Abstract
Plant symbiotic mutants are useful tool to uncover the molecular-genetic mechanisms of nodule senescence. The pea (Pisum sativum L.) mutants SGEFix−-1 (sym40), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) display an early nodule senescence phenotype, whereas the mutant SGEFix−-2 (sym33) does not show premature degradation of symbiotic structures, but its nodules show an enhanced immune response. The nodules of these mutants were compared with each other and with those of the wild-type SGE line using seven marker genes that are known to be activated during nodule senescence. In wild-type SGE nodules, transcript levels of all of the senescence-associated genes were highest at 6 weeks after inoculation (WAI). The senescence-associated genes showed higher transcript abundance in mutant nodules than in wild-type nodules at 2 WAI and attained maximum levels in the mutant nodules at 4 WAI. Immunolocalization analyses showed that the ethylene precursor 1-aminocyclopropane-1-carboxylate accumulated earlier in the mutant nodules than in wild-type nodules. Together, these results showed that nodule senescence was activated in ineffective nodules blocked at different developmental stages in pea lines that harbor mutations in four symbiotic genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Legumes interact with soil bacteria known as rhizobia to form symbiotic nodules. Nodule cells are infected with rhizobia, which differentiate into nitrogen-fixing bacteroids and form organelle-like symbiosomes (Tsyganova et al. 2018). Nodule senescence is the final stage of nodule function, during which symbiosomes and infected cells are degraded and the plant reuses the nutrients accumulated in the nodule (Serova and Tsyganov 2014). Transcriptomic studies of the model legumes Lotus japonicus (Regel) K. Larsen and Medicago truncatula Gaertn. have shown that many genes are up- or down-regulated in senescent nodules (Van de Velde et al. 2006; Maunoury et al. 2010; Cabeza et al. 2014; Chungopast et al. 2014), confirming the importance of this process.
Together with natural senescence, premature nodule senescence can be induced by various unfavorable factors (Swaraj et al. 1993; González et al. 1998; Matamoros et al. 1999; Hernández-Jiménez et al. 2002; Pérez Guerra et al. 2010). In soybean nodules, darkness causes early senescence accompanied by autophagy of symbiosomes (Vauclare et al. 2010). In addition to stressors, various mutations in the symbiotic genes of plants (Kneen et al. 1990; Pladys and Vance 1993; Novák et al. 1995; Morzhina et al. 2000) and rhizobia (Chua et al. 1985; Hirsch and Smith 1987; Chungopast et al. 2014) can cause early nodule senescence.
Genetic analyses of L. japonicus and M. truncatula have revealed that mutations in several genes lead to early nodule senescence. In L. japonicus, these genes include LjSST1, encoding a nodule-specific sulfate transporter located on the symbiosome membrane (Krusell et al. 2005); LjIGN1, encoding a protein with ankyrin repeats and transmembrane domains that is required for bacteroid differentiation and functioning (Kumagai et al. 2007); LjSEN1, encoding an integral membrane protein (Hakoyama et al. 2012); and LjAPN1, encoding a nepenthesin-type aspartic peptidase that is required for nodule functioning in a rhizobial strain-specific manner (Yamaya-Ito et al. 2018). In M. truncatula, the genes whose mutation leads to early nodule senescence include MtDNF2, which produces a main transcript encoding a phosphatidylinositol-specific phospholipase C-like protein, and five different transcripts as a result of alternative splicing (Bourcy et al. 2013); MtRSD encodes a nodule-specific cysteine-2/histidine-2 (C2H2) transcription factor, which is a repressor of VAMP721a that encodes vesicle-associated membrane protein 721a (Sinharoy et al. 2013); MtDNF4 and MtDNF7, which encode the nodule cysteine-rich (NCR) proteins NCR211 and NCR169, respectively (Kim et al. 2015; Horváth et al. 2015); MtSymCRK, which encodes a non-arginine aspartate (nonRD) receptor-like kinase (Berrabah et al. 2014); MtNAD1, which encodes a small uncharacterized protein localized to the endoplasmic reticulum (Wang et al. 2016; Domonkos et al. 2017); and MtNFS1 and MtNFS2 that encode NCR proteins inducing bacterial death and early nodule senescence in rhizobial strain-dependent manner (Yang et al. 2017; Wang et al. 2017).
Studies of genetic collections of pea (Pisum sativum L.) symbiotic mutants have revealed six loci at which mutations lead to the formation of nodules with premature degradation of symbiotic structures and an early senescence phenotype (Borisov et al. 2007). The two loci cloned to date are Sym40, an ortholog of M. truncatula MtEFD (Nemankin 2011), which encodes a transcription factor that negatively regulates the cytokinin response protein RR4 (Vernié et al. 2008), and Sym13, the presumable orthologous LjSEN1 gene (Kulaeva et al. 2017).
Senescence of the symbiotic nodule leads to remobilization of accumulated nutrients to the young developing organs of the plant (Puppo et al. 2005; Van de Velde et al. 2006; Pérez Guerra et al. 2010). During nodule aging, mainly cysteine proteases, known also as thiol proteases, are involved in protein degradation (Pladys and Vance 1993; Kardailsky and Brewin 1996; Van de Velde et al. 2006; Pérez Guerra et al. 2010; Pierre et al. 2014). Senescence of the symbiotic nodule is regulated by transcription factors (D’haeseleer et al. 2010; De Zélicourt et al. 2012; Karmarkar 2014) and phytohormones (Puppo et al. 2005; Van de Velde et al. 2006; Cam et al. 2012; Karmarkar 2014; Guinel 2015). Up-regulation of expression of MtATB2, which encodes a bZIP transcription factor, was observed during the aging of alfalfa nodules (D’haeseleer et al. 2010). In addition, members of the NAC family of transcription factors, MtNAC969 and MtNAC920, are involved in alfalfa nodule senescence (De Zélicourt et al. 2012; Karmarkar 2014). Previous studies showed that ethylene, abscisic acid (ABA), jasmonic acid, and nitric oxide (NO) (Van de Velde et al. 2006; Karmarkar 2014; Serova et al. 2017; Cam et al. 2012) may be positive regulators of nodule aging. Genes encoding enzymes involved in ethylene biosynthesis and jasmonic acid biosynthesis are up-regulated during nodule aging in alfalfa (Van de Velde et al. 2006) and pea (Serova et al. 2017). In addition, ethylene activates the transcription factor MtNAC920 that targets the gene encoding the cysteine protease MtCP2, which is involved in symbiosome degradation (Karmarkar 2014). It is assumed that ABA also is a positive regulator of nodule aging. Puppo et al. (2005) proposed that ABA in conjunction with ethylene orchestrate nutrient recycling during nodule senescence, whereas gibberellins might suppress nodule senescence (Van de Velde et al. 2006; Serova et al. 2017). Moreover, nodule aging is regulated by nitric oxide (II) (NO); an increase in NO concentration causes premature senescence in alfalfa nodules (Cam et al. 2012).
Previously, we selected a set of genes, known to be activated during nodule senescence (Granell et al. 1992; Kardailsky and Brewin 1996; Peck and Kende 1998; Martin et al. 1999; Pariasca et al. 2001; Zdunek-Zastocka 2008; D’haeseleer et al. 2010), as molecular markers of nodule senescence in pea (Serova et al. 2017). These genes encode cysteine protease 1 (Cyp1) and 15a (Cyp15a), a thiol protease (TPP), a bZIP transcription factor (ATB2), and key enzymes for ethylene biosynthesis (ACS2 for 1-aminocyclopropane-1-carboxylate (ACC) synthase, and ACO1 for ACC oxidase) and ABA biosynthesis (AO3 for aldehyde oxidase). Using these markers, we demonstrated the early nodule senescence of the mutants E135F (sym13) and Sprint-2Fix− (sym31) (Serova et al. 2017). In addition, immunolocalization of ACC and expression analysis of 1-aminocyclopropane-1-carboxylate synthase 2 (PsACS2) and 1-aminocyclopropane-1-carboxylate oxidase 1 (PsACO1) indicated the possible involvement of ethylene in pea nodule senescence (Serova et al. 2017). However, because the genetic background of these mutants showed significant differences in ripening (Borisov et al. 1997), it was difficult to compare the results for the different lines. Therefore, comparison of the senescence process among mutants induced in a single wild-type genotype is of great interest.
In the present study, we focused on the mutant lines SGEFix−-1 (sym40), SGEFix−-3 (sym26), and SGEFix−-7 (sym27), which show an early nodule senescence phenotype (Tsyganov et al. 1998, 2013), and the mutant line SGEFix−-2 (sym33), which does not show premature degradation of symbiotic structures, but rather shows strong defense reactions (Ivanova et al. 2015). The common trait of each of these mutants is their ineffective nodules. When ineffective nodules form on plant roots as a result of inoculation with an ineffective rhizobial strain, plants use “sanctions” to prevent further development of ineffective nodules (Kiers et al. 2003). Sanctions lead to reduced nodule growth and limited fitness of the constituent bacteria (Oono et al. 2011; Regus et al. 2017; Westhoek et al. 2017). Sanctions are often realized through induction of senescence (Regus et al. 2017). We suggest that in the pea mutants, senescence is also a widespread reaction to the formation of ineffective nodules, regardless of the developmental stage at which the nodule is blocked. To test this hypothesis, we compared gene transcription in nodules among the mutant lines and between the mutant lines and the wild-type SGE line using seven senescence-associated genes. We also conducted immunolocalization analyses to detect the ethylene precursor, ACC, in nodules of the mutants and wild-type.
Materials and methods
Plant materials, bacterial strain, and plant growth conditions
The pea laboratory line SGE and corresponding mutant lines SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) were analyzed in this study (Table 1). The procedures for seed sterilization, plant inoculation, and plant growth conditions were described previously (Serova et al. 2017). Materials were collected for analyses at 10 days after inoculation (DAI) and at 2, 4, and 6 weeks after inoculation (WAI).
Nodule phenotypes
Photographs of wild-type and mutant nodules were taken with a stereomicroscope SteREO Lumar.V12 (Carl Zeiss, Oberkochen, Germany) equipped with a videocamera Axiocam ICc 1 (Carl Zeiss) and AxioVision Rel. 4.8 software (Carl Zeiss).
Light microscopy
The wild-type and mutant nodules at 2 and 4 WAI were fixated in freshly prepared fixative solution (2.5% (w/v) paraformaldehyde, 0.5% (v/v) glutaraldehyde, 0.1% (v/v) Triton X-100) in PBS and embedded in Steedman’s wax as described previously (Serova et al. 2017). Sections of 10 μm were performed with a НМ360 microtome (Microm, Walldorf, Germany) as described previously (Serova et al. 2017). Sections were stained with 0.1% (w/v) toluidine blue solution (Supplementary Fig. S8) or propidium iodide (0.5 μg ml−1) (Supplementary Fig. S6) in PBS for 10 min and washed in PBS two times for 10 min. Additionally, for mutants SGEFix−-1 (sym40), SGEFix−-2 (sym33) 0.5-μm-thick, resin-embedded sections (see Section below) were cut with a glass knife on a Leica EM UC7 ultramicrotome (Leica Microsystems, Wetzlar, Germany) and collected on IPN multiwell slides. Specimens were stained with 0.5% (w/v) toluidine blue in 0.1 mM sodium borate (Supplementary Fig. S7a, c). Microscopic analyses were carried out using the AxioImagerZ1 (Carl Zeiss) equipped with a microscope camera Axiocam 506 color (Carl Zeiss).
Electron microscopy
Two-week-old nodules of the mutant SGEFix−-7 (sym27) were harvested from roots and placed directly in fixative. A glancing cut on one side of each nodule was made to allow better penetration of the fixative. The whole nodules were fixed in 2.5% (v/v) glutaraldehyde (Sigma-Aldrich, St. Louis, MO, USA) in 0.1 M phosphate buffer, pH 7.2. After fixation, samples were post-fixed in 2% (w/v) osmium tetroxide in phosphate buffer for 2 h. Samples then were dehydrated in a graded series of increasing ethanol concentrations followed by two changes of 100% acetone. Dehydrated samples were progressively embedded in Epon (Honeywell Fluka™, Fisher Scientific, Loughborough, UK) at room temperature. Embedded samples were transferred to blocks in fresh resin and polymerized at 60 °C for 48 h.
Two-week-old nodules of the mutants SGEFix−-1 (sym40), SGEFix−-2 (sym33), and SGEFix−-3 (sym26) were processed using the low-temperature embedding procedure described previously (Tsyganova et al. 2009). Briefly, nodules were fixed in 2.5% (v/v) glutaraldehyde (Sigma-Aldrich) in 0.5 M sodium cacodylate, pH 7.2. Samples then were dehydrated and embedded in London Resin White (Sigma-Aldrich) with benzoin methyl ether as the catalyst for ultraviolet polymerization at − 20 °C using the Leica EM AFS2 automatic freeze substitution system (Leica Microsystems).
For transmission electron microscopy, 90–100-nm-thick ultrathin sections were cut using a diamond knife (Diatome, Bienne, Switzerland) on a Leica EM UC7 ultramicrotome (Leica Microsystems) and collected on gold grids coated with 4% (w/v) pyroxylin and carbon. The grids containing the sections were counterstained with 2% (w/v) aqueous uranyl acetate for 1 h followed by lead citrate for 1 min. All solutions were filtered before use, and filter-sterilized deionized water was used throughout the experiment. The nodule tissues were examined and photographed under a JEM-1400 transmission electron microscope (JEOL Corporation, Tokyo, Japan) at 80 kV.
Gene selection and primer design
The selection of senescence-associated genes and primer design were described previously (Serova et al. 2017).
Total RNA isolation and cDNA synthesis
Total RNA was extracted using the PureZol™ reagent (Bio-Rad, Hercules, CA, USA) in accordance with the manufacturer’s instructions. Samples treated with DNAse I (MBI Fermentas, Vilnius, Lithuania) were loaded onto microchips and then analyzed and quantified with a MultiNA capillary microelectrophoresis system (Shimadzu Corporation, Kyoto, Japan). DNA-free total RNA (1.5 μg) then was used to synthesize cDNA with 200 U RevertAid Reverse Transcriptase (MBI Fermentas) in accordance with the manufacturer’s instructions. The resulting cDNAs were diluted five times for use in real-time PCR.
Relative real-time PCR analysis
Relative real-time PCR experiments were conducted using a С1000™ Thermal Cycler combined with the optical module CFX96™ Real-Time System (Bio-Rad). We used iQ™ SYBR® Green Supermix (Bio-Rad) for reactions and transcript detection in accordance with the manufacturer’s protocol (Serova et al. 2017). The results were processed using CFX Manager™ software (Bio-Rad) and analyzed using the 2−ΔΔCT method. Relative abundance was normalized against that of the reference gene (PsGAPC1). Two-week-old nodules of SGE and zone IV of 4-week-old nodules of SGE were used as calibrators for calculation of relative transcript abundance in experiments with whole samples and laser microdissection and pressure catapulting (LMPC) samples, respectively. For each gene, transcript levels were analyzed with three technical replicates. Data were statistically analyzed using Microsoft Excel software. Gene expression levels were compared using one-way ANOVA; a P value ≤ 0.05 was considered to be significant. Experiments were performed with three replicates of six to eight plants per line.
Laser microdissection and pressure catapulting and RNA extraction
Samples were prepared for LMPC as described previously (Serova et al. 2017). Two- and 4-week-old nodules of wild-type SGE and 2-week-old nodules of the mutant line SGEFix−-7 (sym27) were used in these analyses (Supplementary Figs. S1–S3). Laser microdissection was conducted with a PALM MicroBeam System (Carl Zeiss). Sections were visualized on a computer monitor using an Axiocam ICc 1 video camera (Carl Zeiss). The samples selected with PALM Robo 43 software and isolated using an ultraviolet laser (350 nm) are listed in Table 2. Selected cells were catapulted into the Adhesive Cap (Carl Zeiss) and lysed using the extraction buffer included in the PicoPure™ RNA Isolation Kit (Arcturus Engineering Inc., Mountain View, CA, USA). Total RNA was extracted with the PicoPure™ RNA Isolation Kit (Arcturus Engineering Inc.) in accordance with the manufacturer’s instructions.
In vitro RNA amplification and cDNA synthesis
To generate amplified RNA, in vitro transcription was carried out using the MessageAmp™ II aRNA Amplification Kit (Ambion Inc., Foster City, CA, USA) in accordance with the manufacturer’s protocol. Amplified RNAs were quantified with a Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s guidelines. SuperScript® III reverse transcriptase (Invitrogen) was used to synthesize cDNA from amplified RNA under the conditions recommended by the manufacturer. The cDNA obtained was used for relative real-time PCR with iQ™ SYBR® Green Supermix (Bio-Rad) as described above.
Fixation and 1-aminocyclopropane-1-carboxylate immunolabeling
The protocol used to fix pea nodules was described previously (Serova et al. 2017). Nodules were washed three times with PBS, each wash for 15 min, and then stained with 0.5% toluidine blue solution in PBS for 1 h. The excess dye was removed from the nodules by washing twice with PBS, each wash for 15 min. The samples were embedded in 3% agarose gel, and then sections (50 μm) were cut at room temperature with a НМ650V microtome (Microm). Immunolabeling of ACC and staining of nuclei and bacteria with propidium iodide were conducted as described previously (Serova et al. 2017). The anti-ACC rabbit antibody (Agrisera, Vännäs, Sweden) at 1:100 dilution and goat anti-rabbit IgG Alexa Fluor 488 (Thermo Fisher Scientific, Waltham, MA, USA) at 1:300 dilution were used as primary and secondary antibodies, respectively.
The specificity of the primary anti-ACC antibodies was confirmed using pea nodules saturated with 10 mM ACC (Serova et al. 2017) (Supplementary Fig. S4a–c). As the control, the primary antibodies were omitted from the specific binding of secondary antibodies (Supplementary Fig. S4d). Sections were observed using the LSM 510 META laser scanning confocal system (Carl Zeiss) and images were processed using ZEN2009 software (Carl Zeiss).
Results
Nodule histological and ultrastructural organization in mutants SGEFix−-3 (sym26) and SGEFix−-7 (sym27)
At 2 and 4 WAI, the wild-type SGE formed pink nodules (Supplementary Fig. S5a, b) with typical indeterminate nodule histological and ultrastructural organization (Tsyganov et al. 1998), with three distinguishable zones: the meristem zone, the infection zone, and the nitrogen fixation zone (Supplementary S6a). In wild-type nodules, the fourth senescence zone was apparent at the base of nodules at 4 WAI only (Supplementary Fig. S3a). The mutants SGEFix−-1 (sym40) and SGEFix−-2 (sym33) may form two types of nodules for which the histological and ultrastructural organization has been described previously in detail (Tsyganov et al. 1998; Voroshilova et al. 2009). In the present study, the mutant SGEFix−-1 (sym40) formed only white nodules (Supplementary Fig. S5c), which were filled with cells occupied with hypertrophied infection droplets at 2 (Supplementary Figs. S6b and 7a, b) and 4 WAI (data not shown). The mutant SGEFix−-2 (sym33) also formed white nodules only (Supplementary Fig. S5d), which were filled with a ramified network of “locked” infection threads at 2 (Supplementary Figs. S6c and S7c, d) and 4 WAI (data not shown). The mutants SGEFix−-3 (sym26) and SGEFix−-7 (sym27) formed pinkish nodules (Supplementary Fig. S5e, f). The nodules of SGEFix−-3 (sym26) were dominated by a senescence zone at 2 WAI (Supplementary Fig. S8a), which occupied the main portion of the nodule at 4 WAI (Supplementary Fig. S8b). The senescence zone was smaller in SGEFix−-7 (sym27) nodules than in SGEFix−-3 (sym26) nodules at 2 WAI (Supplementary Fig. S8c) and had increased in size in nodules at 4 WAI (Supplementary Fig. S8d).
In 2-week-old nodules of SGEFix−-3 (sym26), the infection zone in infected cells contained pleomorphic bacteroids that showed clear signs of morphological differentiation (the cells were enlarged and the matrix was less electron-dense compared with bacteria inside infection threads) (Fig. 1a, b). In nodules of the mutant, the zone corresponding to the nitrogen fixation zone in wild-type nodules contained bacteroids with an electron-dense matrix at the tip (Fig. 1c, d). In the senescence zone, the bacteroids inside the symbiosomes had degraded, leaving remnants of bacteroids and symbiosome membranes (Fig. 1e, f).
Ultrastructural organization in nodule of mutant line SGEFix−-3 (sym26). a Infected cell in infection zone, b infection thread and bacteroids in infected cell in infection zone, c infected cell in zone corresponding to nitrogen fixation zone in wild-type, d bacteroid in infected cell in zone corresponding to nitrogen fixation zone in wild-type, e degrading infected cell in senescence zone, f degrading bacteroid in degrading infected cell in senescence zone. IC, infected cell; DIC, degrading infected cell; IT, infection thread; B, bacterium; Ba, bacteroid; DBa, degrading bacteroid; V, vacuole. Large and small arrows indicate electron-dense matrix in bacteroids. Scale bars: a = 5 μm; c, e = 2 μm; b, d, f = 500 nm
In 2-week-old nodules of SGEFix−-7 (sym27), the infected cells in the infection zone contained bacteroids that showed signs of morphological differentiation (Fig. 2a, b). In the zone corresponding to the nitrogen fixation zone in wild-type nodules, small electron-dense inclusions were observed in the bacteroids (Fig. 2c, d). In the senescence zone, bacteroids showed signs of degradation inside the symbiosomes, and the peribacteroid spaces were significantly enlarged (Fig. 2e, f). The tonoplast was ruptured, degraded bacteroids were located in the vacuole (Fig. 2e), and the infected cells contained amyloplasts (Fig. 2c, e). Some infected cells had large lipid inclusions in the cytoplasm (data not shown).
Ultrastructural organization in nodule of mutant line SGEFix−-7 (sym27). a Infected cell in infection zone, b bacteroids in infected cell in infection zone, c infected cell in zone corresponding to nitrogen fixation zone in wild-type, d bacteroids in infected cell in zone corresponding to nitrogen fixation zone in wild-type, e degrading infected cell in senescence zone, f degrading bacteroid in degrading infected cell in senescence zone. IC, infected cell; DIC, degrading infected cell; Ba, bacteroid; DBa, degrading bacteroid; PBS, peribacteroid space; V, vacuole. Large arrows indicate electron-dense inclusions in bacteroids, arrowhead indicates infection thread, asterisks indicate amyloplasts, small arrow indicates degrading symbiosomes in vacuole. Scale bars: a = 5 μm; c, e = 2 μm; b, d, f = 500 nm
Transcription patterns of senescence-associated genes in nodules of the wild-type and mutants
Cysteine protease 1, cysteine protease 15a, and thiol protease
During nodule aging, the relative transcript levels of cysteine protease 1 (PsCyp1), cysteine protease 15a (PsCyp15a), and thiol protease (PsTPP) increased in all tested lines. The transcript levels of these genes were significantly up-regulated already at 4 WAI (which corresponds to the stage of active nitrogen fixation) compared with 2-week-old nodules of the mutants SGEFix−-1 (sym40), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) (Fig. 3a–c). However, a slight increase only of PsCyp1 and PsCyp15 transcript abundance was observed during the aging of the SGEFix−-2 (sym33) mutant nodules (Fig. 3a, b). The relative transcript level of PsTPP was not significantly increased in nodules of SGEFix−-2 (sym33) and even slightly decreased at 6 WAI (Fig. 3c). At 4 WAI, the transcript levels of selected genes encoding cysteine and thiol proteases in nodules of the mutants SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) were significantly higher than those in wild-type nodules (Fig. 3a–c). In wild-type nodules, the transcript levels of the selected genes were up-regulated only at 6 WAI (Fig. 3a–c). The most pronounced (7.4-fold) increase in transcript abundance was detected for PsCyp15a (Fig. 3b).
Relative transcript levels of PsCyp1, PsCyp15a, PsTPP, and PsATB2 in nodules of wild-type SGE and corresponding pea mutants (SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27)) at 2, 4, and 6 weeks after inoculation (WAI). Letters indicate significant differences (one-way ANOVA, P value ≤ 0.05): a, from wild-type SGE at one time point; b, within genotype compared with 2 WAI; c, within genotype compared with 4 WAI
bZIP transcription factor
The bZIP transcription factor (PsATB2) transcript levels increased during nodule aging. In all analyzed mutant genotypes, up-regulation of PsATB2 transcript abundance was detected already at 4 WAI (Fig. 3d). The greatest increase of transcript level was observed in 4-week-old SGEFix−-3 (sym26) mutant nodules compared with 2-week-old nodules (Fig. 3d). At 4 WAI, PsATB2 transcript abundance in nodules was higher in SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27), than in wild-type (Fig. 3d). A significant (6.7-fold) increase in PsATB2 transcript abundance was observed in 6-week-old wild-type nodules only. In contrast, PsATB2 was down-regulated at 6 WAI in SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) (Fig. 3d).
1-Aminocyclopropane-1-carboxylate synthase 2 and 1-aminocyclopropane-1-carboxylate oxidase 1
The transcript abundance of genes encoding key enzymes in ethylene biosynthesis increased during nodule aging. The PsACS2 transcript levels were higher in 4-week-old nodules than in 2-week-old nodules of SGEFix−-1 (sym40), SGEFix−-3 (sym26), and SGEFix−-7 (sym27), respectively (Fig. 4a). However, this increase was not observed in nodules of SGEFix−-2 (sym33) (Fig. 4a). Up-regulation of PsACO1 transcription also was detected in 4-week-old nodules of the analyzed mutants (Fig. 4b). The greatest increase was detected in SGEFix−-3 (sym26) and SGEFix−-7 (sym27) mutants (Fig. 4b). In 4-week-old nodules, PsACS2 and PsACO1 transcript abundance was higher in SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) than in wild-type nodules (Fig. 4a, b). In wild-type nodules, the PsACS2 transcript levels were only significantly up-regulated (15-fold) at 6 WAI and the PsACO1 mRNA level was up-regulated (3-fold) at 4 WAI (Fig. 4a and b).
Relative transcript levels of PsACS2, PsACO1, and PsAO3 in nodules of wild-type SGE and corresponding pea mutants (SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27)) at 2, 4, and 6 weeks after inoculation (WAI). Letters indicate significant differences (one-way ANOVA, P value ≤ 0.05): a, from wild-type SGE at one time point; b, within genotype compared with 2 WAI; c, within genotype compared with 4 WAI
Aldehyde oxidase 3
The transcript levels of aldehyde oxidase 3 (PsAO3), which encodes the enzyme catalyzing the final stage of ABA biosynthesis, increased in most lines during nodule senescence. Up-regulation of PsAO3 transcript abundance was observed in the mutants SGEFix−-1 (sym40), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) at 4 WAI (Fig. 4c). However, the PsAO3 transcript level did not change significantly during nodule aging in SGEFix−-2 (sym33). At 4 WAI, the transcript level of PsAO3 in nodules of SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27) was higher than that in wild-type nodules (Fig. 4c). At 6 WAI, the PsAO3 mRNA level was up-regulated (6.3-fold) in wild-type nodules (Fig. 4c).
It is interesting to note that, unlike the other mutants, in 2-week-old nodules of SGEFix−-2 (sym33) higher transcript abundance was observed for all analyzed genes compared with those in wild-type nodules (Figs. 3a–d and 4a–c).
Transcription patterns of senescence-associated genes in wild-type SGE and SGEFix−-7 (sym27) nodules as determined by laser microdissection and pressure catapulting
The transcription patterns of the seven senescence-associated genes in different zones of wild-type and mutant SGEFix−-7 (sym27) nodules were determined by LMPC. Infected cells from zone III only were excised from 2-week-old wild-type nodules (Fig. S1b–d), whereas infected cells from zone III and the morphologically distinct zone IV were excised from 4-week-old wild-type nodules (Fig. S3b–g). In 2-week-old nodules of the mutant line, infected cells were excised from the zone corresponding to zone III in wild-type nodules (zone III′) and the well-defined zone IV (Supplementary Fig. S2b–g).
Differences in transcription patterns among the protease genes in the nodule zones analyzed were revealed. In the wild-type, PsCyp15a transcript abundance was 3.3-fold higher in zone III of 4-week-old nodules than that in 2-week-old nodules (Fig. 5a). No differences in PsTPP transcript levels were detected (Fig. 5b). However, slight decrease (1.5-fold) and increase (1.6-fold) of PsCyp15a and PsTPP transcript abundance, respectively, were observed in zone IV relative to zone III in wild-type nodules (Fig. 5a, b). In the mutant SGEFix−-7 (sym27), the greatest (13.2- and 14-fold) up-regulation of transcript abundance of PsCyp15a and PsTPP, respectively, was detected in zone IV relative to zone III′ (Fig. 5a, b). The transcript abundance of PsCyp15a and PsTPP was 6.6- and 2.8-fold higher, respectively, in zone III′ of mutant nodules than in zone III of wild-type nodules at 2 WAI (Fig. 5a, b). PsCyp1 transcripts were not detected in infected cells of wild-type and mutant nodules.
Relative transcript levels of PsCyp15a, PsTPP, PsATB2, PsAO3, PsACS2, and PsACO1 in infected cells in zone III (III*) and IV of pea nodules from wild-type SGE and mutant line SGEFix−-7 (sym27) at 2 and 4 weeks after inoculation (WAI). Letters indicate significant differences (one-way ANOVA, P value ≤ 0.05): a, from 2 WAI nodules of wild-type SGE (zone III); b, between zone III and zone IV in 4 WAI nodules of wild-type SGE; c, between zone III* and zone IV in 2 WAI nodules of mutant line SGEFix−-7 (sym27). Breaks in histogram bars indicate change in scale. Zone III*—zone corresponding to nitrogen fixation zone in wild-type nodules
The transcript levels of PsATB2 were increased during nodule cell degradation in the wild-type and mutant nodules. In zone III of wild-type nodules, the transcript abundance of PsATB2 was 2.5-fold higher in 4-week-old nodules than that in 2-week-old nodules (Fig. 5c). In 4-week-old nodules, the PsATB2 transcript level was slightly (1.4-times) higher in zone IV than that in zone III. In the mutant nodules, the greatest (27.2-fold) up-regulation of PsATB2 mRNA level was observed in zone IV relative to that in zone III′. In 2-week-old nodules, the PsATB2 transcript level in zone III′ was significantly (8.7-fold) higher than that in zone III of wild-type nodules (Fig. 5c).
The transcript levels of PsAO3 were increased during the senescence of wild-type and mutant nodules. In wild-type nodules, the transcript level of PsAO3 in zone III was 5.4-fold higher at 4 WAI than that at 2 WAI (Fig. 5d) but did not differ significantly between zones III and IV of 4-WAI nodules. In the mutant nodules, the greatest (40-fold) increase in PsAO3 transcript level was detected in zone IV relative to that of zone III′. At 2 WAI, the PsAO3 transcript level was 2.5 times higher in zone III′ than that in zone III (Fig. 5d).
The transcript levels of PsACS2, which encodes an ethylene precursor (ACC) biosynthesis enzyme, were decreased during the degradation of infected cells in the nodules of the wild-type and mutant. No PsACS2 transcripts were detected in zone III of 4-week-old wild-type nodules (Fig. 5e). In the wild-type nodules, the PsACS2 transcript abundance was significantly (16.2-fold) lower in zone IV in 4-week-old nodules than in zone III in 2-week-old nodules. In mutant nodules, the PsACS2 transcript level was 3.7-fold lower in zone IV than in zone III. The PsACS2 mRNA level in zone III′ of mutant nodules was significantly (31.8-fold) higher than that in zone III of wild-type nodules (Fig. 5e).
During nodule aging and alongside nodule cell degradation, the transcription levels of PsACO1, which encodes an ethylene biosynthesis enzyme, were increased in the wild-type and mutant lines. In the wild-type nodules, PsACO1 transcripts were detected only at 4 WAI. In wild-type nodules, the PsACO1 transcript level was not significantly different between zones IV and III (Fig. 5f). In the mutant nodules, the PsACO1 transcript level was significantly (7.5-fold) higher in zone IV than that in zone III′. The greatest (13.7-fold) increase in PsACO1 transcript level was detected in zone III′ of the mutant nodules relative to that of zone III of the wild-type nodules (Fig. 5f).
Immunolocalization of ACC in wild-type and mutant nodules
We conducted ACC immunolocalization analyses at 10 DAI (Figs. 6 and 7), and at 2 (Fig. 8), 4 (data not shown), and 6 WAI (Fig. 9). In wild-type nodules, the intensity of ACC labeling increased as the nodules aged (Figs. 6a–c, 8a–c, and 9a–f). In nodules of all analyzed mutants, ACC levels peaked at 10 DAI and were significantly higher than those of wild-type nodules (Figs. 6d–o and 7d–o). Compared with wild-type nodules, those of the mutants showed significantly lower ACC labeling intensity at 4 WAI (data not shown). The intensity of ACC labeling in nodules was higher in SGEFix−-3 (sym26) (Figs. 6j–l and 7j–l) and SGEFix−-7 (sym27) (Figs. 6m–o and 7m–o) than in SGEFix−-1 (sym40) (Figs. 6d–f and 7d–f) and SGEFix−-2 (sym33) (Figs. 6g–i and 7g–i).
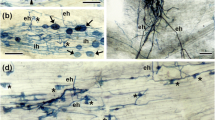
Immunolocalization of 1-aminocyclopropane-1-carboxylate in nodules of wild-type (a–c) and corresponding pea mutants (SGEFix−-1 (sym40) (d–f), SGEFix−-2 (sym33) (g–i), SGEFix−-3 (sym26) (j–l), and SGEFix−-7 (sym27) (m–o)) at 10 days after inoculation (DAI). Zones in nodule are designated by Roman numerals: I—meristem, II—infection zone, III—nitrogen fixation zone, III′—zone corresponding to nitrogen fixation zone in wild-type. Heat map provides color code of fluorescence signal intensities (c, f, i, l, o). Merged differential interference contrast (DIC) and red channel (a, d, g, j, m), merged green and red channels (b, e, h, k, n). Visualization of ACC by the Alexa 488 conjugated secondary antibody (green) and nuclei and bacteria stained with propidium iodide (red). Scale bar = 100 μm
Immunolocalization of 1-aminocyclopropane-1-carboxylate in cells in central part of nodules of wild-type (a–c) and corresponding pea mutants (SGEFix−-1 (sym40) (d–f), SGEFix−-2 (sym33) (g–i), SGEFix−-3 (sym26) (j–l), and SGEFix−-7 (sym27) (m–o)) at 10 days after inoculation (DAI). IC, infected cell; UIC, uninfected cell. Arrows indicate infection threads, arrowheads indicate infection droplets. Heat map provides color code of fluorescence signal intensities (c, f, i, l, o). Merged differential interference contrast (DIC) and red channel (a, d, g, j, m), merged green and red channels (b, e, h, k, n). Visualization of ACC by the Alexa 488 conjugated secondary antibody (green) and nuclei and bacteria stained with propidium iodide (red). Scale bar = 10 μm
Immunolocalization of 1-aminocyclopropane-1-carboxylate in nodules of wild-type (a–c) and corresponding pea mutants (SGEFix−-1 (sym40) (d–f), SGEFix−-2 (sym33) (g–i), SGEFix−-3 (sym26) (j–l), and SGEFix−-7 (sym27) (m–o)) at 2 weeks after inoculation (WAI). Zones of nodule are designated by Roman numerals: I—meristem, II—infection zone, III—nitrogen fixation zone, III′—zone corresponding to nitrogen fixation zone in wild-type, IV—senescence zone. Heat map provides color code of fluorescence signal intensities (c, f, i, l, o). Merged differential interference contrast (DIC) and red channel (a, d, g, j, m), merged green and red channels (b, e, h, k, n). Visualization of ACC by the Alexa 488 conjugated secondary antibody (green) and nuclei and bacteria stained with propidium iodide (red). Scale bar = 100 μm
Immunolocalization of 1-aminocyclopropane-1-carboxylate in nodules of wild-type at 6 WAI (a–f). Cells from nitrogen fixation zone (g–i) and senescence zone (j–l). Zones of nodule are designated by Roman numerals: I—meristem, II—infection zone, III—nitrogen fixation zone, IV—senescence zone. IC, infected cell; UIC, uninfected cell; DIC, degrading infected cell. Heat map provides color code of fluorescence signal intensities (c, f, i, l). Merged differential interference contrast (DIC) and red channel (a, d, g, j), merged green and red channels (b, e, h, k). Visualization of ACC by the Alexa 488 conjugated secondary antibody (green) and nuclei and bacteria stained with propidium iodide (red). Scale bars: a–f = 100 μm, g–l = 10 μm
In 10-day-old wild-type nodules, the strongest ACC signals were detected in meristem cells (Figs. 6a–c and 7a–c) and cells in the infection zone (Fig. 6a–c), whereas weaker ACC signals were detected in the cells of the nitrogen fixation zone (Fig. 7a–c). In nodules of the mutants SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26) (Fig. 6j–l), and SGEFix−-7 (sym27) (Fig. 6m–o), the strongest ACC labeling intensity was in cells in the meristematic zone, the infection zone, and the zone corresponding to the nitrogen fixation zone in wild-type nodules (Fig. 7d–o).
At 6 WAI in the wild-type nodules, the strongest ACC signals were detected in meristematic cells and in cells of the infection and nitrogen fixation zones (Fig. 9a–c, g–i). In addition, strong ACC signals were detected in senescent cells (Fig. 9d–f) that had not yet completely degraded (Fig. 9j–l).
Discussion
To examine the activation of early senescence in ineffective nodules, we analyzed a set of pea mutants in the SGE background in which nodule development is blocked at different stages. The mutants SGEFix−-3 (sym26) and SGEFix−-7 (sym27) form pink-green ineffective nodules in which symbiotic structures prematurely degrade (a phenotype of early senescence). The mutant SGEFix−-1 (sym40) forms white ineffective nodules that also show an early senescence phenotype, but nodule development is blocked at an earlier stage than those of SGEFix−-3 (sym26) and SGEFix−-7 (sym27) (Tsyganov et al. 1998). The mutant in which nodule development is blocked at the earliest stage is SGEFix−-2 (sym33). The early senescence phenotype of the latter mutant had not been described previously (Tsyganov et al. 1998), but the strong defense reactions of its nodules have been reported (Ivanova et al. 2015).
Previous studies have focused on the ultrastructural organization of the allelic mutants RisFixM and RisFixT (with mutations in sym26) and the mutant RisFixQ (with a mutation in sym27) (Novák et al. 1995; Morzhina et al. 2000) in the pea ‘Finale’ background (Engvild 1987). These mutants are characterized by morphologically differentiated bacteroids that undergo premature degradation. In the present study, we provided detailed descriptions of the ultrastructural organization of nodules of allelic mutants in the genes sym26 and sym27 in the SGE background. Both mutants showed similar nodule ultrastructural organization to that of the allelic mutants in the ‘Finale’ background and to those of other mutants exhibiting an early nodule senescence phenotype (Kneen et al. 1990; Novák et al. 1995; Morzhina et al. 2000). We also observed a number of previously undescribed phenotypic features. In the mutant SGEFix−-3 (sym26), we observed an electron-dense matrix that formed a cap at the tip of the bacteroids. Electron-dense inclusions in the bacteroids were also observed in nodules of SGEFix−-7 (sym27). The biological significance of these electron-dense structures is unclear. The mutant SGEFix−-3 (sym26) showed more pronounced bacteroid senescence, with completely degraded bacteroids and symbiosomes in 2-week-old nodules. Bacteroids were not completely degraded in the mutant SGEFix−-7 (sym27), but tonoplast integrity was lost, which is an indicator of programmed cell death (van Doorn 2011), which often occurs during legume senescence (Chua et al. 1985; Banba et al. 2001; Chungopast et al. 2014). In addition, the infected cells accumulated numerous amyloplasts, which is a sign of nodule ineffectiveness (Forrest et al. 1991).
We compared the transcription patterns of seven senescence-associated genes among the mutants and between the mutants and wild-type. These genes encode PsCyp1 and PsCyp15a, PsTPP, PsATB2, PsAO3, PsACS2, and PsACO1. All of these genes are known to participate in nodule senescence. Genes encoding cysteine proteases are the most highly expressed genes during nodule aging (Kardailsky and Brewin 1996; Van de Velde et al. 2006). Nodule aging is also accompanied by an increase in the transcript levels of MtATB2 (D’haeseleer et al. 2010). Abscisic acid (González et al. 2001) and ethylene (Guinel 2015) are both known to play positive roles in nodule senescence.
In nodules of the wild-type SGE, the transcript levels of the seven senescence-associated genes significantly increased at 6 WAI only. Previously, we observed similar activation of these genes in 6-week-old nodules of the pea line ‘Sprint-2’ and cultivar ‘Sparkle’ (Serova et al. 2017). Pea nodules have already formed at 2 WAI, and peak nitrogen fixation occurs at 3 and 4 WAI. In pea, nodule senescence is pronounced at 6 WAI, as confirmed by the transcription of the senescence marker genes. In the mutants for the genes sym26, sym27, and sym40, whose nodules show early senescence, the transcript levels of the senescence marker genes had already increased at 4 WAI. Interestingly, high transcript levels of the senescence marker genes were observed in nodules of the mutant SGEFix−-2 (sym33), which does not show an early nodule senescence phenotype. Previously, we observed increased transcript levels of senescence-associated genes in nodules of the mutants E135F (sym13) and Sprint-2Fix− (sym31) (Serova et al. 2017); the former mutant shows an early senescence phenotype (Kneen et al. 1990; Borisov et al. 1997), but the latter mutant does not (Borisov et al. 1997).
Despite the pronounced activation of senescence-associated genes in the nodules of all of the analyzed mutants, some differences in the transcription patterns of certain genes were observed among the mutants. For example, the transcript levels of PsCyp1, PsCyp15a, PsACS2, and PsACO1 were higher in the nodules of SGEFix−-7 (sym27) than in those of SGEFix−-3 (sym26). The transcript levels of PsTPP and PsATB2 were higher in SGEFix−-3 (sym26) than in the other mutants. The transcript level of PsAO3 was higher in SGEFix−-7 (sym27) than in SGEFix−-3 (sym26) at 4 WAI but was not significantly different at 6 WAI. The biological significance of these differences is difficult to explain. Determining the nucleotide sequences of Sym26 and Sym27 and elucidating the functions of the gene products will help to explain differences in their expression patterns among the mutants. Among the studied mutants, SGEFix−-2 (sym33) showed the highest transcript levels of PsCyp1, PsATB2, PsACO1, and PsAO3 in 2-week-old nodules, and transcription of the genes PsCyp15a, PsTTP, and PsACS2 also increased in 2-week-old nodules compared with those of the wild-type. Such early activation of senescence-associated genes is consistent with the fact that in this mutant nodule development is blocked at the earliest stage among all of the studied mutants. In addition, the transcript level of PsTPP did not change during nodule aging in SGEFix−-2 (sym33). Considering that this mutant lacks bacterial release (Tsyganov et al. 1998), we speculate that the thiol protease PsTPP may be important for symbiosome degradation.
At the cellular level, SGEFix−-7 (sym27) showed increased transcription of senescence-associated genes in the zone corresponding to the nitrogen fixation zone in the wild- type. A similar increase in the transcript levels of senescence-associated genes was observed previously in the mutant E135F (sym13) compared with those of the wild-type ‘Sparkle’ (Serova et al. 2017). However, PsCyp1 transcripts were not detected in cells of nodules from the wild-type and the mutant SGEFix−-7 (sym27). This result suggests that the protease PsCyp1 is not involved in the senescence of the central nodule tissue but may be involved in the senescence of peripheral nodule tissues.
The immunolocalization analysis demonstrated that ACC levels increase with nodule aging in the wild-type. The increase in labeling intensity was correlated with up-regulation of PsACS2 and PsACO1 transcription. Similar patterns of ACC accumulation and PsACS2 and PsACO1 transcription were previously detected in the wild-types ‘Sparkle’ and ‘Sprint-2’ (Serova et al. 2017). In the mutants SGEFix−-1 (sym40), SGEFix−-2 (sym33), SGEFix−-3 (sym26), and SGEFix−-7 (sym27), the ACC concentration peaked in 10-day-old nodules and decreased as the nodules aged. This pattern was indicative of the early activation of nodule senescence in the mutants.
Conclusions
The present results show that senescence-associated genes are activated in the nodules of various mutants in which nodule development is blocked at different stages. Previously, a group of plant and bacterial mutants forming ineffective nodules on M. truncatula roots, which undergo early senescence as a result of their ineffectiveness, was identified (Maunoury et al. 2010). However, nodule development in these mutants is blocked at advanced stages, similar to the pea mutants defective in the genes sym26 and sym27. Given that we observed the activation of early senescence even in the mutant defective in the gene sym33 as well as in mutants defective in the genes sym40, sym26, sym27 (this work), and sym13 and sym31 (Serova et al. 2017) may indicate that early senescence is a general phenomenon in response to ineffective nodule formation and is not restricted to legume mutants that form nodules with morphologically differentiated symbiosomes but lack the ability to fix nitrogen. Previously, sanctions from the plant that restrict nodule size were observed during formation of ineffective nodules after inoculation with ineffective rhizobial strains (Oono et al. 2011; Regus et al. 2017; Westhoek et al. 2017). Sanctions are often accompanied by nodule senescence (Regus et al. 2017). Taken together, these results may indicate that plants use early senescence as a universal mechanism to recycle nutrients that have been allocated to ineffective nodules, similar to developmental senescence (Van de Velde et al. 2006; Pérez Guerra et al. 2010). However, further studies are needed to test this hypothesis.
Abbreviations
- ABA:
-
Abscisic acid
- ACC:
-
1-Aminocyclopropane-1-carboxylate
- WAI:
-
Weeks after inoculation
- DAI:
-
Days after inoculation
References
Banba M, Siddique ABM, Kouchi H, Izui K, Hata S (2001) Lotus japonicus forms early senescent root nodules with Rhizobium etli. Mol Plant-Microbe Interact 14:173–180
Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, Ratet P (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203:1305–1314
Borisov AY, Rozov SM, Tsyganov VE, Morzhina EV, Lebsky VK, Tikhonovich IA (1997) Sequential functioning of Sym-13 and Sym-31, two genes affecting symbiosome development in root nodules of pea (Pisum sativum L.) Mol Gen Genet 254:592–598
Borisov AY, Danilova TN, Koroleva TA, Kuznetsova EV, Madsen L, Mofett M, Naumkina TS, Nemankin TA, Ovchinnikova ES, Pavlova ZB, Petrova NE, Pinaev AG, Radutoiu S, Rozov SM, Rychagova TS, Shtark OY, Solovov II, Stougaard J, Tikhonovich IA, Topunov AF, Tsyganov VE, Vasil’chikov AG, Voroshilova VA, Weeden NF, Zhernakov AI, Zhukov VA (2007) Regulatory genes of garden pea (Pisum sativum L.) controlling the development of nitrogen-fixing nodules and arbuscular mycorrhiza: a review of basic and applied aspects. Appl Biochem Microbiol 43:237–243
Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P (2013) Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol 197:1250–1126
Cabeza R, Koester B, Liese R, Lingner A, Baumgarten V, Dirks J, Salinas-Riester G, Pommerenke C, Dittert K, Schulze J (2014) An RNA sequencing transcriptome analysis reveals novel insights into molecular aspects of the nitrate impact on the nodule activity of Medicago truncatula. Plant Physiol 164:400–411
Cam Y, Pierre O, Boncompagni E, Hérouart D, Meilhoc E, Bruand C (2012) Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol 196:548–560
Chua KY, Pankhurst CE, Macdonald P, Hopcroft DH, Jarvis BD, Scott DB (1985) Isolation and characterization of transposon Tn5-induced symbiotic mutants of Rhizobium loti. J Bacteriol 162:335–343
Chungopast S, Hirakawa H, Sato S, Handa Y, Saito K, Kawaguchi M, Tajima S, Nomura M (2014) Transcriptomic profiles of nodule senescence in Lotus japonicus and Mesorhizobium loti symbiosis. Plant Biotech 31:345–349
D’haeseleer K, De Keyser A, Goormachtig S, Holsters M (2010) Transcription factor MtATB2: about nodulation, sucrose and senescence. Plant Cell Physiol 51:1416–1424
De Zélicourt A, Diet A, Marion J, Laffont C, Ariel F, Moison M (2012) Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J 70:220–230
Domonkos Á, Kovács S, Gombár A, Kiss E, Horváth B, Kováts GZ, Farkas A, Tóth MT, Ayaydin F, Bóka K, Fodor L, Ratet P, Kereszt A, Endre G, Kaló P (2017) NAD1 controls defense-like responses in Medicago truncatula symbiotic nitrogen fixing nodules following rhizobial colonization in a BacA-independent manner. Genes 8:387
van Doorn WG (2011) Classes of programmed cell death in plants, compared to those in animals. J Exp Bot 62:4749–4761
Engvild KC (1987) Nodulation and nitrogen fixation mutants of pea, Pisum sativum. Theor Appl Genet 74:711–713
Forrest SI, Verma DPS, Dhindsa RS (1991) Starch content and activities of starch-metabolizing enzymes in effective and ineffective root nodules of soybean. Can J Bot 69:697–701
González EM, Aparicio-Tejo PM, Gordon AJ, Minchin FR, Royuela M, Arrese-Igor C (1998) Water-deficit effects on carbon and nitrogen metabolism of pea nodules. J Exp Bot 49:1705–1714
González EM, Gálvez L, Arrese-Igor C (2001) Abscisic acid induces a decline in nitrogen fixation that involves leghaemoglobin, but is independent of sucrose synthase activity. J Exp Bot 52:285–293
Granell A, Harris N, Pisabarro AG, Carbonell J (1992) Temporal and spatial expression of a thiolprotease gene during pea ovary senescence, and its regulation by gibberellin. Plant J 2:907–915
Guinel FC (2015) Ethylene, a hormone at the center-stage of nodulation. Front Plant Sci 6:1121
Hakoyama T, Niimi K, Yamamoto T, Isobe S, Sato S, Nakamura Y, Tabata S, Kumagai H, Umehara Y, Brossuleit K (2012) The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol 53:225–236
Hernández-Jiménez MJ, Lucas MM, Rosario de Felipe M (2002) Antioxidant defence and damage in senescing lupin nodules. Plant Physiol Biochem 40:645–657
Hirsch AM, Smith CA (1987) Effects of Rhizobium meliloti nif and fix mutants on alfalfa root nodule development. J Bacteriol 169:1137–1146
Horváth B, Domonkos Á, Kereszt A, Szűcs A, Ábrahám E, Ayaydin F, Bóka K, Chen Y, Chen R, Murray JD, Udvardi MK, Kondorosi É, Kaló P (2015) Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci U S A 112:15232–15237
Ivanova KA, Tsyganova AV, Brewin NJ, Tikhonovich IA, Tsyganov VE (2015) Induction of host defences by Rhizobium during ineffective nodulation of pea (Pisum sativum L.) carrying symbiotically defective mutations sym40 (PsEFD), sym33 (PsIPD3/PsCYCLOPS) and sym42. Protoplasma 252:1505–1517
Kardailsky IV, Brewin NJ (1996) Expression of cysteine protease genes in pea nodule development and senescence. Mol Plant-Microbe Interact 9(8):689–695
Karmarkar V (2014) Transcriptional regulation of nodule development and senescence in Medicago truncatula. Doctoral thesis, Wageningen University, Wageningen, D.C. Available at: http://library.wur.nl/WebQuery/edepot/303161
Kiers ET, Rousseau RA, West SA, Denison RF (2003) Host sanctions and the legume–rhizobium mutualism. Nature 425:78–81
Kim M, Chen Y, Xi J, Waters C, Chen R, Wang D (2015) An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci U S A 112:15238–15243
Kneen BE, LaRue TA, Hirsch AM, Smith CA, Weeden NF (1990) sym 13—a gene conditioning ineffective nodulation in Pisum sativum. Plant Physiol 94:899–905
Kosterin OE, Rozov SM (1993) Mapping of the new mutation blb and the problem of integrity of linkage group I. Pisum Genet 25:27–31
Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17:1625–1636
Kulaeva OA, Zhernakov AI, Afonin AM, Boikov SS, Sulima AS, Tikhonovich IA, Zhukov VA (2017) Pea Marker Database (PMD)—a new online database combining known pea (Pisum sativum L.) gene-based markers. PLoS One 12:e0186713
Kumagai H, Hakoyama T, Umehara Y, Sato S, Kaneko T, Tabata S, Kouchi H (2007) A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol 143:1293–1305
Martin DN, Proebsting WM, Hedden P (1999) The SLENDER gene of pea encodes a gibberellins 2-oxidase. Plant Physiol 121:775–781
Matamoros MA, Baird LM, Escuredo PR, Dalton DA, Minchin FR, Iturbe-Ormaetxe I, Rubio MC, Moran JF, Gordon AJ, Becana M (1999) Stress-induced legume root nodule senescence. Physiological, biochemical, and structural alterations. Plant Physiol 121:97–111
Maunoury N, Redondo-Nieto M, Bourcy M, Van de Velde W, Alunni B, Laporte P, Durand P, Agier N, Marisa L, Vaubert D, Delacroix H, Duc G, Ratet P, Aggerbeck L, Kondorosi E, Mergaert P (2010) Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One 5:e9519
Morzhina EV, Tsyganov VE, Borisov AY, Lebsky VK, Tikhonovich IA (2000) Four developmental stages identified by genetic dissection of pea (Pisum sativum L.) root nodule morphogenesis. Plant Sci 155:75–83
Nemankin NF (2011) Analysis of pea (Pisum sativum L.) genetic system, controlling development of arbuscular mycorrhiza and nitrogen-fixing symbiosis. Dissertation Saint-Petersburg State University (in Russian)
Novák K, Pešina K, Nebesářová J, Škrdleta V, Lisa L, Našinec V (1995) Symbiotic tissue degradation pattern in the ineffective nodules of three nodulation mutants of pea (Pisum sativum L.) Ann Bot 76:303–313
Oono R, Anderson CG, Denison RF (2011) Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc Roy Soc B-Biol Sci 278:2698–2703
Pariasca JA, Sunaga A, Miyazaki T, Hisaka H, Sonoda M, Nakagawa H, Sato T (2001) Cloning of cDNAs encoding senescence-associated genes, ACC synthase and ACC oxidase from stored snow pea pods (Pisum sativum L. var saccharatum) and their expression during pod storage. Postharvest Biol Technol 22:239–247
Peck SC, Kende H (1998) Differential regulation of genes encoding 1-aminocyclopropane-1-carboxylate (ACC) synthase in etiolated pea seedlings: effects of indole-3-acetic acid, wounding, and ethylene. Plant Mol Biol 38:977–982
Pérez Guerra JC, Coussens G, De Keysler A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M (2010) Comparison of developmental and stress-induced nodule senescence in Medicago truncatula. Plant Physiol 152:1574–1584
Pierre O, Hopkins J, Combier M, Baldacci F, Engler G, Brouquisse R, Hérouart D, Boncompagni E (2014) Involvement of papain and legumain proteinase in the senescence process of Medicago truncatula nodules. New Phytol 202:849–863
Pladys D, Vance CP (1993) Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol 103:379–384
Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM (2005) Legume nodule senescence: roles for redox and hormone signaling in the orchestration of the natural aging process. New Phytol 165:683–701
Regus JU, Quides KW, O’Neill MR, Suzuki R, Savory EA, Chang JH, Sachs JL (2017) Cell autonomous sanctions in legumes target ineffective rhizobia in nodules with mixed infections. Am J Bot 104:1299–1312
Serova TA, Tsyganov VE (2014) Symbiotic nodule senescence in legumes: molecular-genetic and cellular aspects. Agr Biol 5:3–15
Serova TA, Tikhonovich IA, Tsyganov VE (2017) Analysis of nodule senescence in pea (Pisum sativum L.) using laser microdissection, real-time PCR, and ACC immunolocalization. J Plant Physiol 212:29–44
Sinharoy S, Torres-Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK (2013) The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25:3584–3601
Swaraj K, Laura JS, Bishnoi NR (1993) Nitrate induced nodule senescence and changes in activities of enzymes scavenging H2O2 in clusterbean (Cyamopsis tetragonaloba Taub.). J Plant Physiol 141:202–205
Tsyganov VE, Borisov AY, Rozov SM, Tikhonovich IA (1994) New symbiotic mutants of pea obtained after mutagenesis of laboratory line SGE. Pisum Genet 26:36–37
Tsyganov VE, Morzhina EV, Stefanov SY, Borisov AY, Lebsky VK, Tikhonovich IA (1998) The pea (Pisum sativum L.) genes sym33 and sym40 control infection thread formation and root nodule function. Mol Gen Genet 256:491–503
Tsyganov VE, Voroshilova VA, Rozov SM, Borisov AY, Tikhonovich IA (2013) A new series of pea symbiotic mutants induced in the line SGE. Russ J Genet: Appl Res 3:156–162
Tsyganova AV, Tsyganov VE, Findlay KC, Borisov AY, Tikhonovich IA, Brewin NJ (2009) Distribution of legume arabinogalactan protein-extensin (AGPE) glycoproteins in symbiotically defective pea mutants with abnormal infection threads. Cell Tissue Biol 3:93–102
Tsyganova AV, Kitaeva AB, Tsyganov VE (2018) Cell differentiation in nitrogen-fixing nodules hosting symbiosomes. Funct Plant Biol 45:47–57
Van de Velde W, Pérez Guerra JC, De Keysler A, De Rycke R, Rombauts S, Maunoury N, Mergaert P (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141:711–720
Vauclare P, Bligny R, Gout E, De Meuron V, Widmer F (2010) Metabolic and structural rearrangement during dark-induced autophagy in soybean (Glycine max L.) nodules: an electron microscopy and 31P and 13C nuclear magnetic resonance study. Planta 231:1495–1504
Vernié T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P (2008) EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20:2696–2713
Voroshilova VA, Demchenko KN, Brewin NJ, Borisov AY, Tikhonovich IA (2009) Initiation of a legume nodule with an indeterminate meristem involves proliferating host cells that harbour infection threads. New Phytol 181:913–923
Wang C, Yu H, Luo L, Duan L, Cai L, He X, Wen J, Mysore KS, Li G, Xiao A, Duanmu D, Cao Y, Hong Z, Zhang Z (2016) NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol 212:176–191
Wang Q, Liu J, Li H, Yang S, Körmöczi P, Kereszt A, Zhu H (2017) Nodule-specific cysteine-rich peptides negatively regulate nitrogen-fixing symbiosis in a strain-specific manner in Medicago truncatula. Mol Plant-Microbe Interact 31:240–248
Westhoek A, Field E, Rehling F, Mulley G, Webb I, Poole PS, Turnbull LA (2017) Policing the legume-Rhizobium symbiosis: a critical test of partner choice. Sci Rep 7:1419
Yamaya-Ito H, Shimoda Y, Hakoyama T, Sato S, Kaneko T, Hossain MS, Shibata S, Kawaguchi M, Hayashi M, Kouchi H, Umehara Y (2018) Loss-of-function of ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1) in Lotus japonicus restricts efficient nitrogen-fixing symbiosis with specific Mesorhizobium loti strains. Plant J 93:5–16
Yang S, Wang Q, Fedorova E, Liu J, Qin Q, Zheng Q, Price PA, Pan H, Wang D, Griffitts JS, Bisseling T, Zhu H (2017) Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc Natl Acad Sci U S A 114:6848–6853
Zdunek-Zastocka E (2008) Molecular cloning, characterization and expression analysis of three aldehyde oxidase genes from Pisum sativum L. Plant Physiol Biochem 46:19–28
Acknowledgements
This work was supported by the Russian Science Foundation [grant number 16-16-10035]. The research was performed using equipment of the Core Centrum “Genomic Technologies, Proteomics and Cell Biology” in ARRIAM, and the “Molecular and Cell Technologies” Research Resource Centre at Saint-Petersburg State University. Jennifer Smith, PhD, and Robert McKenzie, PhD, from Edanz Group (www.edanzediting.com/ac) edited the English version of a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest
Additional information
Handling Editor: Ulrike Mathesius
Electronic supplementary material
ESM 1
(PDF 4038 kb)
Rights and permissions
About this article
Cite this article
Serova, T.A., Tsyganova, A.V. & Tsyganov, V.E. Early nodule senescence is activated in symbiotic mutants of pea (Pisum sativum L.) forming ineffective nodules blocked at different nodule developmental stages. Protoplasma 255, 1443–1459 (2018). https://doi.org/10.1007/s00709-018-1246-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-018-1246-9