Abstract
Ocimum (Lamiaceae) is an important source of essential oils and aroma chemicals especially eugenol, methyl eugenol, linalool, methyl chavicol etc. An elite evergreen hybrid has been developed from Ocimum kilimandscharicum and Ocimum basilicum, which demonstrated adaptive behavior towards cold stress. A comparative molecular analysis has been done through RAPD, AFLP, and ISSR among O. basilicum and O. kilimandscharicum and their evergreen cold-tolerant hybrid. The RAPD and AFLP analyses demonstrated similar results, i.e., the hybrid of O. basilicum and O. kilimandscharicum shares the same cluster with O. kilimandscharicum, while O. basilicum behaves as an outgroup, whereas in ISSR analysis, the hybrid genotype grouped in the same cluster with O. basilicum. Ocimum genotypes were analyzed and compared for their trichome density. There were distinct differences on morphology, distribution, and structure between the two kinds of trichomes, i.e., glandular and non-glandular. Glandular trichomes contain essential oils, polyphenols, flavonoids, and acid polysaccharides. Hair-like trichomes, i.e., non-glandular trichomes, help in keeping the frost away from the living surface cells. O. basilicum showed less number of non-glandular trichomes on leaves compared to O. kilimandscharicum and the evergreen cold-tolerant hybrid. Trichomes were analyzed in O. kilimandscharicum, O. basilicum, and their hybrid. An increased proline content at the biochemical level represents a higher potential to survive in a stress condition like cold stress. In our analysis, the proline content is quite higher in tolerant variety O. kilimandscharicum, low in susceptible variety O. basilicum, and intermediate in the hybrid. Gene expression analysis was done in O. basilicum, O. kilimandscharicum and their hybrid for TTG1, GTL1, and STICHEL gene locus which regulates trichome development and its formation and transcription factors WRKY and MPS involved in the regulation of plant responses to freezing and cold. The analysis showed that O. kilimandscharicum and the hybrid were very close to each other but O. basilicum was more distinct in all respects. The overexpression of the WRKY coding gene showed high expression in the hybrid as compared to O. kilimandscharicum and O. basilicum and the transcription factor microspore-specific (MPS) promoter has also shown overexpression in the hybrid for its response against cold stress. The developed evergreen interspecific hybrid may thus provide a base to various industries which are dependent upon the bioactive constituents of Ocimum species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ocimum is an important member of Lamiaceae, the genus includes about 35 species of aromatic annual and perennial herbs and shrubs. Ocimum genotypes were used for culinary and medicinal purposes. Ocimum basilicum is predominantly herbaceous, having a petiolate bract (Chikkaswamy et al. 2013). Ocimum kilimandscharicum is native to East Africa and introduced and cultivated in India and some parts of Turkey. It is an evergreen aromatic perennial undershrub. It thrives as a natural rounded woody shrub that can reach approximately up to 2 m in height, in warm temperate regions of the tropics, and can be propagated both by seeds as well as vegetatively. The plant has pubescent quadrangular branches with ovate leaves which are densely covered with glandular and non-glandular trichomes originated from epidermal cells (Werker 2000). The indumentums of non-glandular trichomes and the lipophilic substances secreted by glandular trichomes (terpenes, lipids, waxes, and flavonoid aglycones) serve as a barrier against various external factors, including herbivores and pathogens, ultraviolet-B radiation, extreme temperatures, and excessive water loss (Ehleringer 1982; Harborne 1991; Wagner 1991; Werker 2000). They play a similar protective role against biotic and abiotic factors such as water deficit (Ehleringer 1982) and insect herbivores (Wagner 1991). Improving the cold tolerance and developing resistant varieties of Ocimum is very important for the growth areas of the species. In relation to adaptive behavior, O. basilicum and other species are sensitive to cold stress except O. kilimandscharicum; therefore, an evergreen interspecific hybrid was developed by hybridization through breeding efforts from O. basilicum with O. kilimandscharicum for stress tolerance. For the study of molecular polymorphism in various genotypes, the utilization of a molecular marker is the most popular and authentic approach in the present era of molecular biology (Paran et al. 1998; Kolliker et al. 2001; Chikkaswamy et al. 2013). Therefore, to establish the molecular identity and interspecific hybrid pattern at the DNA level and also to understand the genetic relationship of the developed evergreen hybrid genotype with O. basilicum and O. kilimandscharicum, RAPD, AFLP, and ISSR analyses were performed.
In the analyses, RAPD, AFLP, and ISSR markers were used to analyze and evaluate genetic variation existing among these three genotypes/species; RAPD (Willians et al. 1990), AFLP (Vos et al. 1995), and ISSR (Zietkiewicz et al. 1994) marker-based studies were utilized for quantification of genetic variability in several species. The RAPD marker has been used in the analysis of various genotypes due to rapidity and technical simplicity (Grits 1993). For genetic differentiation among Ocimum species/genotypes, 40 decamer primers (20 MAP and 20 Operon M-Series primers) were used in RAPD and 25 primer combinations were selected in AFLP analysis used after multiplexing. Selected ISSR were used for analyzing the evergreen interspecific hybrid of O. basilicum and O. kilimandscharicum. The major advantage of ISSR profiling is its reproducibility and easy handling along with low cost (Jabbarzadeh et al. 2010). The ISSR are a valuable tool for analyzing genetic variability in closely related genotypes (Christopoulos et al. 2010). A comparative analysis of trichomes was performed on abaxial and adaxial surfaces of leaves for the evergreen hybrid and parent species O. kilimandscharicum and O. basilicum. An increased proline content at the biochemical level represents a higher potential to survive in a stress condition like cold stress as reported in different plant species, therefore, proline was also estimated in all the three genotypes. Many genes and transcription factors are involved in the formation and regulation of trichome development and adaptation towards various stresses, thus involved in many physiological processes (Sanchita et al. 2015). We have selected some of the key factors and analyzed them in the parent species and cold-tolerant evergreen hybrid for their expression.
Materials and methods
The Ocimum species and evergreen interspecific hybrid of O. kilimandscharicum and O. basilicum were developed, grown and evaluated in an experimental farm of CSIR-CIMAP, Lucknow, India. The leaf samples of O. basilicum, O. kilimandscharicum and the hybrid were collected from field-grown plants. For the analysis, leaves were harvested from the first node (recently unfurled leaves), second node, and fourth node, respectively.
Trichome analysis
Young, developing and mature fresh leaves (four to five) were collected from all three Ocimum genotypes, and their trichome numbers were counted with the help of a microscope (LEICA KL 200 LED, Schott, Germany) with adjusted 1-mm2 grids at its eyepiece lens at ×40 magnification. Both the types of trichomes, glandular and non- glandular, were counted on abaxial and adaxial surfaces of leaves for O. basilicum, O. kilimandscharicum and the hybrid genotype. Leaves were analyzed at different parts, i.e., tip, middle and base and the total leaf area was also measured with a leaf scanner. Grids covering a 1-mm2 area were fixed at the eyepiece lens; therefore, only those trichomes were counted which were present inside the grid.
Proline estimation
Proline was determined with the help of protocol developed by Bates et al. (1973). Approximately 200 mg fresh leaf sample was homogenized properly in 1 ml of 3 % (w/v) aqueous sulfosalicylic acid. To the aliquots, 96 % acetic acid 3 % sulfosalicylic acid in the ratio 2:1 was added followed by ninhydrin reagent. The mixture was incubated at 96 °C in a water bath for 1 h. The mixture was cooled to room temperature, toluene was added, and the absorbance of the fraction with toluene aspired from the liquid phase was read at a wavelength of 520 nm. Proline concentration was determined using a standard curve and expressed as micromoles of proline per gram fresh weight.
DNA extraction from leaf tissues
DNA extraction in the Ocimum species and hybrid genotype was done with the help of protocol developed by Khanuja et al. (1999). The fresh leaf tissues of the Ocimum genotypes were ground into liquid nitrogen and then transferred to a tube containing extraction buffer (0.5 M CTAB 20 %,100 mM Tris-HCl, 0.5 M EDTA, 5 M NaCl, PVP 1 %, β-mercaptoethanol). During lysis, the suspension was gently mixed and incubated at 65 °C for 2 h with occasional swirling. The suspension was then cooled to room temperature and an equal volume of chloroform: isoamyl alcohol (24:1) was added for 15 min. The mixture was centrifuged at 8000 rpm for 15 min at 25 °C. The clear upper aqueous phase was transferred to a new tube and 1.5 ml 0.5 M NaCl was added and mixed properly with 0.6 volume of ice-cooled isopropanol and incubated at 30 °C for 1–2 h. The nucleic acid was collected by centrifugation at 10,000 rpm for 10 min at 25 °C. The resulting pellet was washed twice with 80 % ethanol. The pellet was air-dried under a sterile laminar hood and the nucleic acid was dissolved in nuclease-free water at room temperature and extracted with an equal volume of chloroform: isoamyl alcohol. The aqueous layer was transferred in a fresh 1.5-ml microfuge tube and 2 volumes of cold ethanol was added then spun at 10,000 rpm for 10 min at 30 °C. The pellet was washed with 80 % ethanol, dried in a vacuum and dissolved in 30 μl of nuclease-free water. DNA concentration and purity were determined by measuring the absorbance of the diluted DNA solution at 260 and 280 nm on a Nanodrop spectrophotometer ND-1000 (Thermo Fisher Scientific, Wilmington, DE, USA). The quality of the DNA was determined using agarose gel electrophoresis stained with ethidium bromide. This method provided good-quality and as well as a good quantity of DNA, which was then subjected to PCR amplification for developing random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP) and ISSR markers.
Development of molecular profiles of Ocimum species and the hybrid genotype
Development of RAPD profiles
The RAPD reaction was performed in 20 μl volume of reaction. The PCR for RAPD analysis consisted of 25 mM MgCl2, dNTPs (mix), 0.5 U Taq DNA polymerase (Bangalore Genei Pvt. Ltd., Bangalore, India), 0.025 μg/μl of each primer, and 0.030 μg/μl plant DNA and deionized water up to 18 μl. DNA amplification was performed by a Veriti thermal cycler (Applied Biosystems, USA). Reaction conditions were as follows: 1 cycle of 4 min at 94 °C, 30 s at 36 °C and 1 min at 72 °C followed by 45 cycles of 1 min at 94 °C, 30 s at 36 °C, and 1 min at 72 °C, then terminated with 4 min at 72 °C. The RAPD fragments were separated on a horizontal gel electrophoresis system of 1.2 % agarose gel by electrophoresis in TAE buffer for 2 h 30 min at 70 V. The gel was stained with ethidium bromide (0.51 × 106 μg/ml) and photographed under ultraviolet light using a UVP gel doc IT™ (Upland, CA, USA). The size of the amplicons was detected using a 1-kb DNA ladder (Fermentas, Life Sciences). The RAPD was performed with 20 MAP (designed and developed at CIMAP) and 20 OPM (procured from Operon Technologies, Alameda, CA, USA) random decamer primers and experiment was repeated to confirm the results obtained.
Development of ISSR profiles
The ISSR reaction was performed in 20 μl volume of reaction. PCR amplification reaction for ISSR analysis consisted of 25 mM MgCl2, 1.25 mM dNTPs (mix), 0.5 U Taq DNA polymerase (Bangalore Genei Pvt. Ltd., Bangalore, India), 0.025 μg/μl of each primer, and 0.030 μg/μl plant DNA and deionized water up to 18 μl. DNA amplification was performed in a Veriti thermal cycler (Applied Biosystems, USA). Reaction conditions were as follows: 1 cycle of 5 min at 94 °C, 30 s at 94 °C, followed by 40 cycles of 30 s at 94 °C, 1.5 min at 45–56 °C, and 1.5 min at 72 °C, 5 min extension at 72 °C, and then terminated with 4 °C on hold. The ISSR fragments were separated on a horizontal gel electrophoresis system of 1.2 % agarose gel by electrophoresis in TAE buffer for 3 h at 70 V. The gel was stained with ethidium bromide (0.51 × 106 μg/ml) and photographed under ultraviolet light using a UVP gel doc IT™ (Upland, CA, USA). Total 14 ISSR were used for analyzing the developed hybrid and parent species. Out of them 11 ISSR primers were selected based on their performance and the experiment was repeated to confirm the results.
Electrophoretic profiles were analyzed for polymorphism based on the presence and absence of DNA bands on the agarose gel. The bands were scored as binary coding, i.e., present (1) and absent (0), each of which was treated as an independent character regardless of its intensity. Only prominent and reproducible bands obtained for each RAPD and ISSR primer were considered and faint or unclear bands were ignored. By comparing the banding patterns of species for a primer, species-specific bands were identified. A dendrogram was constructed using the unweighted pair-group method with arithmetic average (UPGMA).
AFLP profiling of Ocimum species and the evergreen hybrid
AFLP analysis was performed using the method of Vos et al. (1995). Digestion and ligation of 1–3 μg/μl genomic DNA were carried out using 0.5 μl Eco RI (20 U/μl) and 1 μl Mse1 (20 U/μl) enzymes (Bangalore Genei Pvt. Ltd, Bangalore, India), and the reaction volume was made 11 μl by adding 10× DNA ligase buffer that includes ATP and 1 μl 0.5 M NaCl 0.5 μl, 1 mg/ml BSA 1 μl adaptor of both the enzymes, and 1 μl enzyme master mix supplied by Applied Biosystems. The adaptor specific for each restriction enzyme terminus was ligated to the digested DNA terminus by incubation at 37 °C for 12 h. Primary amplification by PCR was done using 1 μl of preselective primer pair and 15 μl AFLP core mix. These primers (Eco RI and Mse I) had no selective nucleotides. The thermo-cycler program for preselective reactions was hold of 72 °C for 2 min; 20 cycles of 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 120 s, followed by 60 °C for 30 s, and then can be stored at 4 °C. Amplification was confirmed by using agarose gel electrophoresis. The diluted primary amplification product was selectively amplified by using 0.25-μl aliquots of the selected primers specific for the frequent cutter restriction enzyme Mse1 (30 ng/μl) and rare cutter restriction enzyme Eco R1 (5 ng/μl). Selective amplification was done with 16 primers which provided 64 possible combinations after multiplexing for each genotype in which 25 primer pair combinations were found appropriate for reproducible results (Table 3). Selective amplification was done using the following touchdown PCR conditions: hold of 94 °C for 2 min and 10 cycles of 94 °C for 30 s, 66 °C for 30 s, and 72 °C for 2 min with a temperature decrease of 1 °C per cycle, followed by 20 cycles at 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 2 min, and finally 60 °C for 30 min. After selective amplification, preparation of loading buffer was done, by adding 1.0 μl Genescan-500 (ROX) size standard and 24.0 μl deionized formamide to each 1.5 μl selective amplification product and heating the tubes for 5 min at 95 °C and for the ABI Prism 310 and electrophoresis was done by Gene Mapper V3.7 at threshold 30. Amplified products were scored for the presence (1) or absence (0) of bands and binary matrices were assembled. The amplification profile obtained for three genotypes was analyzed with 25 primer pair combinations and scored 1140 fragments out of which 619 were polymorphic. The polymorphism percentage obtained by the analysis was 54.29 %. The genetic distances among the samples analyzed by AFLP were calculated by using the Jaccard similarity coefficient.
RNA isolation and cDNA synthesis
The TRIzol method was used for RNA isolation from the leaf tissues. The glassware used for RNA isolation was autoclaved and treated with 0.1 % DEPC to make it RNase free. The qualitative and quantitative estimation of the isolated RNA was determined by agarose gel (1.2 %) electrophoresis as well as by relative absorbance values at 260 and 280 nm (A260/A280) in a Nanodrop spectrophotometer. All RNA samples were stored at -80 °C. The first strand of complementary DNA (cDNA) was synthesized from 3 μg of total RNA with MultiScribe™ Reverse Transcriptase and random primer using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to user’s manual. Total RNA samples were denatured at 65 °C for 5 min and then quickly cooled on ice. Reverse transcriptase and other reaction components were added to the samples. These were then incubated for 10 min at 25 °C (primer annealing), followed by 120 min at 37 °C and finally 10 min at 85 °C to inactivate the enzyme.
Primers for qRT-PCR
For all genes, qRT-PCR primers were designed using the Primer Express 2.0 software. These included predicted annealing temperatures (Tm) of 58–60 °C, primer lengths of 18–24 nucleotides, GC contents of 50–60 %, and PCR amplicon length of 100–120 bp. All primers were customized from a commercial supplier, Integrated DNA Technology (IDT) India Ltd. Transparent testa glabra 1 (TTG1), transcription factor, STICHEL, trihelix DNA-binding protein (GTL1), microspore-specific promoter, and WRKY transcription factor genes were designed based on previously identified genes in Arabidopsis thaliana present in the SRA database of NCBI. These genes are responsible for cold tolerance and trichome development.
Quantitative real-time PCR
qRT-PCR was performed in 96-well plates using the ABI 7900HT RT-PCR detection system. Three different biological replicates for each treatment were used. Each reaction mixture consisted of 2 μl cDNA, 15 μl SYBR green mix (2X) (TaKaRa), 2 μl (5 pmol/μl) of both forward and reverse primers, and 11 μl PCR-grade water equating to a final volume of 30 μl. This reaction mix was dispensed in a 96-well PCR plate in triplicates. The thermal profile of the reaction was an initial denaturation at 95 °C for 2 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 10 s. This was followed by fluorescence acquisition after each cycle (Sanchita et al. 2015). Baseline and Ct values were automatically calculated by the Sequence Detection Systems (SDS) software version 2.2.1 (Applied Biosystems). The relative quantification of transcript abundance was done using the ΔΔCT method with default parameters. The relative quantity was calculated as RQ = 2 − ΔΔCT (Livak and Schmittgen 2001; Pfaffl 2001). A spherisorbs C18 (250 × 4.6 mm i.d.) 10 μm particle size ODS2 column (Waters, Milford, MA, USA) was used with a 40:60 (v/v) mix of acetonitrile/0.1 % (v/v) trifluoroacetic acid in water as the mobile phase. The flow rate was 1.0 ml/min, and the detector wavelength was 220 nm.
Results
Analysis of proline content in O. basilicum, O. kilimandscharicum and the hybrid for comparative behavior
The proline content was highest in the hybrid which indicates tolerant behavior which may be because of hybridization, whereas it was intermediate in tolerant variety O. kilimandscharicum and low in susceptible variety O. basilicum during the stress condition (Fig. 1).
Comparative analysis for trichome density of the cold-tolerant interspecific hybrid, O. kilimandscharicum and O. basilicum
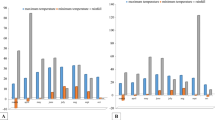
Trichome counting was done for all the three genotypes to compare the Ocimum species to the hybrid. In O. kilimandscharicum young leaves have glandular trichomes on both their surfaces and the hybrid also showed glandular trichomes on both the abaxial and adaxial surfaces of the leaves, but they are less in number than those of O. kilimandscharicum (Fig. 2). O. basilicum possess glandular trichomes approximately equal to that of the hybrid on both its surfaces, but non-glandular trichomes are completely absent on its adaxial surfaces. O. kilimandscharicum has maximum numbers of glandular and non-glandular trichomes on the abaxial as well as adaxial surfaces, but the numbers decreased as the leaves mature (Table 1). The interspecific hybrid showed the same density as O. basilicum had for non-glandular trichomes on the abaxial surface, but unlike O. basilicum, it possesses non-glandular trichomes on its adaxial surface (Fig. 2). However, it is clearly observed that the hybrid has shown both types of trichomes on the abaxial as well as adaxial surfaces as available in O. kilimandscharicum but it differs with O. basilicum in having glandular and non-glandular trichomes on the adaxial surface.
a, b Glandular and non-glandular trichomes present on the abaxial surfaces of leaves in three Ocimum species/genotypes at three developmental stages and c, d glandular and non-glandular trichomes present on the adaxial surface of leaves of three Ocimum species/genotypes at three developmental stages
Analysis of the molecular polymorphism of the cold-tolerant hybrid and parent genotypes as obtained by RAPD profiling
The yield and quantity of DNA extracted was 1451–1500 ng/μl for two to three leaves of all the three genotypes. The DNA obtained was diluted to 25 × 103 pg/μl, and quality was confirmed by analyzing OD values at 260 nm/280 nm on Nanodrop spectrophotometer ND-1000 and also viewed on agarose gel electrophoresis. In this study, 20 MAP and 20 Operon (10 bases long, decamer) primers were used for genetic analysis with all the three Ocimum genotypes. Out of 40 RAPD primers, only 33 primers responded and were screened for reproducible results. Based on the reproducibility of results from MAP primers, a total of 155 bands were obtained in which 56 bands were monomorphic, 57 were polymorphic, and 42 were unique bands (Table 2). Primers MAP 2 (size range of unique bands, 6–8 kb, approximately), MAP 3 (size range of unique bands, 0.5–1 kb, approximately), and MAP 4 (size range of unique bands, 0.5–2 kb, approximately) were found species specific for O. basilicum, the interspecific hybrid, and O. kilimandscharicum, respectively, whereas with Operon primers (M-series), a total of 84 bands were obtained in which 29 monomorphic, 34 polymorphic, and 21 unique bands were present. Primer OPM 2 is species specific for O. kilimandscharicum which produced a 1.9-kb band. All these bands were considered for the precise calculation of genetic diversity. Sufficient polymorphisms to distinguish among genotypes were achieved by DNA profiling through RAPD obtained for all the three genotypes. The hybrid (HYB) showed a common as well as differentiating band pattern with both the parents, i.e., O. kilimandscharicum (OK) and O. basilicum (OB) with primers MAP 4 and 5 at the molecular level.
A dendrogram was constructed using the unweighted pair-group method with arithmetic average (UPGMA). The dendrogram (Fig. 4a) revealed the maximum similarity between O. kilimandscharicum and the interspecific hybrid (0.601) as compared to O. basilicum (0.598). Calculation of genetic distances showed that O. kilimandscharicum and the hybrid were in close proximity compared to O. basilicum which behaves as an out-grouped member. The same type of experiment was performed earlier to get the genetic diversity among the Gymnema sylvestre genotypes by Osman et al. (2013). Willians et al. (1990) had already used RAPD markers for detecting polymorphism. The Gerbera jamesonii bolus line polymorphism was also confirmed by RAPD markers (Rusinowski and Domeradzka 2012).
Molecular analysis of the evergreen cold-tolerant interspecific hybrid and parent genotypes by ISSR
ISSR profiles were developed by utilizing 11 selected primers which produced a scorable banding pattern (Fig. 3). The selected primers ISSR 1, 2, 7, 8, 9, 10, 11, 13, 14, 15, and 16 performed better. The most informative among them was ISSR 9 which showed 66.66 % polymorphism, whereas the least polymorphic was ISSR 14. The results showed that ISSR 9 (GAG)5 and ISSR 7 (CAC)7 T may be utilized further to analyze the closely related genotypes of Ocimum. A dendrogram was constructed based on UPGMA clustering (Fig. 4b), which showed the close grouping of the evergreen hybrid genotype with O. kilimandscharicum, whereas O. basilicum was an outgroup member as resulted from RAPD and AFLP analyses.
Genetic analysis of the cold-tolerant interspecific evergreen hybrid and parent species of Ocimum by AFLP
Sixty-four primer pair combinations of Eco R1 and Mse 1 primers were used for analyzing all the three Ocimum genotypes in which 25 primer pairs were screened for analysis of genetic variability among the genotypes, with the AFLP marker generating a total 1140 bands, out of which 619 were polymorphic. Out of 25 primer combinations, the most polymorphic primer pair was CTC + AAG, being highest in providing 117 polymorphic bands out of the 162 total number of bands produced (Table 3). The polymorphism generated by AFLP analysis clearly demonstrated the distinctiveness of the cold-tolerant hybrid with other genotypes. The genetic distances among the genotypes analyzed by AFLP were calculated by using the Jaccard similarity coefficient. The degree of similarity among O. kilimandscharicum and the hybrid was 0.0339. The analysis indicated that genetic divergence found between O .basilicum and the hybrid was 0.01818. The construction of the dendrogram was done by using the unweighted pair-group method (UPGMA), and all the three genotypes were grouped into two clusters (Fig. 5). The dendrogram revealed the maximum similarity between O. kilimandscharicum and the hybrid. Calculation of genetic distances showed that O. kilimandscharicum and the hybrid were close to each other and O. basilicum behaved as an out group as obtained also by RAPD analysis. Osman et al. (2011) analyzed seed-raised progeny of Gymnema sylvestre for genetic diversity analysis through AFLP.
Gene expression analysis from leaf tissues of O. basilicum, O. kilimandscharicum, and their hybrid
The expression profiling of key genes of different categories has been performed using leaf tissues of O. basilicum, O. kilimandscharicum, and their hybrid. All the genes considered in this study were transparent testa glabra 1 (TTG1), transcription factor, STICHEL, and trihelix DNA-binding protein (GTL1), genes responsible for trichome development and its formation, which showed 6, 8.2, and 7 times increased expression, respectively, in O. kilimandscharicum, whereas 0.5, 1.5, and 0.1, respectively, in O. basilicum and 1, 7.5, and 0.2 up-regulated expression, respectively, in comparison to their hybrid (Sanchita et al. 2015). Microspore-specific promoter 2, a cold responsible gene, has shown 2.2, 7.5, and 0 times increased expression, respectively, in O. kilimandscharicum, hybrid and O. basilicum genotypes and in the case of the WRKY transcription factor gene, shows down-regulation with factor −0.5, −2, and 0 times overexpressions, respectively. (Fig. 6).
Discussion
The meticulous study indicated that the RAPD, AFLP and ISSR analyses of Ocimum genotypes yielded similar results, although difficulties are associated with the reproducibility of RAPD, which limits the applicability of this technique (Perez et al. 1998). RAPD may nevertheless be useful for preliminary assessments of intraspecific, interspecific, and intergeneric variations (Garcia et al. 2004; Dhawan et al. 2011). In contrast, the greater accuracy and information content of AFLP make this technique more useful for detailed investigation (Kolliker et al. 2001). ISSR is an unequivocal identification method which provides the multilocus pattern which is reproducible and polymorphic in plant genomes (Bornet and Branchard 2004). Trichome density also gave similar results as obtained by molecular markers, i.e., O. kilimandscharicum and the hybrid are very close to each other but O. basilicum was more distinct in all respects, either the trichome count or molecular level. Genetic diversity was studied among Ocimum species based on ISSR, RAPD, and SRAP (Chen et al. 2013), and they found that the SRAP dendrogram was correlated most with the combined dataset.
Notwithstanding the fact that in this study the developed evergreen Ocimum elite interspecific hybrid showed a distinct identity as well as three markers system analyze the genetic relatedness in similar way and they were correlated with trichome density in the same pattern. Ocimum species vary greatly in their ability to tolerate cold temperatures, only O. kilimandscharicum can tolerate low temperatures in winters. The ability of this species to withstand low temperatures may involve the complex mechanisms that contribute to cold tolerance as described for other species including the accumulation of low molecular weight cryo-protective molecules and alterations in membrane lipid composition (Nanjo et al. 1999; Taji et al. 2002). Cold-tolerant behavior showed by the interspecific hybrid may be possible due to the transfer of few genes related with cold tolerance from the parent plant. A number of genes have been reported to be induced by low temperature stress (Shinozaki and Yamaguchi-Shinozaki 2000). The analysis of proline was indicative of tolerance in O. kilimandscharicum and the hybrid compared to susceptible species O. basilicum, which is further validating the adaptive behavior of the developed hybrid.
The result demonstrated that RAPD, AFLP, and ISSR markers provide effective tools for the detection and evaluation of genetic variation existing among O. kilimandscharicum, O. basilicum and their interspecific hybrid. Markers determined that O. kilimandscharicum and the hybrid were close and are being utilized for integration of useful characters. A significant aspect of the study was that the RAPD analysis has achieved to discriminate Ocimum species by using only 19 MAP primers and 14 OPM primers. Trichome count also differentiated these three genotypes according to the RAPD, AFLP and ISSR profiles. This study clearly establishes the hybrid pattern of cold-tolerant genotypes and also showed the divergence and similarity from both the parents (Fig. 5), which was also clearly discerned by analyzing trichomes. This study may provide a basic understanding of the genetic makeup of the evergreen hybrid genotype developed from both the parents. In this analysis, young, developing and adult leaves are three developmental stages in which the hybrid showed both types of trichomes on the abaxial and adaxial surfaces similar to O. kilimandscharicum, but it differs from O. basilicum in having glandular and non-glandular trichomes on the adaxial surface. The developed evergreen interspecific hybrid of O. kilimandscharicum and O. basilicum demonstrated adaptive behavior towards cold stress.
The transparent testa glabra 1 (TTG1) locus regulates several trichome developmental and biochemical pathways in Arabidopsis, including the formation of hairs on leaves, stems and roots (Walker et al. 1999). Trihelix DNA-binding protein (GTL1) is reported to be involved in various processes of leaf trichomes in Arabidopsis thaliana developed through several distinct cellular processes, such as patterning, differentiation, and growth (Breuer et al. 2009) and the STICHEL (STI) gene, which plays an important role in the regulation of the branch number of the unicellular trichomes in Arabidopsis. O. kilimandscharicum and the hybrid are very close to each other, but O. basilicum was more distinct in all respects and plays an important role in trichome development. Therefore, the TTG1, GTL1, and STI genes were showing up expression in response against several trichome development and its formation. WRKY transcription factors are key regulators in plant responses to abiotic and biotic stresses. These genes also regulate plant responses to freezing and cold (Huang and Duman 2002). It has been reported to show its role in cold tolerance in A. thaliana (Zou et al. 2010). In our study, WRKY was found to be down-regulated and the WRKY coding gene showed high expression in the hybrid as compared to O. kilimandscharicum and O. basilicum. Another transcription factor microspore-specific (MPS) promoter has also shown a high expression in the hybrid for its response against cold stress (Sanchita et al. 2013). Therefore, WRKY and MPS genes responded against cold stress. This stress tolerance of the hybrid may involve changes at the whole-plant, tissue, cellular, physiological, and molecular levels. This distinctiveness and combination of intrinsic changes define the capacity of the hybrid genotype to sustain under unfavorable environmental conditions which may involve many changes at the physiological and biochemical levels. In response to cold, many genes may be regulated differentially and their gene products function in providing stress tolerance to plants (Sanchita et al 2013). Therefore, in this way, the traditional methods were being complemented by molecular techniques for enabling and supporting breeding programs. The developed evergreen hybrid could become a boon to various industries which are dependent upon the bioactive constituents of Ocimum species.
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- RAPD:
-
Random amplified polymorphic DNA
- GT:
-
Glandular trichomes
- NGT:
-
Non-glandular trichomes
- ISSR:
-
Interspecific simple sequence repeats
References
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39(1):205–207
Bornet B, Branchard M (2004) Use of ISSR fingerprints to detect microsatellites and genetic diversity in several related Brassica taxa and Arabidopsis thaliana. Hereditas 140:245–247
Breuer C, Kawamura A, Ichikawa T, Tominaga-Wada R, Wada T, Kondou Y, Muto S, Matsui M, Sugimoto K (2009) The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant Cell 21(8):2307–2322
Chen YS, Dai TX, Chang YT, Wang SS, Ou SL, Chuang WL, Cheng CY, Lin HY, Lin LY, Ku HM (2013) Genetic diversity among Ocimum species based on ISSR, RAPD and SRAP markers. AJCS 7:1463–1471
Chikkaswamy BK, Paramanik RC, Varadaraj N, Paramanik A, Ramesh HL, Shivashankar M, Sivaram VR (2013) Determination of genetic variation in Piper species using 4C nuclear DNA and RAPD marker. Int J Res Pharm Sci 4:58–64
Christopoulos MV, Rouskas D, Tsantili E, Bebeli PJ (2010) Germplasm diversity and genetic relationships among walnut (Juglans regia L.) cultivars and Greek local selections revealed by inter-simple sequence repeat (ISSR) markers. Sci Hortic 125:584–592
Dhawan SS, Rai GK, Darokar MP, Lal RK, Mishra HO, Khanuja SPS (2011) Comparative genetic analysis of trichome less and normal pod genotype of Mucuna pruriens. Genet Mol Res 10(3):2049–2056
Ehleringer J (1982) The influence of water stress and temperature on leaf pubescence development in Encelia farinoza. Am J Bot 69:670–675
Garcia AAF, Benchimol LL, Barbosa AMM, Geraldi IO, Souza CL Jr, Souza AP (2004) Comparison of RAPD, RFLP, AFLP SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Grits P (1993) The use of molecular and biochemical markers in crop evolution studies. In: Hecth MK (ed) Evolutionary biology, vol 27. Plenum Press, New York, pp 51–54
Harborne JB (1991) Flavonoids pigments. In: Rosenthal GA, Berenbaum MR (eds) Herbivores. Their interactions with secondary plant metabolites. Academic, San Diego, pp 389–429
Huang T, Duman JG (2002) Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Mol Biol 48:339–350
Jabbarzadeh Z, Khosh-khui M, Salehi H, Saberivand A (2010) Inter simple sequence repeat (ISSR) markers as reproducible and specific tools for genetic diversity analysis of rose species. Afr J Biotechnol 37:6091–6095
Khanuja SPS, Shasany AK, Darokar MP, Kumar S (1999) Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Mol Biol Report 17:1–7
Kolliker R, Jones ES, Jahufer MZZ, Forster JW (2001) Bulked AFLP analysis for assessment of genetic diversity in white clover (Tritilium repens L.). Euphytica 121:305–315
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Nanjo, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett 461:205–210
Osman MA, Dhawan SS, Bahl JR, Darokar MP, Khanuja SPS (2011) AFLP marker and polymorphism among progenies of Gymnema sylvestre—an important medicinal plant of India. Nat Prod Commun 6:1679–1682
Osman MA, Dhawan SS, Bahl JR, Darokar MP (2013) Genetic diversity analysis in Gymnema sylvestre R. Br. by RAPD. Int J Int Sci Inn Tech 2:50–54
Paran L, Afergoot E, Shifriss C (1998) Variation in Capsicum annum revealed by RAPD and AFLP markers. Euphytica 99:167–173
Perez T, Albornoz J, Dominguez A (1998) An evolution of RAPD fragments reproducibility and nature. Mol Ecol I7:1347–1358
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29(9):1–6
Rusinowski Z and Domeradzka O (2012) Evaluation of the utility of the random amplified polymorphic DNA method and of the semi-specific PCR to assess the genetic diversification of the Gerbera jamesonii bolus line. Sci World J. 2012:1–5. doi:10.1100/2012/450920
Sanchita, Dhawan SS, Sharma A (2013) Analysis of differentially expressed genes in abiotic stress response and their role in signal transduction pathways. Protoplasma. doi:10.1007/s00709-013-0528-5
Sanchita, Singh R, Mishra A, Dhawan SS (2015) Physiological performance, secondary metabolite and expression profiling of genes associated with drought tolerance in Withania somnifera. Protoplasma. doi:10.1007/s00709-015-0771-z
Shinozaki K, Yamaguchi- Shinozaki Y (2000) Molecular responses to dehydration and low temperature: difference and cross talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417–426
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wagner GJ (1991) Secreting glandular trichomes: more than just hairs. Plant Physiol 96:675–679
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11:1337–1350
Werker E (2000) Trichome diversity and development. Adv Bot Res 31:1–35
Willians JGK, Kuelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Zou C, Jiang W, Yu D (2010) Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot 61(14):3901–3914
Acknowledgments
The authors are thankful to the Director, CSIR-CIMAP, for the support and encouragement. This study was supported by CSIR-CIMAP Supra-Institutional Project CHEMBIO-BSC203.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Peter Nick
Rights and permissions
About this article
Cite this article
Dhawan, S.S., Shukla, P., Gupta, P. et al. A cold-tolerant evergreen interspecific hybrid of Ocimum kilimandscharicum and Ocimum basilicum: analyzing trichomes and molecular variations. Protoplasma 253, 845–855 (2016). https://doi.org/10.1007/s00709-015-0847-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-015-0847-9