter HG, Jungblut PR (2008) Finding one’s way in proteomics: a protein species nomenclature. Chem Cent J 3:11
Schultz J, Allison H, Grice M (1962) Specificity of the cleavage of proteins by dilute acid. I. Release of aspartic acid from insulin, ribonuclease, and glucagon. Biochemistry 1:694–698
Sharma S, Simpson DC, Tolic N, Jaitly N, Mayampurath AM, Smith RD, Pasa-Tolic L (2007) Proteomic profiling of intact proteins using WAX-RPLC 2-D separations and FTICR mass spectrometry. J Proteome Res 6:602–610
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860
Simo-Alfonso E, Gelfi C, Sebastiano R, Citterio A, Righetti PG (1996a) Novel acrylamido monomers with higher hydrophilicity and improved hydrolytic stability: I. Synthetic route and product characterization. Electrophoresis 17:723–731
Simo-Alfonso E, Gelfi C, Sebastiano R, Citterio A, Righetti PG (1996b) Novel acrylamido monomers with higher hydrophilicity and improved hydrolytic stability: II. Properties of N-acryloylaminopropanol. Electrophoresis 17:732–737
Simpson DC, Ahn S, Pasa-Tolic L, Bogdanov B, Mottaz HM, Vilkov AN, Anderson GA, Lipton MS, Smith RD (2006) Using size exclusion chromatography-RPLC and RPLC-CIEF as two-dimensional separation strategies for protein profiling. Electrophoresis 27:2722–2733
Sims RJ 3rd, Reinberg D (2008) Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol 9:815–820
Smith BJ (2003) Chemical cleavage of polypeptides. Methods Mol Biol 211:63–82
Speers AE, Wu CC (2007) Proteomics of integral membrane proteins—theory and application. Chem Rev 107:3687–3714
Speicher K, Kolbas O, Harper S, Speicher D (2000) Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J Biomol Tech 11:74–86
Spross J, Sinz A (2009) Immobilized monolithic enzyme reactors for application in proteomics and pharmaceutics. Anal Bioanal Chem 395:1583–1588
Spross J, Sinz A (2010) A capillary monolithic trypsin reactor for efficient protein digestion in online and offline coupling to ESI and MALDI mass spectrometry. Anal Chem 82:1434–1443
Stewart II, Thomson T, Figeys D (2001) 18O labeling: a tool for proteomics. Rapid Commun Mass Spectrom 15:2456–2465
Strader MB, Verberkmoes NC, Tabb DL, Connelly HM, Barton JW, Bruce BD, Pelletier DA, Davison BH, Hettich RL, Larimer FW, Hurst GB (2004) Characterization of the 70S ribosome from Rhodopseudomonas palustris using an integrated “top-down” and “bottom-up” mass spectrometric approach. J Proteome Res 3:965–978
Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ (2007) Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem 79:477–485
Swaney DL, McAlister GC, Coon JJ (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods 5:959–964
Swaney DL, Wenger CD, Coon JJ (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res 9:1323–1329
Swatkoski S, Russell SC, Edwards N, Fenselau C (2006) Rapid chemical digestion of small acid-soluble spore proteins for analysis of Bacillus spores. Anal Chem 78:181–188
Swatkoski S, Gutierrez P, Ginter J, Petrov A, Dinman JD, Edwards N, Fenselau C (2007a) Integration of residue-specific acid cleavage into proteomic workflows. J Proteome Res 6:4525–4527
Swatkoski S, Russell S, Edwards N, Fenselau C (2007b) Analysis of a model virus using residue-specific chemical cleavage and MALDI-TOF mass spectrometry. Anal Chem 79:654–658
Swatkoski S, Gutierrez P, Wynne C, Petrov A, Dinman JD, Edwards N, Fenselau C (2008) Evaluation of microwave-accelerated residue-specific acid cleavage for proteomic applications. J Proteome Res 7:579–586
Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA 101:9528–9533
Sze SK, Ge Y, Oh H, McLafferty FW (2002) Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc Natl Acad Sci USA 99:1774–1779
Taouatas N, Drugan MM, Heck AJ, Mohammed S (2008) Straightforward ladder sequencing of peptides using a Lys-N metalloendopeptidase. Nat Methods 5:405–407
Tran JC, Doucette AA (2008) Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal Chem 80:1568–1573
Trimpin S, Inutan ED, Herath TN, McEwen CN (2010a) Matrix-assisted laser desorption/ionization mass spectrometry method for selectively producing either singly or multiply charged molecular ions. Anal Chem 82:11–15
Trimpin S, Inutan ED, Herath TN, McEwen CN (2010b) Laserspray ionization, a new atmospheric pressure MALDI method for producing highly charged gas-phase ions of peptides and proteins directly from solid solutions. Mol Cell Proteomics 9:362–367
Tsugita A, Takamoto K, Kamo M, Iwadate H (1992) C-terminal sequencing of protein. A novel partial acid hydrolysis and analysis by mass spectrometry. Eur J Biochem 206:691–696
van Breukelen B, Georgiou A, Drugan MM, Taouatas N, Mohammed S, Heck AJ (2010) LysNDeNovo: an algorithm enabling de novo sequencing of Lys-N generated peptides fragmented by electron transfer dissociation. Proteomics 10:1196–1201
van Montfort BA, Doeven MK, Canas B, Veenhoff LM, Poolman B, Robillard GT (2002a) Combined in-gel tryptic digestion and CNBr cleavage for the generation of peptide maps of an integral membrane protein with MALDI-TOF mass spectrometry. Biochim Biophys Acta 1555:111–115
van Montfort BA, Canas B, Duurkens R, Godovac-Zimmermann J, Robillard GT (2002b) Improved in-gel approaches to generate peptide maps of integral membrane proteins with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom 37:322–330
Vellaichamy A, Tran JC, Catherman AD, Lee JE, Kellie JF, Sweet SMM, Zamdborg L, Thomas PM, Ahlf DR, Durbin KR, Valaskovic GA, Kelleher NL (2010) Size-sorting combined with improved nanocapillary liquid chromatography-mass spectrometry for identification of intact proteins up to 80 kDa. Anal Chem 82:1234–1244
VerBerkmoes NC, Bundy JL, Hauser L, Asano KG, Razumovskaya J, Larimer F, Hettich RL, Stephenson JL Jr (2002) Integrating ‘top-down” and “bottom-up” mass spectrometric approaches for proteomic analysis of Shewanella oneidensis. J Proteome Res 1:239–252
Vestling MM, Kelly MA, Fenselau C (1994) Optimization by mass spectrometry of a tryptophan-specific protein cleavage reaction. Rapid Commun Mass Spectrom 8:786–790
Vorm O, Roepstorff P (1994) Peptide sequence information derived by partial acid hydrolysis and matrix-assisted laser desorption/ionization mass spectrometry. Biol Mass Spectrom 23:734–740
Wall DB, Kachman MT, Gong S, Hinderer R, Parus S, Misek DE, Hanash SM, Lubman DM (2000) Isoelectric focusing nonporous RP HPLC: a two-dimensional liquid-phase separation method for mapping of cellular proteins with identification using MALDI-TOF mass spectrometry. Anal Chem 72:1099–1111
Wall DB, Kachman MT, Gong SS, Parus SJ, Long MW, Lubman DM (2001) Isoelectric focusing nonporous silica reversed-phase high-performance liquid chromatography/electrospray ionization time-of-flight mass spectrometry: a three-dimensional liquid-phase protein separation method as applied to the human erythroleukemia cell-line. Rapid Commun Mass Spectrom 15:1649–1661
Wang B, Malik R, Nigg EA, Körner R (2008) Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal Chem 80:9526–9533
Washburn MP, Wolters D, Yates JR 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19:242–247
Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphrey-Smith I (1995) Progress with gene-product mapping of Mollicutes: Mycoplasma genitalium. Electrophoresis 16:1090–1094
Whitelegge JP, Halgand F, Souda P, Zabrouskov V (2006) Top-down mass spectrometry of integral membrane proteins. Exp Rev Proteomics 3:585–596
Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 13:19–50
Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW (2006) STED-microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 440:935–939
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362
Wu J, Gage DA, Watson JT (1996) A strategy to locate cysteine residues in proteins by specific chemical cleavage followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem 235:161–174
Wu CC, MacCoss MJ, Howell KE, Yates JR 3rd (2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol 21:532–538
Wu SL, Kim J, Hancock WS, Karger B (2005) Extended range proteomic analysis (ERPA): a new and sensitive LC–MS platform for high sequence coverage of complex proteins with extensive post-translational modifications-comprehensive analysis of beta-casein and epidermal growth factor receptor (EGFR). J Proteome Res 4:1155–1170
Wu SL, Hühmer AF, Hao Z, Karger BL (2007) On-line LC–MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J Proteome Res 6:4230–4244
Wu S, Lourette NM, Tolić N, Zhao R, Robinson EW, Tolmachev AV, Smith RD, Pasa-Tolić L (2009a) An integrated top-down and bottom-up strategy for broadly characterizing protein isoforms and modifications. J Proteome Res 8:1347–1357
Wu S, Yang F, Zhao R, Tolić N, Robinson EW, Camp DG 2nd, Smith RD, Pasa-Tolić L (2009b) Integrated workflow for characterizing intact phosphoproteins from complex mixtures. Anal Chem 81:4210–4219
Yamagami T, Ishiguro M (1998) Complete amino acid sequences of chitinase-1 and -2 from bulbs of genus Tulipa. Biosci Biotechnol Biochem 62:1253–1257
Yan F, Subramanian B, Nakeff A, Barder TJ, Parus SJ, Lubman DM (2003) A comparison of drug-treated and untreated HCT-116 human colon adenocarcinoma cells using a 2-D liquid separation mapping method based upon chromatofocusing PI fractionation. Anal Chem 75:2299–2308
Yang Y, Zhang S, Howe K, Wilson DB, Moser F, Irwin D, Thannhauser TW (2007) A comparison of nLC–ESI-MS/MS and nLC–MALDI-MS/MS for GeLC-based protein identification and iTRAQ-based shotgun quantitative proteomics. J Biomol Tech 18:226–237
Yergey JA, Cotter RJ, Heller D, Fenselau C (1984) Resolution requirements for middle molecule mass spectrometry. Anal Chem 56:2262–2263
Yoo C, Zhao J, Pal M, Hersberger K, Huber CG, Simeone DM, Beer DG, Lubman DM (2006) Automated integration of monolith-based protein separation with on-plate digestion for mass spectrometric analysis of esophageal adenocarcinoma human epithelial samples. Electrophoresis 27:3643–3651
Zabrouskov V, Whitelegge JP (2007) Increased coverage in the transmembrane domain with activated-ion electron capture dissociation for top-down Fourier-transform mass spectrometry of integral membrane proteins. J Proteome Res 6:2205–2210
Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL (2007) ProSight PTM 2.0: improved protein identification and characterization for top-down mass spectrometry. Nucleic Acids Res 35:W701–W706
Zhang J, Wu SL, Kim J, Karger BL (2007) Ultratrace liquid chromatography/mass spectrometry analysis of large peptides with post-translational modifications using narrow-bore poly(styrene-divinylbenzene) monolithic columns and extended range proteomic analysis. J Chromatogr A 1154:295–307
Zhang N, Shaw AR, Li N, Chen R, Mak A, Hu X, Young N, Wishart D, Li L (2008) Liquid chromatography electrospray ionization and matrix-assisted laser desorption ionization tandem mass spectrometry for the analysis of lipid raft proteome of monocytes. Anal Chim Acta 627:82–90
Zheng S, Yoo C, Delmotte N, Miller FR, Huber CG, Lubman DM (2006) Monolithic column HPLC separation of intact proteins analyzed by LC–MALDI using on-plate digestion: An approach to integrate protein separation and identification. Anal Chem 78:5198–5204
Zhong H, Marcus SL, Li L (2005) Microwave-assisted acid hydrolysis of proteins combined with liquid chromatography MALDI MS/MS for protein identification. J Am Soc Mass Spectrom 16:471–481
Zhu K, Kim J, Yoo C, Miller FR, Lubman DM (2003) High sequence coverage of proteins isolated from liquid separations of breast cancer cells using capillary electrophoresis-time-of-flight MS and MALDI-TOF MS mapping. Anal Chem 75:6209–6217
Zubarev RA, Chivanov VD, Hakansson P, Sundqvist BU (1994) Peptide sequencing by partial acid hydrolysis and high resolution plasma desorption mass spectrometry. Rapid Commun Mass Spectrom 8:906–912
Zubarev RA, Kelleher NL, McLafferty FW (1998) Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc 120:3265–3266
Zvonok N, Xu W, Williams J, Janero D, Krishnan S, Makriyannis A (2010) Mass spectrometry-based GPCR proteomics: comprehensive characterization of the human cannabinoid 1 receptor. J Proteome Res 9:1746–1753
Acknowledgments
We would like to thank the Cluster of Excellence “Macromolecular Complexes” at the Goethe-University Frankfurt for the financial support and our colleagues Thorsten W. Jaskolla, Stavroula Markoutsa and Dorota Urbanek.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meyer, B., Papasotiriou, D.G. & Karas, M. 100% protein sequence coverage: a modern form of surrealism in proteomics. Amino Acids 41, 291–310 (2011). https://doi.org/10.1007/s00726-010-0680-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-010-0680-6