Abstract
The present study aims to investigate the roles of TCF4 and its underlying mechanism in colorectal cancer (CRC). Doxorubicin-resistant DLD-1 (DLD1 DR), TCF4 overexpression, and TCF4 knockdown cell lines were constructed. A flow cytometer was used to analyze frequencies of CD133+ cell in the DLD1 and DLD1 DR cells. Quantitative real-time PCR (qPCR) was used to determine the expressions of cancer stem cell (CSC) makers. Stemness of CRC cells were determined using tumorsphere formation assay. The correlation between TCF4 and ZEB1/ZEB2 were determined using public data from The Cancer Genome Atlas (TCGA) datasets. ZEB1/ZEB2 overexpression cell lines were constructed and cell viabilities were then determined using MTT and colony formation assays. TCF4 overexpression promoted proliferation of CRC cell lines and relative expressions of TCF4 were significantly increased in the DLD1 DR cells. TCF4 overexpression promoted CRC cell doxorubicin resistance, whereas TCF4 knockdown significantly decreased doxorubicin resistance. Additionally, TCF4 overexpression also significantly increased frequencies of CSC cells, expressions of CSC markers, and CRC ability to form tumorsphere. Furthermore, TCF4 promoted ZEB1 and ZEB2 expression, leading to CRC proliferation and doxorubicin resistance. TCF4 promoted CRC doxorubicin resistance and stemness by regulating expressions of ZEB1 and ZEB2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most commonly occurring cancer worldwide. There were at least 1.8 million new diagnosed CRC patients in 2018(Siegel et al. 2017). The incidence and mortality rates of CRC have declined in the past few decades. However, more than 40% mortality rates in the CRC patients are still a major challenge (Siegel et al. 2017). Most of the CRC-related deaths are caused by recurrence and metastasis of CRC and its chemotherapy resistance (Ishii et al. 2011; Soveri et al. 2019). Therefore, understanding the molecular mechanisms of CRC chemotherapy resistance is crucial for the development of therapeutic and preventive strategies against CRC.
Cancer stem cells (CSCs) are a subpopulation of tumorigenic cells with abilities to self-renew and differentiate into a variety of lineages (Clarke and Hass 2006; Lodestijn et al. 2019). As a driving force for the progression of cancer, CSCs have been identified to play a crucial role in many types of cancer including ovarian, melanoma, hepatoma, breast cancer, and CRC (Clarke and Hass 2006; Reya et al. 2001). Due to their ability for DNA repair and multidrug resistance ATP-binding cassette transporters expression, CSCs are naturally resistant to chemotherapy. Those CSCs confer drug resistance to their progeny, leading to the tumors eventually become less sensitive to the following chemotherapy (Reya et al. 2001). Therefore, regulation of CSC stemness and thereby increasing the sensitivities of the tumor to chemotherapy are an important strategy to overcome chemotherapy resistance.
Transcription factors (TFs) are proteins binding with DNA helix at a specific region, leading to activation or inhibition of transcription of downstream target genes by trans-activation or trans-repression domains (Darnell Jr 2002; Libermann and Zerbini 2006). Many studies have demonstrated that TFs are deregulated in the occurrence and development of many types of cancer including breast cancer, hepatoma, CRC, and melanoma (Darnell Jr 2002; Yang and Hung 2009). By comparison of 1571 identified oncogenic protein with 1988 identified transcription factors, there are 19% transcription factors which are also oncogenes (Lambert et al. 2018). Therefore, the regulation of functions of transcription factors might provide a promising method for cancer therapy. TCF4 is also called immunoglobulin transcription factor, belonging to the basic helix-loop-helix family (de Pontual et al. 2009). It is broadly expressed and plays a crucial role in the development of the nervous system. TCF4 defection has been identified to be associated with Pitt-Hopkins syndrome, a rare neurological disorder disease (Amiel et al. 2007). In 2015, for the first time, Liu and colleagues have identified the activation of TCF4 leading to tumor angiogenesis and growth by recruitment of tumor-associated macrophages (Liu et al. 2015). This study has demonstrated that overexpression of TCF4 is associated with the development of breast cancer. However, the roles of TCF in other types of cancer are still unknown.
In the present study, for the first time, we investigated the roles of TCF in the CRC. Interestingly, we found that TCF4 plays an important role in the regulation of CRC drug resistance and stemness. Furthermore, the underlying mechanisms of TCF4 were investigated and target genes of TCF4 were identified.
Materials and methods
Cell lines
Colorectal adenocarcinoma cell lines including DLD-1 and HT29 were purchased from the Cell Bank of the Chinese Academic of Sciences (Shanghai, China). Cells were verified by STR analysis before use. The cells were cultured in Roswell Park Memorial Institute 1640 medium supplemented with 0% fetal bovine serum (FBS) (Life Technologies, Gaithersburg, MD, USA) and 100 U/ml penicillin and 100 μg/ml streptomycin (Beyotime, Shanghai, People’s Republic of China) at 37 °C in the presence of 5% CO2 at constant humidity.
Construction of doxorubicin-resistant DLD-1 cell lines
DLD-1 doxorubicin-resistant (DLD-1 DR) cell lines were constructed according to the previously reported methods (Chu et al. 1991; Riganti et al. 2005). DLD-1 cells were cultured with complete medium supplemented with doxorubicin (0.5 μM, 1 μM, 2 μM, 4 μM) for 26 weeks. Stepwise selection of DLD-1 cells was performed and DLD-1R cells were examined using an MTT assay.
Construction of gene overexpression cell lines
To construct TCF4 overexpression cell lines, TCF4 was amplified and sub-cloned into p-Sin 3 × flag vector. The DLD1 cells were transfected with p-Sin 3 × flag vector containing TCF4 and followed by the selection of antibiotics. Expressions of TCF4 were determined by either quantitative real-time (PCR) qPCR or Western blotting, to ensure the stable cell lines were successfully established. Similarly, to construct zinc finger E-box binding homeobox (ZEB) 1 and ZEB2 overexpression cell lines, ZEB1 and ZEB2 were amplified and sub-cloned into p-Sin 3 × flag vector. The DLD1 cells were transfected with p-Sin 3 × flag vector containing ZEB1 or ZEB2 and followed by the selection of antibiotics.
MTT assay
Cell viability was determined using MTT assay according to the previously reported methods (Yang et al. 2015). The cells were seeded in the plate and followed with different treatments at different time points. After that, an MTT solution was added into each well and the microplate was incubated at 37 °C for 4 h. DMSO solution was then added and the optical density was read at 570 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Colony formation assay
Colony formation assay was performed according to the previously reported method (Borowicz et al. 2014). DLD1 and HT29 cells with different treatments were seeded in a 6-well plate and grown for indicated days. Next, the colonies were fixed with 4% paraformaldehyde and then stained with crystal violet (0.05% w/v) overnight. The numbers of colonies were counted using Image J.
Tumorsphere formation assay
Tumorsphere medium is consisted of Dulbecco’s modified Eagle medium/F12, epidermal growth factor (20 ng/ml), insulin (5 μg/ml), 2% B-27, and 0.4% FBS. Tumorsphere formation assay was performed according to the previously reported method (Johnson et al. 2013). The DLD1 and DLD1 DR cells were harvested and suspended in phosphate-buffered saline. Next, dissociated cells were then seeded in tumorsphere medium. The spheres were observed under an inverted microscope.
Flow cytometry analysis
Cells were detached using trypsin and then centrifuged. Cells were re-suspended in the staining buffer containing fluorescent labeled CD133 antibody, according to previously reported method (Shang et al. 2018). After that, the cell suspension was run through flow cytometry (BD Biosciences, CA, USA) and the results were analyzed using FlowJo (Ashland, OR, USA).
Isolation of total RNA and quantitative polymerase chain reaction
Isolation of total RNA was according to the previously reported method (Liu et al. 2016). Trizol reagent was used to extract total RNA from cell lines, according to the manufacturer’s instructions (Takara, Japan). To remove DNA contamination, RNase-free DNase I was used. Primers for TCF4, SOX4, KLF4, NANOG, OCT4, ZEB1, and ZEB2 and internal control GADPH were used for amplification of these genes. To analyze the accuracy of the PCR reaction, the melt curves were used. To evaluate the expressions of genes, 2−△△Ct values were calculated. The mRNA expression values of TCF4, SOX4, KLF4, NANOG, OCT4, ZEB1, and ZEB2 were normalized to that of GADPH. The primers used for qPCR were listed below: TCF4-F: TGAGAACCTGCAAGACACGAA, TCF4-R: CTGCCTTCTGCTCTGGTGTCA, SOX4-F: CAAGCGACCCATGAACGCCTT, SOX4-R: CAGCCGCTCCGCCTCTCGAA, KLF4-F: TACCCATCCTTCCTGCCCGAT, KLF4-R: TGAGCATCATCCCGTGTGTCC, NANOG-F: TCCACCAGTCCCAAAGGCAAA, NANOG-R: CTGTTTCTTGACCGGGACCT, OCT4-F: ATATGCAAAGCAGAAACCCTCGT, OCT4-R: CTCCAGGTTGCCTCTCACTCG, ZEB1-F: AGGCTATAAACGCTTTACCTC, ZEB1-R: ATTACACCCAGACTGCGTCA, ZEB2-F: TCCAATGCAGCACTTAGGTGT, ZEB2-R: TTTCTTCAGCCTTGCAGTCCA.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer. Next, the lysate was centrifuged at 13000g to remove insoluble material. After that, bicinchoninic acid protein assay kit (Thermo Fisher, Waltham, MA, USA) was used to qualify protein concentrations.
Equal amounts of protein from cell lysates were separated by 10% SDS gel. Next, the SDS gel was then transferred to a polyvinylidene fluoride membrane and 5% non-fat milk was used to block the membrane. After the membrane was probed with antibodies against TCF4, and actin overnight at 4 °C, the membrane was then incubated with the secondary antibodies. ChemiDoc MP imaging system (Bio-Rad, Hercules, CA, USA) was used to examine chemiluminescence. The expressions of TCF4 were normalized to the internal control actin.
Statistical analysis
SPSS (SPSS, Chicago, IL, USA) was used to statistical analysis. All data were expressed as mean ± S.D. To evaluate the significance, the Student t test, a one- or two-way analysis of variance with multiple comparisons, and the Student-Newman-Keuls (SNK) test were performed. A P value less than 0.05 was considered as a statistical significance.
Results
TCF4 promoted CRC cell proliferation
To investigate the effects of TCF4 on the CRC cell proliferation, we constructed the TCF4 overexpressed DLD1 cell line. After the plasmid containing TCF4 was transfected into the cells, Western blotting and qPCR were used to examine transfection efficiency. As shown in Fig. 1a, b, mRNA and protein levels of TCF4 were significantly increased when compared with those in the normal DLD1 cell lines. We then determined the effects of TCF4 on the DLD1 cell proliferation using cell counting and MTT assay. As shown in Fig. 1c, e, the cell viabilities of DLD1 cell line transfected with TCF4 were significantly increased when compared with those in the normal DLD1 cell lines. The population doubling time (PDT) of the DLD1 cells transfected with vector or TCF4 was 20.5 h or 17.8 h, respectively. Similarly, we observed increased cell viabilities of HT29 cell line transfected with TCF4 (Fig. 1d, f). The population doubling time (PDT) of the HT29 cells transfected with vector or TCF4 was 20.4 h or 17.7 h, respectively. Additionally, to evaluate the effects of TCF4 overexpression on CRC cellular clonogenic potential, colony formation assay was performed. As shown in Fig. 1g, h, the colony numbers of CRC cell line transfected with TCF4 were significantly increased when compared with those in the normal CRC cell line. To exclude the effects of transfection on the colony, we observed the size of CRC cells after transfection. The results showed that the size of CRC cells was not changed after the cells were transfected with TCF4. These results suggested that TCF4 promoted CRC cell proliferation.
TCF4 promoted CRC proliferation. a, b The mRNA and protein expressions of TCF4 in DLD1 cells transfected with the plasmid containing TCF4 were determined using qPCR and Western blotting. c, d Cell viabilities of DLD1 and HT29 cells transfected with the plasmid containing TCF4 was determined by cell counts assay. e, f Cell viabilities of DLD1 and HT29 cells transfected with the plasmid containing TCF4 was determined by MTT assay. g, h Cell viability of DLD1 and HT29 cells transfected with the plasmid containing TCF4 was determined by soft agar colony formation assay. Data was shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance
TCF4 promoted CRC cell doxorubicin resistance
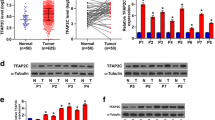
To investigate the effects of TCF4 on CRC cell chemotherapy resistance, a DLD1 doxorubicin-resistant (DLD1 DR) cell line was constructed. As shown in Fig. 2a, DLD1 DR demonstrated lower sensitivities to doxorubicin treatment in comparison with DLD1 cell line. The IC50 of DLD1 or DLD1 DR cells were 1.63 μM and 4.21 μM, respectively. Next, mRNA expressions of TCF4 were examined in DLD1 and DLD1 DR cell lines. As shown in Fig. 2b, mRNA expressions of TCF4 in the DLD1 DR cell line were significantly increased when compared with that in the DLD1 cell line.
TCF4 promoted CRC cell doxorubicin resistance. a The DLD1- and Dox-resistant DLD1 DR cells was treated with different concentrations of Dox for 5 days, and then cell viability was determined by MTT assay. b The mRNA expressions of TCF4 in DLD1 and DLD1 DR cells were determined using qPCR. c The mRNA expressions of TCF4 in DLD1 cells treated with different concentrations of Dox were determined using qPCR. d The mRNA expressions of TCF4 in DLD1 cells treated 2 μM Dox for different time points were determined using qPCR. e The DLD1 cells were transfected with TCF4 expression plasmid and treated with different concentrations of Dox for 5 days; cell viability was determined using MTT assay. f The mRNA expression of TCF4 in DLD1 DR cells transfected with TCF4 siRNA was determined using qPCR. g The DLD1 DR cells were transfected with TCF4 siRNA and treated with different concentrations of doxorubicin for 5 days; cell viability was determined using MTT assay. Data was shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance
To confirm the relationship between the expressions of TCF4 and doxorubicin resistance, we further determined relative expressions of TCF4 in doxorubicin-treated DLD1 cells. As shown in Fig. 2c, an increase of TCF4 mRNA expression in the DLD1 cells were accompanied by the increase of doxorubicin concentrations. Additionally, as shown in Fig. 2d, mRNA expressions of TCF4 were significantly increased with prolonged doxorubicin treatment time. When the DLD1 cells treated with doxorubicin for 48 h, mRNA expressions of TCF4 were increased over 3-fold as compared with that in normal DLD1 cells.
To clarify the effects of TCF4 on doxorubicin resistance, a TCF4 overexpression cell line was constructed and then treated with doxorubicin. As shown in Fig. 2e, after treatment of doxorubicin (0.5 μM, 1 μM, 2 μM, 4 μM), cell viabilities of DLD1 cells transfected with TCF4 were higher when compared with that of DLD cells transfected with the vector. The IC50 values of the cells transfected with vector or TCF4 were 1.34 μM and 2.42 μM, respectively. Next, we further evaluated the effects of TCF4 knockdown on doxorubicin resistance in DLD1 DR cells. As shown in Fig. 2f, the relative expressions of TCF4 in the DLD1 DR cells were significantly decreased after siRNA TCF4 silencing. Interestingly, as shown in Fig. 2g, DLD1 DR demonstrated increased sensitivities to doxorubicin treatment after TCF4 silencing. The IC50 of the DLD1 DR cells transfected with NC or siTCF4 were 3.78 μM and 2.04 μM, respectively. These results suggested that TCF4 promoted CRC cell doxorubicin resistance.
TCF4 promoted CRC cell stemness
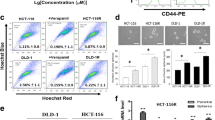
To investigate the effects of TCF4 on the CRC cell stemness, flow cytometer was used to analyze the populations of cancer stem cells. First, the frequency of CD133+ cancer stem cells was evaluated. As shown in Fig. 3a, the frequency of CD133+ cells dramatically increased in the DLD1 DR cells when compared with that in the DLD1 cells. Additionally, we observed an increase of the frequency of CD133+ cells in DLD1 cells transfected with TCF4, indicating that TCF4 was correlated with CRC cell stemness (Fig. 3b).
TCF4 promoted CRC cell stemness properties. a Flow cytometry was used to analyze the frequency of CD133+ cancer stem cell in DLD1 and DLD1 DR cells. b Frequency of CD133+ cancer stem cell in DLD1 cells transfected with plasmid containing TCF4. c The expressions of cancer stem cell markers in DLD1 cells transfected with TCF4 were determined using qPCR. d, e Tumorsphere formation abilities were evaluated in the DLD1, DLD1 DR, and DLD1 cells transfected with plasmid containing TCF4. Data was shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance
We next evaluated expressions of cancer stem cell markers including SOX4, KLF4, NANONG, and OCT4. As shown in Fig. 3c, the relative expressions of SOX4, KLF4, NANONG, and OCT4 were significantly increased in the DLD1 cells transfected with TCF4 when compared with those in the DLD1 cells transfected with the vector. As shown in Fig. 3d, tumorsphere formation assay demonstrated a significant increase of the ability to form tumorsphere in the DLD1 DR cells. Additionally, overexpression of TCF4 significantly increased the capacity to form tumorsphere in the DLD1 cells (Fig. 3e).
TCF4 promoted ZEB1 and ZEB2 expression
We further explored the target genes of TCF4 using The Cancer Genome Atlas (TCGA) database. As shown in Fig. 4a, b, the expressions of TCF4 in the CRC patients were correlated with ZEB1 and ZEB2. To confirm the relationship between TCF4 and ZEB1/ZEB2, we determined expressions of ZEB1 and ZEB2 in DLD1 cells. As shown in Fig. 4c, the relative expressions of ZEB1 and ZEB2 were significantly increased in DLD1 cells transfected with TCF4 when compared with those in the DLD1 cells transfected with the vector. Furthermore, we further evaluated the effects of TCF4 knockdown on expressions of ZEB1 and ZEB2 in DLD1 cells. We found that TCF4 knockdown significantly decreased expressions of ZEB1 and ZEB2 in DLD1 cells (Fig. 4d). These results suggested that TCF4 is correlated with ZEB1 and ZEB2 in CRC cells.
TCF4 promoted ZEB1 and ZEB2 expression. a, b The correlation between TCF4 and ZEB1/ZEB2 in colon cancer patients was analyzed in TCGA dataset. c The expressions of ZEB1 and ZEB2 in DLD1 cells transfected with plasmid containing TCF4 were determined using qPCR. d The expressions of ZEB1 and ZEB2 in DLD1 cells transfected with TCF4 siRNA were determined using qPCR. Data was shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance
ZEB1 and ZEB2 promoted CRC cell proliferation and doxorubicin resistance
To explore the effects of ZEB1 and ZEB2 on the CRC cell proliferation and doxorubicin resistance, ZEB1 and ZEB2 overexpression cell lines were constructed. As shown in Fig. 5a, the relative expressions of ZEB1 and ZEB2 were significantly increased in the DLD1 cells, indicating their overexpression cell lines were successfully constructed. Interestingly, we found the cell viabilities were significantly increased in DLD1 cells transfected with ZEB1 or ZEB2 (Fig. 5b, c). We further evaluated CRC cell doxorubicin resistance in ZEB1 and ZEB2 overexpressed DLD1 cells. As shown in Fig. 5d, DLD1 cells transfected with ZEB1 or ZEB2 showed lower sensitivities to doxorubicin treatment. The IC50 values of the DLD1 cells transfected with vector containing ZEB1 and ZEB2 were 1.63 μM, 2.87 μM, and 2.65 μM, respectively. These results supported that ZEB1 and ZEB2 promoted CRC cell proliferation and doxorubicin resistance.
ZEB1 and ZEB2 promoted colon cancer proliferation and doxorubicin resistance. a The mRNA expressions of ZEB1 or ZEB2 in DLD1 cells transfected with plasmids containing ZEB1 or ZEB2 were determined using qPCR. b Cell viability of DLD1 cells transfected with plasmids containing ZEB1 or ZEB2 was determined using cell counting assay. c Cell viability of DLD1 cells transfected with ZEB1 or ZEB2 expression plasmid was determined using MTT assay. d The DLD1 cells were transfected with plasmids containing ZEB1 or ZEB2 and then treated with different concentrations of doxorubicin for 5 days; cell viability was determined using MTT assay. Data was shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001; ns, no significance
Discussion
Chemotherapy resistance is one of the major hurdles for CRC treatment (Dylla et al. 2008; Longley et al. 2006). Most of the CRC-related deaths are caused by recurrence and metastasis of CRC and its chemotherapy resistance (Longley et al. 2006). CSCs are a subpopulation of tumorigenic cells which are naturally resistant to chemotherapy (Clarke and Hass 2006). ABC transporters are highly expressed in CSCs and responsible for pumping chemotherapeutic drug outside the cells. Moreover, CSCs confer drug resistance to their progeny, resulting in the tumors eventually becoming less sensitive to chemotherapy (Clarke and Hass 2006; Reya et al. 2001). Therefore, targeting CSCs are thought to be a promising method to treat CRCs.
TCF 4 has been identified as a crucial factor in the development of the nervous system. In 2014, Marc and colleagues have implicated novel functions of TCF4 based on genome-wide association studies (Forrest et al. 2014). Except that TCF4 mutations are associated with Pitt-Hopkins syndrome, its mutations are also correlated with some other neurological diseases including primary sclerosing cholangitis, Fuchs’ endothelial corneal dystrophy, and schizophrenia (Amiel et al. 2007; Forrest et al. 2014). By interaction with basic helix-loop-helix proteins, TCF4 is responsible for the regulation of cell differentiation. Additionally, TCF4 has also been revealed to be associated with epithelial-mesenchymal transition (EMT) (Forrest et al. 2014). In 2015, Liu and colleagues have identified that the activation of TCF4 induces tumor angiogenesis and growth by recruitment of tumor-associated macrophages (Liu et al. 2015). This study is for the first time to investigate the roles of TCF4 in breast cancer. However, the roles of TCF in other types of cancer and its underlying molecular mechanisms are still unclear. Therefore, in this study, we investigated the role of TCF4 and its underlying mechanisms in the CRC.
To investigate the roles of TCF4 in the CRC, we constructed a TCF4 overexpressed CRC cell line. Interestingly, we found that TCF4 overexpression significantly increased viabilities and colony formation abilities of CRC cells. We then investigated the relationship between TCF overexpression and chemotherapy resistance. Doxorubicin is a classic chemotherapeutic drug for many types of cancer. Additionally, doxorubicin-resistant cell lines are widely used as a model for investigation underlying mechanism of chemotherapy resistance (Strumberg et al. 1996). Therefore, in the present study, a doxorubicin-resistant DLD1 cell lines called DLD R1 was constructed to explore the roles of TCF4 in the chemotherapy. The results demonstrated that TCF4 was overexpressed in the DLD R1 cells. Interestingly, when TCF expression was inhibited, the cells became sensitive to doxorubicin treatment. These results suggested that TCF overexpression resulted in the CRC cell line resistant to doxorubicin treatment.
CSC stemness is one of the major obstacles to decrease the sensitivities of the tumor to chemotherapy. Therefore, we further investigated the frequency of CSCs and its relationship with TCF4 expression. CD133 is widely used to mark CSC in various tumor types including CRC (Dalerba et al. 2007). In this study, the results demonstrated a higher frequency of CD133+ cells in DLD R1 as well as in TCF4 overexpressed cells. To confirm TCF4 is correlated with CSC population, the relative expressions of several important CSC markers including SOX4, KLF4, NANONG, and OCT4 were determined. For instance, KLF4 and OCT4 have been identified as oncogenes in CRC stem cell–enriched spheroids (Leng et al. 2013). Activation of NANONG and SOX4 promotes CSC stemness and EMT (Chiou et al. 2010). The results demonstrated that the relative expressions of SOX4, KLF4, NANONG, and OCT4 were significantly increased in the DLD1 cells transfected with TCF4 when compared with those in the DLD1 cells transfected with the vector. In the present study, we also observed that SOX4 and KLF4 with higher expressions when compared with those of NANONG and OTC4. However, we did not compare the expression change among those target genes considering qPCR is a relative qualification method to detect the expression change of target genes.
Additionally, overexpression of TCF4 significantly increased the capacity to form tumorspheres in the DLD1 cells. Interestingly, we observed a higher rate of sphere formation in the resistant cells than in the transfected cells, and this does not correspond to their relative expression of TCF4 markers. We speculated that the underlying mechanism of DOX resistance in DLD1 DR cells is complicated, which is not only determined by the upregulation of TCF4 but also regulated by other mechanisms. Therefore, although the expression levels of TCF4 in DLD1 DR cell were lower than the cells transfected with TCF4 overexpression plasmid, the stem cell characteristics and DOX resistance ability of DLD1 DR cells are still stronger than the cells transfected with TCF4 overexpression plasmid. Overall, these results suggested that overexpression of TCF4 promoted CSC stemness in CRC cells.
We further explored the target genes of TCF4 based on TCGA dataset. Notably, we identified that ZEB1 and ZEB2 are associated with TCF4 in CRC patients. In this study, we found that TCF4 promoted expressions of ZEB1 and ZEB2 in CRC cells. Interestingly, when TCF4 siRNA was used to knock down the TCF4, expressions of ZEB1 and ZEB2 were significantly decreased. ZEB1 acts as a transcription repressor and has been implicated to induce EMT by recruitment of ATP-dependent chromatin remodeler SMARCA4 (Dave et al. 2011). ZEB2 is a paralog of ZEB1, which is capable of regulation microRNAs in cancers (Yoshihara et al. 2009). In this study, for the first time, we found that ZEB1 and ZEB2 promoted colon cancer proliferation as well as doxorubicin resistance. Furthermore, we revealed that expressions of ZEB1 and ZEB2 are regulated by TCF4 in CRC. However, the underlying mechanisms of ZEB1 and ZEB2 for the regulation of CRC are still unknown.
Conclusion
In summary, we identified TCF4 as a crucial factor in the regulation of CRC drug resistance and stemness. TCF4 overexpression could promote CRC cells resistant to doxorubicin therapy and increase the stemness of CRC cells. However, TCF4 knockdown significantly decreased abilities of CRC cells to resistant doxorubicin therapy. Furthermore, we found that the underlying mechanisms of TCF4 promoting CRC drug resistance and stemness are in part due to regulating expressions of ZEB1 and ZEB2. All of these imply that TCF4 might serve as a potential target for CRC therapy.
References
Amiel J et al (2007) Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet 80:988–993
Borowicz S, Van Scoyk M, Avasarala S, Rathinam MKK, Tauler J, Bikkavilli RK, Winn RA (2014) The soft agar colony formation assay. J Visual Exper 92:e51998
Chiou SH et al (2010) Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 70:10433–10444. https://doi.org/10.1158/0008-5472.CAN-10-2638
Chu E, Drake JC, Koeller DM, Zinn S, Jamis-Dow CA, Yeh GC, Allegra CJ (1991) Induction of thymidylate synthase associated with multidrug resistance in human breast and colon cancer cell lines. Mol Pharmacol 39:136–143
Clarke MF, Hass AT (2006) Cancer stem cells reviews in cell biology and molecular medicine
Dalerba P et al (2007) Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci 104:10158–10163
Darnell JE Jr (2002) Transcription factors as targets for cancer therapy. Nat Rev Cancer 2:740
Dave N, Guaita-Esteruelas S, Gutarra S, Frias À, Beltran M, Peiró S, de Herreros AG (2011) Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J Biol Chem 286:12024–12032
de Pontual L, Mathieu Y, Golzio C, Rio M, Malan V, Boddaert N, Soufflet C, Picard C, Durandy A, Dobbie A, Heron D, Isidor B, Motte J, Newburry-Ecob R, Pasquier L, Tardieu M, Viot G, Jaubert F, Munnich A, Colleaux L, Vekemans M, Etchevers H, Lyonnet S, Amiel J (2009) Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum Mutat 30:669–676
Dylla SJ et al (2008) Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One 3:e2428
Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ (2014) The emerging roles of TCF4 in disease and development. Trends Mol Med 20:322–331. https://doi.org/10.1016/j.molmed.2014.01.010
Ishii Y, Hasegawa H, Endo T, Ochiai H, Kitagawa Y (2011) Current view and possibilities of chemotherapy sensitivity and resistance assays and metabolic enzyme analysis in individualized chemotherapy for the patients with colorectal cancer. Nihon Rinsho 69 Suppl 3:487–493
Johnson S, Chen H, Lo P (2013) In vitro tumorsphere formation assays. Bio-protocol 3:e325
Lambert M, Jambon S, Depauw S, David-Cordonnier MH (2018) Targeting transcription factors for cancer treatment. Molecules. https://doi.org/10.3390/molecules23061479
Leng Z et al (2013) Kruppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells. PLoS One 8:e56082. https://doi.org/10.1371/journal.pone.0056082
Libermann TA, Zerbini LF (2006) Targeting transcription factors for cancer gene therapy. Curr Gene Ther 6:17–33
Liu C et al (2015) Activating transcription factor 4 promotes angiogenesis of breast cancer through enhanced macrophage recruitment. Biomed Res Int 2015:974615. https://doi.org/10.1155/2015/974615
Liu C, Zhao J, Liu Y, Huang Y, Shen Y, Wang J, Sun W, Sun Y (2016) A novel pentacyclic triterpenoid, ilexgenin A, shows reduction of atherosclerosis in apolipoprotein E deficient mice. Int Immunopharmacol 40:115–124. https://doi.org/10.1016/j.intimp.2016.08.024
Lodestijn SC, Lenos KJ, Miedema DM, Bijlsma MF, Vermeulen L (2019) Cancer stem cells: here, there, and everywhere. Mol Cell Oncol 6:1540235. https://doi.org/10.1080/23723556.2018.1540235
Longley DB, Allen WL, Johnston PG (2006) Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim Biophys Acta (BBA)-Rev Cancer 1766:184–196
Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414:105
Riganti C, Miraglia E, Viarisio D, Costamagna C, Pescarmona G, Ghigo D, Bosia A (2005) Nitric oxide reverts the resistance to doxorubicin in human colon cancer cells by inhibiting the drug efflux. Cancer Res 65:516–525
Shang W et al (2018) Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc Natl Acad Sci 115:E4051–E4060
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A (2017) Colorectal cancer statistics. CA Cancer J Clin 67:177–193. https://doi.org/10.3322/caac.21395
Soveri LM, Lamminmaki A, Hanninen UA, Karhunen M, Bono P, Osterlund P (2019) Long-term neuropathy and quality of life in colorectal cancer patients treated with oxaliplatin containing adjuvant chemotherapy. Acta Oncol. https://doi.org/10.1080/0284186X.2018.1556804
Strumberg D, Harstrick A, Doll K, Hoffmann B, Seeber S (1996) Bendamustine hydrochloride activity against doxorubicin-resistant human breast carcinoma cell lines. Anti-Cancer Drugs 7:415–421
Yang J-Y, Hung M-C (2009) A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res 15:752–757
Yang H et al (2015) Ilexgenin A induces B16-F10 melanoma cell G1/S arrest in vitro and reduces tumor growth in vivo. Int Immunopharmacol 24:423–431. https://doi.org/10.1016/j.intimp.2014.12.040
Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, Kudo Y, Inoue I, Tanaka K (2009) Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci 100:1421–1428
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent
Not applicable.
Additional information
Handling Editor: Jörn Bullerdiek
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, S., Yang, X., Qin, X. et al. TCF4 promotes colorectal cancer drug resistance and stemness via regulating ZEB1/ZEB2 expression. Protoplasma 257, 921–930 (2020). https://doi.org/10.1007/s00709-020-01480-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-020-01480-6