Abstract
A survey was performed on a Vitis cultivar collection in Stellenbosch, South Africa. Metaviromes were generated for each cultivar, using an RNAtag-seq workflow. Analysis of assembled contigs indicated the presence of two putatively novel members of the genus Vitivirus, provisionally named "grapevine virus N" (GVN) and "grapevine virus O" (GVO). Comparisons of amino acid sequences showed that GVN and GVO are most closely related to grapevine virus G and grapevine virus E, respectively. The incidence of these novel viruses within the sampling site was low, with GVO and GVN associated with only five and two cultivars, respectively, of the 229 sampled.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Vitis vinifera L. (grapevine) is a significant crop in the South African agricultural sector, with the country being in the top ten for both wine and table grape production [1]. Vegetative propagation and global trade of cultivars has led to grapevine being disproportionately affected by more viruses than any other crop [2]. The exponential increase in the use of metaviromics has accelerated the discovery of grapevine pathogens, with the number of currently known viruses approaching 90 [3]. South Africa has a long history of viticulture and, consequently, a diverse set of viruses circulating within the industry.
Members of the genus Vitivirus (family Betaflexiviridae) typically have genomes of approximately 7,500 nt with five open reading frames coding for a replication-associated protein (RAP), a 22-kDa protein associated with vector transmission, a movement protein (MP), a coat protein (CP), and a nucleic acid binding protein (NABP) that functions as a suppressor of RNA silencing [4]. Grapevine vitiviruses are common components of viral populations in South African vineyards [5], with grapevine virus A (GVA), grapevine virus B, and grapevine virus E (GVE) having been reported previously [6,7,8]. In general, single infections with grapevine vitiviruses are associated with very mild symptoms [9], but synergistic coinfections with other viruses result in several severe and economically important disease phenotypes [10]. GVA and GVB are part of a complex of viruses associated with rugose wood [11] and in South Africa are associated with Shiraz decline and corky bark disease, respectively [7, 12]. In recent years, the application of metaviromics approaches has led to the discovery of a number of new grapevine vitiviruses, including grapevine virus F [11], grapevine virus G (GVG) [13], grapevine virus H [14], and grapevine virus L [15].
In December 2019, a total of 229 samples were collected from the Vitis cultivar collection at the Nietvoorbij Campus of the Agricultural Research Council (ARC), as described previously [16] (GPS coordinates: -33.912053, 18.862291). Total RNA was isolated according to White et al. [17], and RNAtag-seq libraries were prepared as described [18] and sequenced using an Illumina HiSeq 2500 instrument (Illumina, San Diego, CA, USA; ARC - Biotechnology Platform, Onderstepoort, Pretoria, South Africa) as paired ends (2 × 125 nt) using TruSeq V4 chemistry (Illumina, San Diego, CA, USA).
Datasets were demultiplexed using Je software [19]. Trimming was performed with CLC Genomics Workbench 9 (QIAGEN Bioinformatics, Aarhus, Denmark), using the following parameters: minimum read length, 20 nt; quality limit, 0.05; adapter trimming with Illumina universal (5’-AGATCGGAAGAG-3’) and RNAtag-seq (5’-TACACGACGCTCTTCCGATCTNNNNNNNNT-3’) adapters. Assembly of trimmed reads into contigs was done using SPAdes 3.14.0 [20] with the meta option. The cultivars and datasets associated with this study are listed in Supplementary Table S1. These datasets are associated with NCBI BioProject PRJNA763365. Contigs of putative viral origin were identified using BLASTn [21] and the viral fraction of the NCBI Refseq database. Plant virus contigs were then submitted to the browser-based version of BLASTx, using the NCBI nr database, for further confirmation of their identity.
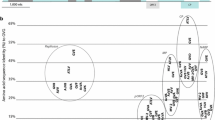
A novel vitivirus, provisionally named "grapevine virus N" (GVN), was associated with the V. vinifera cultivars Azal tinto, Bourboulenc, Cinsault noir, Crouchen blanc, and Palamino, while the provisionally named "grapevine virus O" (GVO) was associated with Roter Zierfandler and Royal Molenberg. The genome organization of GVN and GVO and associated amino acid sequences of each gene was determined using ORF Finder [22], showing the typical arrangement of members of the genus Vitivirus [11]. The average amino acid sequence identity (AAI) between each gene product and that of the closest relative was determined using the AAI calculator from the enveomics collection [23] and is shown in Table 1. The amino acid sequences of ORF1 (RAP) of both GVN and GVO were aligned with cognate NCBI RefSeq sequences of extant vitiviruses, using BioEdit 7.2.5 [24]. A best-fit maximum-likelihood phylogeny, based on the Le Gascuel model + G + I + F, was generated using MEGA X [25] and is shown in Figure 1.
Maximum-likelihood phylogeny based on the amino acid sequences derived from ORF 1 (replicase-associated proteins; RAP) of grapevine virus N and grapevine virus O from this study (indicated by solid circles) and references derived from other extant vitiviruses. The cognate RAP sequence from potato virus T was used as an outgroup. The phylogeny represents the tree with the highest log likelihood and was generated in MEGA X using the Le Gascuel model with frequencies, invariant sites and gamma distribution (n = 5). Bootstrapping was applied (1000 replicates), and the percentage of trees in which the associated taxa clustered together is shown next to the branches. Bootstrap percentages lower than 50 are not shown.
Portions of the GVN and GVO CP genes from each sample were amplified using RT-PCR. Primers with the following sequences were used to set up two-step RT-PCR reactions: GVN-CP-F (5’-TCGCTGAGATAATAAGGAGGATTGAG-3); GVN-CP-R (5’-GACTTGAATCACACTGGCTTCAGA-3’); GVO-CP-F (5’-GGTGTGATAGAGGAT AACCACAGT-3’); and GVO-CP-R (5’-TACACTCTAAACGACCACAACAGT-3’). Two-step RT-PCR reactions were carried using Promega GoScriptTM Reverse Transcriptase and GoTaq® Taq Polymerase (Promega, Madison, WI, USA) according to the manufacturer's instructions. Amplicons of the expected size were visualized on an agarose gel (572 and 789 bp for GVO and GVN, respectively), and their identities were confirmed using bidirectional Sanger sequencing (Inqaba Biotechnical Industries, Pretoria, South Africa).
The 5’-terminal nucleotides of GVN and GVO were confirmed using the 5' RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s specifications. Total RNA from Palamino and Roter Zierflander was used as the input for the GVN and GVO 5’ RACE reactions, respectively. The following gene-specific primers were used: For GVN, GVN-GSP1 (5’- CACTATATCTTAACTCATCT-3’) and GVN-GSP2 (5’- CCTCTACAATATGACTAGATATGCT-3’). For GVO, GVO-GSP1 (5’- AGGGTCGTCTTATCTTCATC-3’) and GVO-GSP2 (5’-TCCCTATACCTTAGGTTATC CTTAGC-3’). 3’ RACE amplification was not performed, as all of the GVN and GVO contigs contained a poly-A tract between 48 and 52 nt in length (data not shown). Confirmation of the 5’ ends showed that the GVN and GVO genomes are 7,486 and 7,560 nt in length, respectively, excluding the poly-A tract.
Considering the RAP phylogeny, GVN and GVO are most closely related to GVG and GVE, respectively, which was also confirmed by the AAI values obtained for the various genes of these viruses. Phylogenetically, GVN and GVO both clustered within the recently described GVE superclade [26]. The AAI values indicate that GVN and GVO are novel members of the family Betaflexiviridae, for which the species demarcation limit is 80% AAI for either the RAP or CP gene [27]. However, some additional criteria may need to be implemented in order to determine whether GVN and GVO are representatives of two new species or distinct members of already established taxa. GVN and GVO showed limited incidence and genetic diversity within the virus population of the vineyard under study, which could suggest a recent introduction. However, more widespread surveys of South Africa’s viticultural regions are needed in order to get a complete view of their distribution nationwide. Finally, the possible role of GVO and GVN in grapevine disease requires further investigation, as both viruses were found in coinfections with other viruses, including grapevine leafroll-associated virus 3, grapevine leafroll-associated virus 2, and at least two additional grapevine vitiviruses (data not shown). Given the potential for grapevine vitiviruses to be associated with synergistic coinfections [9], GVN and GVO should be considered potentially harmful pathogens for South African viticulture.
References
Gbejewoh O, Keesstra S, Blancquaert E (2021) The 3Ps (Profit, Planet, and People) of Sustainability amidst Climate Change: A South African Grape and Wine Perspective. Sustainability 13:2910
Martelli GP (2018) Where grapevine virology is heading to. In: Proceedings of the 19th congress of the International Council for the Study of virus and virus-like diseases of the grapevine (ICVG), Santiago, Chile, 9-12 April 2018
Fuchs M (2020) Grapevine viruses: a multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard. J Plant Pathol 102:643–653
Galiakparov N, Tanne E, Sela I, Gafny R (2003) Functional analysis of the grapevine virus a genome. Virology 306:42–50
du Preez J, Stephan D, Mawassi M, Burger JT (2011) The grapevine-infecting vitiviruses, with particular reference to grapevine virus A. Arch Virol 156:1495–1503
Engelbrecht DJ, Kasdorf GGF (1987) Occurrence and transmission of grapevine virus A in South African grapevines. South African J Enol Vitic 8(1):23–29
Goszczynski DE (2010) Divergent molecular variants of Grapevine virus B (GVB) from corky bark (CB)-affected and CB-negative LN33 hybrid grapevines. Virus Genes 41:273–281
Goszczynski DE, Jooste AEC (2003) Identification of divergent variants of Grapevine virus A. Eur J Plant Pathol 109:397–403
Rowhani A, Daubert S, Arnold K, Al Rwahnih M, Golino KV, D, Uyemoto JK, (2018) Synergy between grapevine vitiviruses and grapevine leafroll viruses. Eur J Plant Pathol 151:919–925
Rowhani A, Uyemoto JK, Golino D, Daubert SD, Al Rwahnih M (2016) Viruses involved in graft- incompatibility and decline. In Meng B, Fuchs M, Martelli G Golino D (eds) Grapevine viruses: Molecular biology, diagnostics, and management, Chapter 12. Springer, New York, pp 289–302
Al Rwahnih M, Sudarshana MR, Uyemoto JK, Rowhani A (2012) Complete genome sequence of a novel vitivirus isolated from grapevine. J Virol 86(17):9545
Goszczynski DE, du Preez J, Burger JT (2008) Molecular divergence of Grapevine virus A (GVA) variants associated with Shiraz disease in South Africa. Virus Res 138:105–110
Blouin AG, Keenan S, Napier KR, Barrero RA, MacDiarmid RM (2018) Identification of a novel vitivirus from grapevines in New Zealand. Arch Virol 163:281–284
Candresse T, Theil S, Faure C, Marais A (2018) Determination of the complete genomic sequence of grapevine virus H, a novel vitivirus infecting grapevine. Arch Virol 163:277–280
Debat HJ, Zavallo D, Brisbane RS, Voncina D, Almeida RPP, Blouin AG, Al Rwahnih M, Gomez TG, Asurmendi S (2019) Grapevine virus L: A novel vitivirus in grapevine. Eur J Plant Pathol 155:319–328
Read DA, Thompson GD, Swanevelder D, Pietersen G (2021) Detection and diversity of grapevine virus L from a Vitis cultivar collection in Stellenbosch, South Africa. Eur J Plant Pathol https://doi.org/10.1007/s10658-021-02380-y
White EJ, Venter M, Hiten NF, Burger JT (2008) Modified cetyltrimethylammonium bromide method improves robustness and versatility: the benchmark for plant RNA extraction. Biotechnol J 3:1424–1428
Shishkin AA, Giannoukos G, Kucukural A et al (2015) Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Meth 12:323–325
Girardot C, Scholtalbers J, Sauer S et al (2016) Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinformatics 17(1):419
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27(5):824–834
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, Wagner L (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res 31:28–33
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr 4:e1900v1
Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Stecher G, Tamura K, Kumar S (2020) Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol 37:1237–1239
Maree HJ, Blouin AG, Diaz-Lara A, Mostert I, Al Rwahnih M, Candresse T (2020) Status of the current vitivirus taxonomy. Arch Virol 165:451–458
Adams MJ, Antoniw JF, Bar-Joseph M, Brunt AA, Candresse T, Foster GD, Martelli GP, Milne RG, Zavriev SK, Fauquet CM (2004) The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch Virol 149:1045–1060
Funding
David Read is grateful for the financial support provided by the National Research Foundation of South Africa under grant UID 104901.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Data availability
The data that support the findings of this study are openly available in NCBI public databases.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Read, D.A., Thompson, G.D., Cordeur, N.L. et al. Genomic characterization of grapevine viruses N and O: novel vitiviruses from South Africa. Arch Virol 167, 611–614 (2022). https://doi.org/10.1007/s00705-021-05333-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05333-2