Abstract
The medicinal plant Withania somnifera is researched extensively to increase the quantity of withanolides and specifically withaferin A, which finds implications in many pharmacological activities. Due to insufficient knowledge on biosynthesis and unacceptability of transgenic approach, it is preferred to follow alternative physiological methods to increase the yield of withanolides. Prior use of elicitors like salicylic acid, methyl jasmonate, fungal extracts, and even mechanical wounding have shown to increase the withanolide biosynthesis with limited success; however, the commercial viability and logistics of application are debatable. In this investigation, we tested the simple nitrogeneous fertilizers pertaining to the enhancement of withaferin A biosynthesis. Application of ammonium sulfate improved the sterol contents required for the withanolide biosynthesis and correlated to higher expression of pathway genes like FPPS, SMT1, SMT2, SMO1, SMO2, and ODM. Increased expression of a gene homologous to allene oxide cyclase, crucial in jasmonic acid biosynthetic pathway, suggested the involvement of jasmonate signaling. High levels of WRKY gene transcripts indicated transcriptional regulation of the pathway genes. Increase in transcript level could be correlated with a corresponding increase in the protein levels for WsSMT1 and WsWRKY1. The withaferin A increase was also demonstrated in the potted plants growing in the glasshouse and in the open field. These results implicated simple physiological management of nitrogen fertilizer signal to improve the yield of secondary metabolite through probable involvement of jasmonate signal and WRKY transcription factor for the first time, in W. somnifera besides improving the foliage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Withania somnifera (L.) Dunal commonly known as ashwagandha (family Solanaceae) is an important medicinal plant used traditionally for its diverse activities in various indigenous systems of medicine. The plant is known for its aphrodisiac, rejuvenating, anti-inflammatory, and anti-tumorigenic properties (Naidu et al. 2003). Though the plant contains alkaloids, withanolides, flavanoids, glycosides, and saponins, it is mostly recognized for the triterpene steroidal lactones of ergostane skeleton referred as “withanolides” (Elsakka et al. 1990). Withaferin A, the major withanolide of this plant, is described to be very active pharmacologically and hence, is the most studied molecule (Dhuley 2000; Jayaprakasam et al. 2003; Choudhary et al. 2005; Kaileh et al. 2007). Due to its importance, the plant and the genes involved in the biosynthetic pathway have been the target of research by various groups with the ultimate goal to increase the metabolites (withanolides) in planta. With the aim to identify better genotypes containing higher withanolides, Gupta et al. (2011) have studied the expression pattern of FPPS gene in response to different elicitors (salicylic acid, methyl jasmonate, and mechanical wounding), and Pandey et al. (2010) have developed transgenic protocol for a future improvement. Similarly, several genes were cloned and characterized from the biosynthetic pathway of withanolides including 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) (Akhtar et al. 2012; Gupta et al. 2013a; Singh et al. 2014), and squalene epoxidase (SE) (Razdan et al. 2013). Three branch point oxidosqualene cyclases from W. somnifera were also analyzed to understand the metabolic flux towards specific secondary metabolites (Dhar et al. 2014). Gupta et al. (2013b) functionally annotated and compared the leaf and root transcriptome of W. somnifera. Recently, the role of squalene synthase (SQS) with a reference to phytosterols biosynthesis and withanolides accumulation was studied by virus-induced gene-silencing approach in W. somnifera (Singh et al. 2015).
Although, transgenic plants are good models to understand the biochemical function of genes and could be a better alternative to obtain high withanolide concentrations in the plant as the acceptability of transgenic plants is still a matter of dispute. Further, generation of transgenic plant is a tedious and time-consuming process. Hence, most of the researchers focus on the elicitor-treated callus or hairy root (Doma et al. 2012) to increase the metabolite in tissue culture. Reports with limited success using elicitation approach for increasing the secondary metabolite content in some plants are available. However, this approach was of less significance in terms of withanolides production in W. somnifera. The generally used elicitors for increasing withaferin A content are salicylic acid, methyl jasmonate, fungal extracts or mechanical wounding, and hairy root culture, but the mechanism of enhancement via hairy root culture is poorly understood (Doma et al. 2012). On the contrary, the use of “nitrogenous fertilizer” to increase the biomass is a well-known phenomenon and has an apparent effect on protein synthesis. Present investigation for the first time tends to reveal the role of the nitrogenous fertilizers in increasing the accumulation of withaferin A with simultaneous enhancement of the related gene(s) expression, through jasmonate-signaling pathway with probable involvement of WRKY transcription factors in W. somnifera. In addition to the indicative mechanism of withaferin A increment, this finding will help in increasing the biosynthesis of economical therapeutic compound in planta.
Materials and methods
Plant material and withanolide analysis
W. somnifera var. “CIM-Poshita” seeds and plants were obtained from the National Gene Bank for Medicinal and Aromatic Plants (NGBMAP) maintained at CSIR-Central Institute of Medicinal and Aromatic Plants in Lucknow, India. Seeds were sown in mid July for raising a nursery. After sowing, a mixture of farm yard manure (FYM) and soil was thinly spread over the seeds and irrigated with a sprinkler hose. The seeds took 8–10 days to germinate, and seedlings were ready for transplantation in 5 weeks. Transplantation of seedlings was carried out in plots of 10.0 m × 10.0 m size with 10 cm × 20 cm row to row and plant to plant spacing. A light irrigation just after the planting ensured the establishment of the seedlings. The established plants were also maintained under glasshouse conditions i.e., 60–75 % relative humidity and 25 ± 2 °C temperature. Withanolide extraction and the contents were estimated following the protocol described earlier by Singh et al. (2014). The soil characteristics of field and pot trails are given in Table S1.
Genes, ESTs, and quantitative RT-PCR
RNA isolation and quantitative reverse transcription polymerase chain reaction (RT-PCR) experiments were carried out following Misra et al. (2012). First strand cDNA was synthesized by RT-PCR approach using the ThermoScriptTM RT-PCR system (Invitrogen, https://www.lifetechnologies.com). The expression levels of withanolide biosynthetic pathway genes, cytochrome P450 (CYPs) and TF genes under various conditions were quantified using real-time PCR with SYBR green I chemistry (Applied Biosystems, https://www.lifetechnologies.com) using the primers described in Table S2. Threshold cycle (Ct) values obtained after real-time PCR were used for calculation of ΔCt (target-endogenous control). The quantification was carried out by calculation of ΔΔCt to determine the fold difference in gene expression [Δ Ct target–Δ Ct calibrator]. Relative quantity (RQ) was determined by 2−ΔΔCT. W. somnifera cyclophilin gene was used as endogenous control. The genes and expressed sequence tags (ESTs) included in the expression analysis are sterol methyltransferase 1 (SMT1, GQ921847) and obtusifoliol-4α-demethylase (ODM, GQ921846) (isolated earlier and described by Pal et al. 2011); partial sequences resembling to sterol methyltransferase 2 (SMT2) and sterol methyl oxidase 1 and 2 (SMO1 and 2) were amplified from W. somnifera leaf cDNA using degenerate primers. Primers for farnesyl pyrophosphate synthase (FPPS, HM855234) and squalene epoxidase (SE, GU574803) were designed from the respective sequences available in NCBI database. Real-time primers were also designed for unknown CYP genes (GR923492, GR923679, GR923693, and GR923780) and a non CYP EST GR923598 which was homologous to “allene oxide cyclase” a crucial Jasmonic acid biosynthesis pathway gene. GR923677 showing homology to deacetoxyvindoline 4-hydroxylase was also included in this analysis. In addition, the transcripts of transcription factors WRKY1 (GR923578), WRKY2 (GR923530), WRKY3 (GR923374), MYB (GR923616), bHLH (GR923360), and bZIP (GR923446) were investigated for ammonium sulfate stimulation. All the ESTs were isolated earlier either from leaf or root cDNA libraries of W. somnifera (Pal et al. 2011).
Feeding assays to ascertain the influence of fertilizer elements
Fresh twigs (four leaves) from a 12-week-old single plant of W. somnifera were used in this experiment. For each treatment, fresh twig samples were dipped in different concentrations of DMSO (5, 10, 15, 20, and 100 %) and ammonium sulfate (0.05, 0.1, 0.2, 0.4, 0.5, 0.6, 1, 1.5, and 2 M) solutions in sterile Milli-Q water. To check the effect of NPK (nitrogen, phosphorus, and potassium) on withanolide biosynthesis, a separate experiment was performed in which fresh twigs were dipped overnight in a very dilute concentration of ammonium sulfate (NH4(SO4)2), ammonium hydroxide (NH4OH), nitric acid (HNO3), and Urea (NH2CONH2) for N effect, phosphoric acid (H3PO4) for P effect, and potassium hydroxide (KOH) for K effect (all the acids were of 0.1 % v/v solutions; bases and salts were of 0.1 mg/mL solutions). To check the effect of sulfur (sulfate), sulfuric acid (H2SO4, 0.1 % v/v solutions) was included in the investigation. Experiment in potted plants (6-week old) was carried out in three replicates with ammonium sulfate and urea, where 6.6 and 1.5 % solutions (1000 mL to have 7.0 g nitrogen) respectively, were added to the pot soil to saturation initially, and the pots were subsequently irrigated with 100 mL of Milli-Q water at an interval of 7 days. Ammonium sulfate application was repeated 15 days after the first application. For field experiments, 0.5 M ammonium sulfate and 0.05 M urea were used. One liter each of these solutions were carefully applied to the root zone of individual plant. The field plants were irrigated with same amount of water at weekly interval and ammonium sulfate/ urea application was repeated at 15 days interval. All these concentrations were finalized after proper standardization experiments, and always Milli-Q water was taken as control. Subsequently, samples were collected, washed thoroughly with Milli-Q water, dried for 2 days at 42 ° C, and ground to fine powder. Extraction of withanolides, sterols, and their quantitative analysis was performed following our earlier publication (Singh et al. 2014). All these analysis were carried out with three biological replicates.
Western blot analysis of WsSMT1 and WSCPL336 proteins
W. somnifera twigs were treated with 0.5 M ammonium sulfate solution along with suitable control in which twigs were dipped in sterile Milli-Q water; leaf samples were collected from treated as well as control plants at different intervals (0 h, 6 h, 9 h, 12 h, and 24 h), weighed, frozen immediately in liquid nitrogen, and preserved in a deep freezer (−80 °C). Total protein was isolated from collected leaf tissues separately. Two gram of leaf tissues was ground in liquid nitrogen and was added in protein extraction buffer (50 mM bis-tris (pH −7.5), 1 % PVP, and 10 mM DTT), incubated in ice for 30 min, and centrifuged at 4 °C for 30 min at 12,000×g. Total protein from supernatant was quantified using Bradford reagent and BSA as standard. WsSMT1 protein was analyzed to be a membrane targeted using ChloroP (http://www.cbs.dtu.dk/services/ChloroP/), and hence the first 69 bases were removed from WsSMT1 sequence. The truncated WsSMT1 (38 kDa protein) and normal WSCPL336 (WsWRKY1, 35 kDa) were cloned in pET 28 a(+) vector in-frame with the His6-tag and expressed in E. coli [BL21(DE3)] following Koeduka et al. (2006). Protein was extracted from exponentially growing bacteria (3 l of LB medium) after 6 h of induction at 37 °C with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG; A600 = 0.7). His6-tagged WsSMT1 and WSCPL336 protein were purified using Ni-NTA spin columns (Novagen), analyzed by SDS–PAGE, and estimated using Bradford Reagent (Sigma).
Purified protein samples were injected separately in mice through standard procedures to raise the hyper immune sera against them and specificity was confirmed through immune–diffusion. Total protein isolated from treated as well as control leaves were run on 12 % SDS PAGE gel along with PiNK Plus prestained protein ladder (175 kDa) (BIOCHEM, USA). Western blotting was performed by transferring resolved protein from acrylamide gel to PVDF membrane using iBlot 2 Dry blotting system (Invitrogen, USA). The membrane with transferred protein was blocked using super blocker (Pierce, USA) overnight at 4 °C, followed by washing membrane thrice with washing buffer and hybridization with hyper immune sera (primary antibody) for an hour at 37 °C. This was again followed by the washing step and incubated with the secondary antibody (goat anti-mouse IgG-HRPO) at 37 °C for 2 h. The membrane was then treated with 1 mg mL−1 of di-aminobenzidine (DAB) in PBS for 10–15 min until the bands became visibly clear. The membrane was washed with distilled water and dried before capturing the images. The antibody-raising protocol was pre-approved by the CPCSEA through institutional animal ethics committee.
Reactive oxygen species assay
For reactive oxygen species (ROS) assay, modified procedure of Thordal-Christensen et al. (1997) was used. Fresh leaves from treated twig as well as from potted plant were plucked and placed in a solution of 1 mg mL−1 (4.67 mM) 3,3′-diaminobenzidine-HCl (DAB, Sigma, cat #D-8001), pH 3.8 (a low pH is necessary in order to solubilize DAB) and incubated in the dark at 25 °C for 7 h. To visualize the ROS stain, leaf chlorophyll was bleached by submersion in 96 % (v/v) boiling ethanol for 5 min. DAB is specific for hydrogen peroxide (H2O2), which was visualized in the leaves as reddish brown spots under a light microscope. A 7-h-incubation time is described to be optimum for visualizing H2O2 stain.
Results
Effect of DMSO on withaferin A biosynthesis
While investigating the effect of mevinolin and fosmidomycin (Singh et al. 2014) on HMGR and DXR proteins on withanolide biosynthesis pathway, the solvent DMSO used to dissolve mevinolin was observed to be inducing the withanolide biosynthesis. Hence, the effect of DMSO on withanolide biosynthesis was initially checked. Withaferin A content was observed to be maximum (five times compared to control) in the leaf of twigs dipped in 10 and 15 % DMSO concentrations while the increase was approximately four times in the twigs treated with 5 and 20 % DMSO. A minor increase was detected in the twigs treated with 100 % DMSO (Fig. 1a, Figure S1). At the same time, gene expression study was also performed to check the expression of cytochrome genes which were earlier reported to be expressing either in a leaf or a root (Pal et al. 2011). Neither of the selected CYP ESTs nor the allene oxide cyclase EST got induced by DMSO treatment (Fig. 1b). All pathway-related genes (FPPS, SE, SMT1, SMT2, SMO1, SMO2, and ODM) showed highly suppressed gene expression in 10 % DMSO-treated-leaf cDNAs (Fig. 1c).
Levels of withaferin A and gene expression after DMSO treatment. a Withaferin A content (%) in W. somnifera twigs at different concentrations of DMSO. b Expression of CYPs and AOC in 10 % DMSO-treated twigs of W. somnifera and c expression pattern of withanolide pathway-related genes in 10 % DMSO-treated twigs. RQ relative quantity. Tukey-Kramer multiple comparisons test for significance analysis was carried out for comparing withaferin A content (P < 0.001 represented by only small alphabets, P < 0.01 by single quotation mark, P < 0.05 by double quotation mark on small alphabets), and Dunnett’s multiple comparisons test was carried out for gene expression analysis (P < 0.01). Data are means ± SE (n = 3 biological replicates)
Evaluation of nutrient effects on withaferin biosynthesis
Effect of NPK on withaferin A biosynthesis
Since DMSO contains sulfur, dilute solution of sulfuric acid was initially taken gradually extending the experiment to other fertilizer elements. Effect of nitrogen, phosphorus, and potassium (NPK) on withaferin A content was detected by dipping the W. somnifera twig in solvents containing the salts/acid/base of respective elements (Fig. 2, Figure S2). A decrease in withaferin A was observed in case of the treatment with dilute solution of sulfuric acid and phosphoric acid compared to control, while a significant increase was observed in case of treatments with dilute solutions of liquid ammonia, nitric acid, ammonium sulfate, and urea in decreasing order. Also, an improvement in withaferin A content was detected for potassium hydroxide solution. Since liquid ammonia and nitric acid are strong base and acid, while ammonium sulfate is a common fertilizer, the latter was taken further for experimentation.
Levels of withaferin A when treated with different compounds. Withaferin A content (%) in the twigs of W. somnifera treated with sulfuric acid (H2SO4), urea (NH2CONH2), ammonium hydroxide (NH4OH), nitric acid (HNO3), ammonium sulfate ((NH4)2SO4), phosphoric acid (H3PO4), and potassium hydroxide (KOH). Tukey-Kramer multiple comparisons test for significance analysis was carried out for comparing withaferin A content (P < 0.001 represented by only small alphabets, P < 0.01 by single quotation mark, P < 0.05 by double quotation mark on small alphabets). Data are means ± SE (n = 3 biological replicates
Ammonium sulfate treatment influencing withaferin A and sterol content vis a vis gene expression
Maximum increase in withaferin A content was found in the W. somnifera twigs treated with 0.5 M ammonium sulfate solution. A significant increase in withaferin A content with increase in ammonium sulfate concentration up to 0.5 M followed by a gradual decrease was evident (Fig. 2 a, Figure S3). At this concentration (0.5 M ammonium sulfate), the transcript abundance of FPPS, SMT1, SMT2, SMO1, SMO2, and ODM was notably higher compared to control with no change in SE expression was observed (Fig. 3 c). A slight improvement in expression patterns of CYP72A (GR923693), CYP86A2 (GR923766), and CPR (GR923780) transcripts whereas, 8-fold increase in the expression of transcript homologous to allene oxide cyclase (GR923598) after ammonium sulfate treatment was determined (Fig. 3 b). Comparing all the transcription factors induced upon ammonium sulfate treatment, the transcripts of WRKY1 (GR923578) increased about 90-folds while of WRKY3 (GR923374) increased 8-fold (Fig. 4). Since, sterols are the precursor of withaferin A biosynthesis (Singh et al. 2014), the relevant sterol content in plant in response to different treatments was also estimated. Significant increase in cycloartenol, sitosterol, stigmasterol, and campesterol was detected in the leaf tissues of ammonium sulfate-treated twigs (Fig. 5). The increase in transcript levels for different genes due to ammonium sulfate treatment was validated by analyzing the protein content for WsSMT1 and WsWRKY1. Compared to the control, an increase in the expression of the WsSMT1 (38 kDa) isoform was detected at all the time points in the leaves of ammonium sulfate-treated twigs and was absent in the western blot of untreated leaf protein. Similar result was obtained for the 35 kDa WsWRKY1 protein. The western blot supports the above observations (see Figure S4), but there are methodological limitations concerning the specificity of the antibodies.
Withaferin A and gene expression after ammonium sulfate treatment. a Withaferin A content (%) in W. somnifera twigs at different concentrations of ammonium sulfate. b Expression of CYPs and AOC in 500-mM ammonium sulfate-treated twigs of W. somnifera and c expression pattern of withanolide pathway-related genes in 500-mM ammonium sulfate-treated twigs. RQ relative quantity. Tukey-Kramer multiple comparisons test for significance analysis was carried out for comparing withaferin A content (P < 0.001 represented by only small alphabets, P < 0.01 by single quotation mark, P < 0.05 by double quotation mark on small alphabets), and Dunnett multiple comparisons test was carried out for gene expression analysis (P < 0.01). ns not significant. Data are means ± SE (n = 3 biological replicates)
Expression profile of transcription factors due to ammonium sulfate treatment. RQ relative quantity. WRKY1 (GR923578), WRKY2 (GR923530), WRKY3 (GR923374), MYB (GR923616), bHLH (GR923360), and bZIP (GR923446). Dunnett’s multiple comparisons test was carried out for gene expression analysis (P < 0.01). ns not significant (P > 0.05). Data are means ± SE (n = 3 biological replicates)
Effect of ammonium sulfate and urea on potted and field plants
Experiment was performed on rooted plants in pots to check the effect of ammonium sulfate on withaferin A content. Though the withaferin A did not increase significantly in the control plants after 7 and 30 days, a significant increase was detected in the plants treated with urea and ammonium sulfate. The increase was more in the leaf of plants treated with ammonium sulfate compared to urea at both the stages (Fig. 6a, Figure S5). A similar trend was observed in the well-established plants in the field. The increase was more prominent in the ammonium sulfate-treated plants after 30 days (Fig. 6b, Figure S6).
Effect of ammonium sulfate and urea treatment on withaferin A content. a Plant in greenhouse shows increase in withaferin A content, 7 and 30 days after treatment. b Plant in the field shows increase in withaferin A content, 7 and 30 days after treatment. Gray color-filled box and dark box represent urea and ammonium sulfate treatments respectively compared to empty box representing the control. Tukey-Kramer multiple comparisons test for significance analysis was carried out for comparing withaferin A content (P < 0.001 represented by only small alphabets, P < 0.01 by single quotation mark, P < 0.05 by double quotation mark on small alphabets). Data are means ± SE (n = 3 biological replicates
Reactive oxygen species analysis
ROS are formed in stressed plants, and secondary metabolites are also described to be synthesized under stress conditions. Leaf of DMSO (10 %)-treated twigs showed to be under severe stress with highest H2O2 formation as identified by the amount and intensity of brown spots on the leaves. Ammonium sulfate- and urea-treated leaves also showed peroxide accumulation but very less compared to 10 % DMSO treatment (Fig. 7). When the twigs were dipped in DMSO, these did not survive after the second day of treatment and wilted, whereas the twigs dipped in urea or ammonium sulfate continued to survive and flourish.
Discussion
Sangwan et al. (1993) and Mannan et al. (2010) have hypothesized earlier that DMSO application in vitro enhances secondary metabolite production in Artemisia annua. Earlier, an increase in withaferin A by DMSO (solvent control) treatment in W. somnifera was also observed (Singh et al. 2014.). To investigate this event, withaferin A was targeted for the analysis as it is the major compound biosynthesized de novo in the leaf (Kaul et al. 2009) and in different parts of the plant (Sangwan et al. 2008). The content of this compound in the leaf increased with an increased concentration of DMSO treatment until 10–15 % and decreased thereafter. Though withanolide biosynthetic pathway is not fully understood, still the pathway route has been fairly described (Singh et al. 2014) and some of the gene sequences are available in NCBI database. Using these available resources, it was observed that the increase in withaferin A due to DMSO treatment is not supported by the expression of genes of the biosynthetic pathway (analyzed in this investigation). Hence, these interesting observations propelled a series of experimentations to understand the basic reasons affecting withanolide biosynthesis in planta. DMSO is an organosulfur with good skin penetration properties and has been used as a co-solvent to assist absorption of the flavonol glycoside icariin into nematode C. elegans (Cai et al. 2011). Increase in artemisinin content observed earlier in rooted plants due to DMSO treatment indicated the transport of the chemical from root to leaf, which is the site of artemisinin biosynthesis (Mannan et al. 2010). In this investigation, twigs without root could also transport DMSO and affect withaferin A biosynthesis as the increase in withaferin A in the leaf is indicative of it. Though ROS (H2O2) is described to be playing a major role to increase secondary metabolite in in vitro systems (Sangwan et al. 1993; Zhang et al. 2003; Mannan et al. 2010), the mechanism is yet to be explained. In the present investigation, severe peroxide formation in DAB assay was observed in the twigs treated with 10 % DMSO which turned necrotic after the treatment.
Initially, the element sulfur in DMSO was thought to be involved in increasing the withaferin A content, dilute sulfuric acid, and ammonium sulfate, both containing sulfur were checked for their influence on withaferin A accumulation. Interestingly, dilute sulfuric acid did not increase the withaferin A content, whereas ammonium sulfate increased the withaferin A accumulation, thus eliminating the effect of sulfur on withaferin A biosynthesis. As nitrogen is an important fertilizer element and a part of ammonium sulfate, different nitrogen-containing compounds (ammonium sulfate, dilute nitric acid, dilute ammonium hydroxide, and urea) were compared for their effect on withaferin A biosynthesis. All these compounds were observed to significantly increase the accumulation of withaferin A compared to control, indicating the involvement of N, either in nitrate or ammonium form. Ammonium nitrate was not analyzed because of its explosive nature. Effects of P (phosphorous) and K (potassium) were probed by including dilute phosphoric acid and potassium hydroxide in the analysis and K significantly enhanced withaferin A accumulation compared to control. However, a significant decrease in withaferin A content was observed in phosphoric acid application. Dilute sulfuric and phosphoric acid treatments splitted the withaferin A peak into two, thereby reducing the withaferin A content (Figure S2). Though dilute nitric acid (a strong acid) and dilute ammonium hydroxide (a strong base) demonstrated highest increase in withaferin A but is not used as fertilizer in the field, only ammonium sulfate and urea were finally included for further investigation. Similarly, though potassium hydroxide (a strong base) also significantly improved the withaferin A level, the mechanism due to potassium application may be studied in isolation differently by including fertilizer salts of potassium.
The transcripts of FPPS, generating FPP for triterpene biosynthesis increased significantly when the twigs were treated with 500 mM ammonium sulfate. Increased expression of SMT1, SMT2, SMO1, SMO2, and ODM in the triterpene biosynthesis pathway (Fig. 8) correlated with increased synthesis of sterols and withaferin A. Previously, predominant route for withanolide biosynthesis has been assigned to be through campesterol/stigmasterol (Singh et al. 2014). In this analysis also, significant increase in withaferin A, cycloartenol, sitosterol, stigmasterol, and campesterol levels indicated the influence of ammonium sulfate in driving the metabolites towards withaferin A. Of all the cytochrome ESTs tested for expression, an increase of a lesser degree was observed for the transcripts of GR923693 (CYP72A) and GR923780 (CPR). But transcript abundance of another EST GR923598 homologous to the allene oxide cyclase (AOC) believed to be crucial in jasmonic acid (JA) biosynthetic pathway by preferentially synthesizing cis-(+)-12-oxo-phytodienoic acid (OPDA) (Stenzel et al. 2003) was highest (more than 8-fold compared to control). Also, AOC expression and accumulation correlated temporally upon external stimuli such as wounding (Ziegler et al. 2000). In A. thaliana, AOC is found abundantly in the leaf (Stenzel et al. 2003), whereas in tomato, AOC is confined to vascular bundles (Hause et al. 2000) and also the sieve elements (Hause et al. 2003). Specific roles for JA and other oxylipins are also suggestive by the abundant occurrence of AOC protein in ovules of tomato flowers and by the distinct oxylipin signature in different tomato-flower organs (Hause et al. 2000). Oxylipin signals such as allene oxide synthase-dependent jasmonate, in addition to other signal components, are employed directly or indirectly by elicitors for induction of plant secondary metabolite accumulation (Zhao et al. 2005). The role of stress-responsive jasmonates in the induction of secondary metabolite is also well-known (Chen et al. 2006) though the exact mechanism yet needs a clear demonstration. The most convincing example is ORCA3, a jasmonate-responsive APETALA2 (AP2)-domain transcription factor from Catharanthus roseus. Overexpression of this gene resulted in enhanced expression of several metabolite biosynthetic genes, consequently, increasing accumulation of terpenoid indole alkaloids (Fits and Memelink 2000). Elicitation conditions with methyl jasmonate and salicylic acid to improve the productivity of withanolides in the adventitious root (Sivanandhan et al. 2012), hairy root (Sivanandhan et al. 2013), and cell suspension cultures (Sivanandhan et al. 2014) of W. somnifera has been described. Gupta et al. (2011) demonstrated significant elevation of FPPS gene expression in different chemotypes of W. somnifera in response to salicylic acid, methyl jasmonate, and mechanical injury. In the present investigation, the pathway genes, having significant increase in expression in relation to a very high AOC gene expression related to the jasmonate biosynthesis pathway, were demonstrated for the first time in W. somnifera. Most important is the induction by nitrogenous fertilizer for high AOC and withaferin A biosynthesis pathway gene transcripts. Although different phenotypic impacts of N supply/source in plants have been identified, the N sensors and signaling pathways mediating these effects have yet to be fully characterized (Kraiser et al. 2011). As the involvement of MAD-box transcription factor gene family has been implicated in the regulating pathway of localized nitrate application (Zhang and Forde 1998), the role of similar transcription factors in influencing the transcription of genes related to jasmonate or withaferin A cannot be ruled out. This observation is supported by jasmonate-responsive ORCA3 example. In this investigation, we also detected a high level of induction for WRKY type transcription factors due to ammonium sulfate treatment. These observations were supported by the increase in WsWRKY1 and WsSMT1 proteins due to ammonium sulfate treatment (Figure S4). WRKY TFs are indicated to be key components in the induction process for natural product biosynthesis (Schluttenhofer et al. 2014). These are implicated for benzylisoquinoline alkaloid biosynthesis in Coptis japonica (Kato et al. 2007), sesquiterpene in Gossypium arboretum (Xu et al. 2004), Artemisia annua (Ma et al. 2009), paclitaxel in Taxus chinensis (Li et al. 2013), terpene indol alkaloids in Catharanthus roseus (Suttipanta et al. 2011). Dombrecht et al. (2007), and proposed regulatory controls mediated by JIN1/MYC2 in the jasmonate-signaling pathway by coordinating a transcriptional cascade involving a number of transcription factors like AP2/ERFs, MYBs, zinc fingers, and WRKYs with proven roles in regulating downstream gene expression including the biosynthesis of secondary metabolites. Though, the importance of WRKY TFs in jasmonate-signaling network is relatively less studied, some of the examples are available in Arabidopsis. These are AtWRKY6, AtWRKY8, AtWRKY11, AtWRKY17, AtWRKY25, AtWRKY28, AtWRKY38, AtWRKY60, AtWRKY62, and AtWRKY70 expressing differentially in response to jasmonate signal for plant defense (Schluttenhofer et al. 2014). The WRKY transcription factors, WRKY33, WRKY40, WRKY53, and WRKY70 implicated previously in the regulation of plant defense responses (Pandey and Somssich 2009), were also demonstrated to be specifically upregulated by ammonium (Patterson et al. 2010). Hence, very high expression of allene oxide cyclase due to ammonium sulfate signal can be correlated to high level of jasmonate biosynthesis which in turn induces high levels of WsWRKY1 expression leading to pathway gene expression vis a vis the sterols and withaferin A.
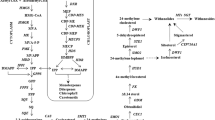
Putative biosynthetic pathway of withanolide biosynthesis in W. somnifera. The genes showing overexpression due to ammonium sulfate treatment are shown as up arrows. BAS β-amyrin synthase, CAS cycloartenol synthase, CYPs cytochrome P450 monooxygenases, DXR 1-deoxy-d-xylulose 5-phosphate reductoisomerase, DXS 1-deoxy-d-xylulose 5-phosphate synthase, FPPS farnesylpyrophosphate synthase, HMGR 3-hydroxy-3-methyl-glutaryl-CoA reductase, LAS lanosterol synthase, ODM obtusufoliol 14-demethylase, SE squalene epoxidase, SMO methyl sterol monooxygenase, SMT sterol methyl transferase, SS squalene synthase
The response due to ammonium sulfate is also in contrast to the DMSO treatment, where expression of almost all the genes were suppressed, and the treated twigs turned necrotic on the third day leading to senescence. This indicated the conversion of the flux by the existing enzymes until senescence, instead of forming new enzymes to increase the concentration of withaferin A. This phenomenon is transient and unsustainable as the twigs do not survive the DMSO treatment in the long run. The stress factor from these experiments can be linked, as DMSO-treated leaves are highly stressed leading to necrosis because of membrane rupture, whereas the ammonium sulfate- and urea-treated leaves showed mild stress as observed by the ROS analysis. With this knowledge, the simple nitrogenous fertilizers were applied to the potted plants grown in the greenhouse and the field grown plants. Significant increase in withaferin A content was detected at both after 7 and 30-day stages of the plants. The increase in withaferin A was less in the field-grown plants compared to the potted plants as expected, because the application of ammonium sulfate was in a controlled condition with a defined quantity of soil in the greenhouse experiments in contrast to the plants in the field where environmental and soil parameters are difficult to control. Fertilizer application to the plant in the field is also easier and cost-effective, compared to the application of the elicitors like jasmonate/methyl jasmonate.
In summary, this investigation demonstrated the possible linkage of mild stress, jasmonate biosynthesis, WsWRKY1 TF, and pathway gene expression leading to the increased biosynthesis of withaferin A in W. somnifera as induced by nitrogenous fertilizers. Though general effect of nitrogen assimilation is well-documented in plant health and yield, this probable linkage was indicated for the first time for secondary metabolite biosynthesis and will have greater consequences on secondary metabolism by managing fertilizer elements. As the mechanisms of fertilizer application to the soil in the field already exists, this finding will be helpful in designing a simpler cost-effective method to improve the yield of secondary metabolites in planta.
Accession numbers
Sequence data present in this study can be found in GenBank database (www.ncbi.nlm.nih.gov) with the help of accession numbers provided in the text.
References
Akhtar N, Gupta P, Sangwan NS, Sangwan RS, Trivedi PK (2012) Cloning and functional characterization of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Withania somnifera: an important medicinal plant. Protoplasma 250:613–622
Cai WJ, Huang JH, Zhang SQ, Wu B, Kapahi P, Zhang XM, Shen ZY (2011) Icariin and its derivative icariside II extend healthspan via insulin/IGF-1 pathway in C. elegans. Plos One 6(12):e28835. doi:10.1371/journal.pone.0028835
Chen H, Jones AD, Howe GA (2006) Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett 580:2540–2546
Choudhary MI, Hussain S, Yousuf S, Dar A, Mudassar, Atta ur R (2005) Chlorinate and di-epoxy withanolides from Withania somnifera and their cytotoxic effects against human lung cancer cell line. Phytochem 71:2205–2209
Dhar N, Rana S, Razdan S, Bhat WW, Hussain A, Dhar RS, Vaishnavi S, Hamid A, Vishwakarma RA, Lattoo SK (2014) Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) Dunal. J Biol Chem 289(24):17249–67
Dhuley JN (2000) Adaptogenic and cardioprotective action of ashwagandha in rats and frogs. J Ethnopharmacol 70:57–63
Doma M, Abhaynkar G, Reddy VD, Kavi Kishor PB (2012) Carbohydrate and elicitor enhanced withanolide (withaferin A and withanolide A) accumulation in hairy root cultures of Withania somnifera (L.). Ind J Expt Biol 50:484–490
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM et al (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245
Elsakka M, Grigorescu E, Stanescu U, Stanescu U, Dorneanu V (1990) New data referring to chemistry of Withania somnifera species. Rev Med Chir Soc Med Nat Iasi 94:385–387
Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289:295–297
Gupta P, Agarwal AV, Akhtar N, Sangwan RS, Singh SP, Trivedi PK (2013a) Cloning and characterization of 2-C-methyl-D-erythritol-4-phosphate pathway genes for isoprenoid biosynthesis from Indian ginseng, Withania somnifera. Protoplasma 250:285–295
Gupta P, Akhtar N, Tewari SK, Sangwan RS, Trivedi PK (2011) Differential expression of farnesyl diphosphate synthase gene from Withania somnifera in different chemotypes and in response to elicitors. Plant Growth Regul 65:93–100
Gupta P, Goel R, Pathak S, Srivastava A, Singh SP, Sangwan RS, Asif MH, Trivedi PK (2013b) De novo assembly, functional annotation and comparative analysis of Withania somnifera leaf and root transcriptomes to identify putative genes involved in the withanolides biosynthesis. PLoS ONE 8:doi:10.1371/journal.pone.0062714.
Hause B, Hause G, Kutter C, Miersch O, Wasternack C (2003) Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant Cell Physiol 44:643–648
Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C (2000) Tissue-specific oxylipin signature of tomato flowers—allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J 24:113–126
Jayaprakasam B, Zhang Y, Seeram NP, Nair MG (2003) Growth inhibition of human tumor cell lines by withanolides from Withania somnifera leaves. Life Sci 74:125–132
Kaileh M, Berghe WV, Heyerick A, Horion J, Piette J, Libert C, De Keukeleire D, Essawi T, Haegeman G (2007) Withaferin A strongly elicits IKK hyperphosphorylation concomitant with potent inhibition of its kinase activity. J Biol Chem 282:4253–4264
Kato N, Dubouzet E, Kokabu Y, Yoshida S, Taniguchi Y, Dubouzet JG, Yazaki K, Sato F (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol 48(1):8–18
Kaul MK, Kumar A, Ahuja A, Mir BA, Suri KA, Qazi GN (2009) Production dynamics of withaferin A in Withania somnifera (L.) Dunal complex. Nat Prod Res 23:1304–11
Kraiser T, Gras DE, Gutierrez AG, Gonzalez B, Gutierrez RA (2011) A holistic view of nitrogen acquisition in plants. J Exp Bot 62:1455–1466
Koeduka T, Fridman E, Gang DR, Vassao DG, Jackson BL, Kish CM et al (2006) Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc Natl Acad Sci U S A 103:10128–10133
Li S, Zhang P, Zhang M, Fu C, Yu L (2013) Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol 15(1):19–26
Ma D, Pu G, Lei C, Ma L, Wang H, Guo Y, Chen J, Du Z, Wang H, Li G, Ye H, Liu B (2009) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol 50(12):2146–2161
Mannan A, Liu C, Arsenault PR, Towler MJ, Vail DR, Lorence A, Weathers PJ (2010) DMSO triggers the generation of ROS leading to an increase in artemisinin and dihydroartemisinic acid in Artemisia annua shoot cultures. Plant Cell Rep 29(2):143–152. doi:10.1007/s00299-009-0807-y
Misra A, Chanotiya CS, Gupta MM, Dwivedi UN, Shasany AK (2012) Characterization of cytochrome P450 monooxygenases isolated from trichome enriched fraction of Artemisia annua L. leaf. Gene 510:193–201
Naidu PS, Singh A, Kulkarni SK (2003) Effect of Withania somnifera root extract on haloperidol induced orofacial dyskinesia: possible mechanism of action. J Med Food 6:107–114
Pal S, Singh S, Shukla AK, Gupta MM, Khanuja SPS, Shasany AK (2011) Comparative withanolide profiles, gene isolation, and differential gene expression in the leaves and roots of Withania somnifera. J Hort Sci Biotechnol 86:391–397
Pandey V, Misra P, Chaturvedi P, Mishra MK, Trivedi PK, Tuli R (2010) Agrobacterium tumefaciens-mediated transformation of Withania somnifera (L.) Dunal: an important medicinal plant. Plant Cell Rep 29:133–141
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA (2010) Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ 33:1486–501
Razdan S, Bhat WW, Rana S, Dhar N, Lattoo SK, Dhar RS, Vishwakarma RA (2013) Molecular characterization and promoter analysis of squalene epoxidase gene from Withania somnifera (L.) Dunal. Mol Bio Rep 40(2):905–916
Sangwan RS, Agarwal K, Luthra R, Thakur RS, Sangwan NS (1993) Biotransformation of arteannuic acid into arteannuin-B and artemisinin in Artemisia annua. Phytochem 34:1301–1302
Sangwan RS, Chaurasiya ND, Lal P, Misra LN, Tuli R, Sangwan NS (2008) Withanolide A is inherently de novo biosynthesized in roots of the medicinal plant ashwagandha (Withania somnifera). Physiol Plant 133:278–287
Schluttenhofer C, Pattanaik S, Patra B, Yuan L (2014) Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics 15:502. doi:10.1186/1471-2164-15-502
Singh AK, Dwivedi V, Rai A, Pal S, Reddy SGE, Rao DKV, Shasany AK, Nagegowda DA (2015) Virus induced gene silencing of Withania somnifera squalene synthase negatively regulates sterol and defence related genes resulting in reduced withanolides and biotic stress tolerance. Plant Biotechnol J. doi:10.1111/pbi.12347
Singh S, Pal S, Shanker K, Chanotiya CS, Gupta MM, Dwivedi UN, Shasany AK (2014) Sterol partitioning by HMGR and DXR for routing intermediates towards withanolide biosynthesis. Physiol Plant. doi:10.1111/ppl.12213
Sivanandhan G, Arun M, Mayavan S, Rajesh M, Jeyaraj M, Dev GK, Manickavasagam M, Selvaraj N, Ganapathi A (2012) Optimization of elicitation conditions with methyl jasmonate and salicylic acid to improve the productivity of withanolides in the adventitious root culture of Withania somnifera (L.) Dunal. Appl Biochem Biotechnol 168(3):681–96
Sivanandhan G, Kapil DG, Jeyaraj M, Rajesh M, Manickavasagam M, Selvaraj N, Ganapathi A (2013) Increased production of withanolide A, withanone and withaferin A in hairy root cultures of Withania somnifera (L.) Dunal elicited with methyl jasmonate and salicylic acid. Plant Cell Tissue Organ Cult 114:121–129
Sivanandhan G, Selvaraj N, Ganapathi A, Manickavasagam M (2014) Enhanced biosynthesis of withanolides by elicitation and precursor feeding in cell suspension culture of Withania somnifera (L.) Dunal in shake-flask culture and bioreactor. PLoS ONE 9(8):e104005. doi:10.1371/journal.pone.0104005
Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C (2003) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51:895–911
Suttipanta N, Pattanaik S, Kulshrestha M, Patra B, Singh SK, Yuan L (2011) The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol 157(4):2081–2093
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194
Xu Y-H, Wang J-W, Wang S, Wang J-Y, Chen X-Y (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol 135:507–515
Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279:407–409
Zhang Y, Ye H, Li G (2003) Effect of horseradish peroxidase on the biosynthesis of artemisinin in Artemisia annua in vitro. China J Appl Environ Biol 9:616–618
Zhao J, Davis LC, Verpoorte R (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23:283–333
Ziegler J, Stenzel I, Hause B, Maucher H, Miersch O, Hamberg M, Grimm M, Ganal M, Claus Wasternack C (2000) Molecular cloning of allene oxide cyclase: the enzyme establishing the stereochemistry of octadecanoids and jasmonates. J Biol Chem 275:19132–19138
Acknowledgments
The authors express their sincere gratitude to the Director, CIMAP for his keen interest and providing facilities for the experiments. The seed and plant material was provided by the National Gene Bank for Medicinal and Aromatic Plants. This work was supported by CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow-226015 from the Twelfth Five Year Plan project of CSIR (BSC0203). S. P. (AcSIR) was supported by CSIR-SRF.
Author contributions
S. P. and A. K. Y. have helped in the experimentation and analysis; M. M. G. has helped in withanolide and sterol experiments and analysis; R. K. V has helped in the soil and data analysis. A. K.S. and D.A.N. have helped in the analysis of the transcription factors; A.P. and S. R. have helped in raising the polyclonal antibodies for western blot experiments. A. K. S. has helped in the planning, experimentation, analysis, interpretation, and manuscript writing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Handling Editor: Peter Nick
Rights and permissions
About this article
Cite this article
Pal, S., Yadav, A.K., Singh, A.K. et al. Nitrogen treatment enhances sterols and withaferin A through transcriptional activation of jasmonate pathway, WRKY transcription factors, and biosynthesis genes in Withania somnifera (L.) Dunal. Protoplasma 254, 389–399 (2017). https://doi.org/10.1007/s00709-016-0959-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-016-0959-x