Abstract
Five epidemic waves of human infection with influenza A (H7N9) virus have emerged in China since spring 2013. We previously described the epidemiological characterization of the fifth wave in Jiangsu province. In this study, 41 H7N9 viruses from patients and live-poultry markets were isolated and sequenced to further elucidate the genetic features of viruses of the fifth wave in Jiangsu province. Phylogenetic analysis revealed substantial genetic diversity in the internal genes, and 18 genotypes were identified from the 41 H7N9 virus strains. Furthermore, our data revealed that 41 isolates from Jiangsu contained the G186V and Q226L/I mutations in their haemagglutinin (HA) protein, which may increase the ability of these viruses to bind the human receptor. Four basic amino acid insertions were not observed in the HA cleavage sites of 167 H7N9 viruses from Jiangsu, which revealed that highly pathogenic avian influenza (HPAI) H7N9 viruses did not spread to Jiangsu province in the fifth wave. These findings revealed that multiple genotypes of H7N9 viruses co-circulated in the fifth wave in Jiangsu province, which indicated that the viruses have undergone ongoing evolution with genetic mutation and reassortment. Our study highlights the need to constantly monitor the evolution of H7N9 viruses and reinforce systematic influenza surveillance of humans, birds, and pigs in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human infections with avian influenza viruses frequently have occurred since 18 individuals were first infected with highly pathogenic avian influenza (HPAI) H5N1 viruses in Hong Kong in 1997. Several subtypes of avian influenza viruses have since been reported to infect humans, including H5, H9, H10, H6 and H7 [1,2,3,4,5,6]. In spring 2013, a novel reassortant H7N9 avian influenza virus causing severe respiratory disease in humans emerged in the Yangtze River Delta region of China [3, 7, 8]. Since then, five epidemic waves of human infection with H7N9 viruses have been observed [9, 10]. As of 26 October 2017, worldwide, 1564 confirmed cases of human infection with H7N9 viruses have been reported to the World Health Organization (WHO), with at least 607 deaths. In the fifth wave (1 October 2016-30 September 2017), the cumulative number of human infection cases was 764, which is markedly higher than in each of the previous four waves (135, 320, 226 and 119, respectively) [10].
Jiangsu province, located in the Yangtze River Delta of China, had the highest cumulative numbers of reported human infection (n = 150) with H7N9 viruses in the fifth wave. We previously described the epidemiological characterization of the current fifth wave in Jiangsu province [11]. In this study, 41 H7N9 viruses from patients and live-poultry markets were isolated and sequenced to further investigate the genetic features of viruses of the fifth wave in Jiangsu province. Compared with the haemagglutinin and neuraminidase genes, the internal genes of the 41 H7N9 viruses showed a higher degree of diversity. Based on phylogenetic analysis, at least 18 genotypes were identified. The co-existence of multiple genotypes of H7N9 viruses indicated that the viruses have undergone ongoing evolution with genetic mutation and reassortment in the fifth wave in Jiangsu province.
Materials and methods
Virus surveillance and isolation
Since 2013, surveillance for influenza A (H7N9) viruses has been conducted in human and live-poultry markets (LPMs) in Jiangsu province. Samples collected from patients with suspected infection and LPMs were tested for H7N9, virus using real-time PCR, by local municipal centers for disease control and prevention (CDC). All H7N9-positive samples were submitted to Jiangsu CDC for virus isolation. Two hundred µl of each original specimen was inoculated allantoically into 9- to 11-day-old specific-pathogen-free (SPF) embryonated chicken eggs for 48 to 72 hours at 37 °C in a biosafety level 3 (BSL-3) facility (BSL-3 Lab of Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China).
Genome sequencing
Viral RNA extraction was performed using an RNeasy Plus Mini Kit (QIAGEN, Germany). The primer Uni12 (5’-AGCGAAAGCAGG-3’) was used for reverse transcription [12]. PCR was performed with a set of 28 primer pairs specific for H7N9 influenza virus. All primer sequences are available upon request. PCR products were purified using a QIAamp Gel Extraction Kit (QIAGEN) and sequenced using an ABI 3735 DNA Analyzer (Applied Biosystems, USA) using an ABI BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA).
Sequence alignment and phylogenetic analysis
The incoming sequences were compiled using the Lasergene sequence analysis software package (DNAStar, Madison, WI, USA). Nucleotide BLASTn analysis (http://www.ncbi.nlm.nih.gov/BLAST) was used to identify related reference viruses. Whole genome sequences of 54 H7N9 viruses isolated from 2013 to 2017 were included for phylogenetic analysis, all of which were obtained from the GenBank and Global Initiative on Sharing All Influenza Data (GISAID) databases. Among them, four viruses, A/Shanghai/2/2013, A/Anhui/1/2013, A/Hong Kong/125/2017 (A/Hunan/02650/2016-like virus) and A/Guangdong/17SF003/2016) were H7N9 candidate vaccine viruses (CVVs) proposed by WHO. Pairwise sequence alignments were also performed with the MegAlign program (DNASTAR) to investigate nucleotide and amino acid sequence similarities. Phylogenetic analysis of the aligned sequences for eight genomic segments was performed by the maximum composite likelihood method using MEGA6 software [13]. The reliability of the unrooted neighbor-joining tree was assessed by bootstrap analysis with 1,000 replications; only bootstrap values ≥ 70% are shown. Horizontal distances are proportional to genetic distance. Alignments of each influenza virus sequence were created using the program ClustalX 1.83.
Based on the bootstrap value and branch length, each gene segment was classified into different clades. The genotypes of the isolates were identified based on the combination of clades to which their eight gene segments belonged.
Results
Virus isolation
Between October 2016 and March 2017, a total of 152 H7N9 viruses were isolated from human samples (n = 103) or environmental samples (n = 34) in a BSL-3 facility. Eight gene segments of 41 H7N9 viruses (human, n = 35, environment, n = 6) isolated from December 2016 to February 2017 were sequenced. All 35 human cases were admitted to hospital, and 16 (46%) died. Of the 35 cases, male patients accounted for 74% (n = 26), and the median age was 56 years (23-89).
Phylogenetic analysis
Pairwise alignment of all 41 H7N9 isolates showed that the lowest nucleotide sequence identity in the eight genes ranged from 93.5% to 98.9% (PB2, 97.7%; PB1, 94.7%; PA, 95.2%; HA, 95.9%; NP, 93.5%; NA, 98.8%; M, 96.3%; NS, 97.2%). Among the four CVVs, the HA segments of 41 viruses shared the highest nucleotide sequence similarity with A/Hunan/02650/2016 (97.4%-98.7% identity). For the HA genes, A/Xuzhou/550/ 2017 shared low nucleotide sequence similarity with the other 40 viruses (95.9%-96.5% identity), while the other 40 viruses shared 97.9%-99.2% nucleotide sequence identity. These findings showed that all of the H7N9 viruses were very similar, although their genes showed different levels of diversity (Table 1).
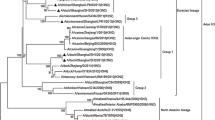
The Yangtze River Delta region (including Jiangsu province, the city of Shanghai and Zhejiang province) has been identified as the original source of H7N9 viruses, and since the second H7N9 epidemic wave, the Pearl River Delta region (mainly Guangdong province) has been identified as an additional H7N9 outbreak source [14]. In the fifth wave, HA genes of H7N9 viruses formed two distinct lineages: the Yangtze River Delta lineage and the Pearl River Delta lineage [9]. In this study, HA genes of all 41 H7N9 isolates from Jiangsu province belonged to the former (Fig. 1). In the Yangtze River Delta lineage of the HA tree, 40 out of 41 viruses from Jiangsu province clustered in one clade (clade 1.1). However, A/Xuzhou/550/ 2017 formed an independent clade (clade 1.4). The NA tree exhibited a topology similar to that of the HA tree, with viruses also forming the Yangtze River Delta lineage and Pearl River Delta lineage, and 41 H7N9 isolates from Jiangsu clustered in clade 1.1 (Fig. 1). Interestingly, A/Xuzhou/550/2017 was located at the root of clade 1.1. Together with the low sequence identities from pairwise alignment, we could deduce that A/Xuzhou/550/ 2017 was separated from the other 40 viruses. Six internal genes of 41 H7N9 viruses could be classified into more than one clade, with one major clade and at least one minor clade (Fig. 1). In all 41 H7N9 viruses, the PB2 genes clustered in four clades, and the PB1, PA and M genes were grouped into three clades. The NP and NS genes were divided into two clades. These results illustrate the genetic diversity of the H7N9 viruses circulating during this outbreak in Jiangsu, and this mainly resulted from the high level of genetic heterogeneity of the internal genes of the H7N9 viruses.
Phylogenetic trees for the HA, NA, NP, M, NS, PB2, PB1, and PA gene segments of 41 H7N9 and related reference viruses. The unrooted neighbor-joining phylogenetic trees were generated by the maximum composite likelihood model in MEGA 6 software. The reliability of the tree was assessed by bootstrap analysis with 1,000 replications. Bootstrap values are shown for selected nodes (only for those with a frequency greater than 70%). Horizontal distances are proportional to genetic distance. The 41 H7N9 viruses from Jiangsu in 2016-2017 are indicated by a black circle (●), and the four H7N9 candidate vaccine viruses are indicated by a square (■)
Genotype identification
Based on phylogenetic analysis of the eight genes, we classified the 41 H7N9 viruses into 18 genotypes, of which G2 (n = 7) and G8 (n = 5) were the most frequent (Table 2). Except for G4 and G9, all of the genotypes were detected in H7N9 isolates from humans. Geographically, 13 genotypes co-circulated in the southern part of Jiangsu Province (including Suzhou, Wuxi, Changzhou and Nanjing city). In contrast, only five genotypes co-circulated in the northern part of Jiangsu (including Taizhou, Yancheng, Huaian and Xuzhou city). The co-existence of multiple genotypes of H7N9 viruses in Jiangsu province indicated that the viruses have undergone ongoing evolution with genetic mutation and reassortment in the fifth wave.
Molecular characterization
Molecular markers associated with host adaptation, virulence, and drug resistance were analyzed (Table 3). HA proteins of all 41 viruses contained the mutations G186V and Q226L/I (H3 numbering), which suggested that they may have dual receptor affinity for both human-type 2,6-linked sialic acid and avian-type 2,6-linked sialic acid receptors [15,16,17]. Multiple basic amino acids were not observed in the HA cleavage sites of any of the 41 viruses, which is a molecular feature of low-pathogenic avian influenza (LPAI) viruses. More than half of human-origin H7N9 viruses (n = 18) possessed the PB2-E27K substitution, which may increase viral replication and pathogenicity in mammalian hosts [18, 19]. Some mutations in the NA protein, such as E119V, R152K, I222K/R, and R292K, can confer resistance to neuraminidase inhibitors (NAIs) [20, 21]. In this study, the isolate A/Suzhou/88/2016 possessed the NA-R292K mutation, implying that it has lost sensitivity to NAIs. All 41 isolates had a five-amino-acid deletion in the NA stalk, which may be associated with adaptation to territory-based poultry. The substitution S31N in the M2 protein suggests resistance to adamantine antiviral drugs.
Discussion
Jiangsu province was one of the most strongly impacted regions in the fifth wave of human infections with H7N9, and most of the infections in Jiangsu occurred from December 2016 to February 2017 (n = 124). In this study, phylogenetic analysis of 41 H7N9 viruses showed that at least 18 genotypes co-circulated in the fifth wave in Jiangsu, illustrating the genetic diversity of these viruses and their continuous evolution. Compared to the prior four waves, the H7N9 viruses in the fifth wave displayed antigenic divergence, and WHO therefore proposed two new CVVs, A/Hong Kong/125/2017 and A/Guangdong/17SF003/2016 [9]. In this study, all of the viruses from Jiangsu province, except for the virus A/Xuzhou/550/ 2017, clustered in one clade together with A/Hunan/02650/2016-like CVVs in the HA tree. However, the antigenicity of the 41 H7N9 viruses needs further investigation.
The marked increase in the number of human infections with H7N9 virus in the fifth wave appears to be due to the extensive geographic spread and high prevalence of H7N9 viruses in poultry [9, 10, 22], and there is little virological evidence that the viruses themselves have a higher transmissibility to humans. Our sequence data revealed that the 41 isolates from Jiangsu had G186V and Q226L/I mutations in their HA proteins, which means they may possess dual-receptor-binding properties. However, the receptor-binding specificity of the 41 viruses needs to be further tested in binding assays using sialylglycopolymers and glycan arrays. For H7N9 influenza prevention and control strategies, it is necessary to further monitor the genetic changes known to be associated with antigenic mutation, host adaption, virulence, and transmissibility based on viral whole-genomic sequencing.
In December 2016, highly pathogenic avian influenza (HPAI) H7N9 viruses were detected in humans and poultry in Guangdong province that contained four amino acid insertions in the HA cleavage site [9, 17, 22]. Since then, at least 12 provinces in China have reported HPAI H7N9 viruses in poultry. Phylogenetic analysis showed that the HPAI H7N9 virus is likely to have emerged in late May 2016, and their ancestor virus originated from LPAI H7N9 viruses introduced from the Yangtze Delta region [22]. Because of their increased virulence, HPAI H7N9 viruses pose a greater threat to public health and poultry health than LPAI H7N9 viruses. In this study, in addition to the 41 isolates for which the full genome was sequenced, the HA genes of other H7N9 viruses from humans (n = 91) and the environment (n = 32) were also sequenced (data not shown). Four-basic-amino-acid insertions were not observed in the HA cleavage sites of any of the 167 viruses, which revealed that HPAI H7N9 viruses did not spread to Jiangsu province in the fifth wave.
Since the late 1990s, H5, H9, H6, H7 and H10 viruses of multiple subtypes/genotypes have become enzootic in poultry (including chickens, ducks, quail, etc.) in China [23,24,25,26], which provide an abundant gene pools for further inter- or intro-subtype reassortment [27]. Furthermore, the co-circulation of these viruses in southern China, along with their ability to infect humans and their potential for future reassortment with human H3N2 and/or H1N1 viruses clearly raises concern about their pandemic potential [28, 29]. Our study highlights the need to constantly monitor the evolution of H7N9 virus and reinforce systematic influenza surveillance in humans, birds and pigs in China.
References
Belser JA, Bridges CB, Katz JM, Tumpey TM (2009) Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis 15(6):859–865
Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J et al (2014) Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383(9918):714–721
Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K et al (2013) Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368(20):1888–1897
Pan M, Gao R, Lv Q, Huang S, Zhou Z, Yang L, Li X, Zhao X, Zou X, Tong W et al (2016) Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: Virological and clinical findings. J Infect 72(1):52–59
Peiris JS, de Jong MD, Guan Y (2007) Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 20(2):243–267
Wei SH, Yang JR, Wu HS, Chang MC, Lin JS, Lin CY, Liu YL, Lo YC, Yang CH, Chuang JH et al (2013) Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med 1(10):771–778
Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, Ma C, Lycett SJ, Leung CY, Chen X et al (2013) The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 502(7470):241–244
Zhou L, Tan Y, Kang M, Liu F, Ren R, Wang Y, Chen T, Yang Y, Li C, Wu J et al (2017) Preliminary epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus, China, 2017. Emerg Infect Dis 23(8):1355–1359
Kile JC, Ren R, Liu L, Greene CM, Roguski K, Iuliano AD, Jang Y, Jones J, Thor S, Song Y et al (2017) Update: increase in human infections with novel asian lineage avian influenza A(H7N9) viruses during the fifth epidemic - China, October 1, 2016-August 7, 2017. MMWR Morb Mortal Wkly Rep 66(35):928–932
Wang X, Jiang H, Wu P, Uyeki TM, Feng L, Lai S, Wang L, Huo X, Xu K, Chen E et al (2017) Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis 17(8):822–832
Huo X, Chen L, Qi X, Huang H, Dai Q, Yu H, Xia Y, Liu W, Xu K, Ma W et al (2017) Significantly elevated number of human infections with H7N9 virus in Jiangsu in eastern China, October 2016 to January 2017. Euro Surveill 22(13):30496
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR (2001) Universal primer set for the full-length amplification of all influenza A viruses. Adv Virol 146(12):2275–2289
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Wang D, Yang L, Zhu W, Zhang Y, Zou S, Bo H, Gao R, Dong J, Huang W, Guo J et al (2016) Two outbreak sources of influenza A (H7N9) viruses have been established in China. J Virol 90(12):5561–5573
Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z et al (2013) Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342(6155):243–247
Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA (2013) Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342(6163):1230–1235
Zhu W, Zhou J, Li Z, Yang L, Li X, Huang W, Zou S, Chen W, Wei H, Tang J et al (2017) Biological characterisation of the emerged highly pathogenic avian influenza (HPAI) A(H7N9) viruses in humans, in mainland China, 2016 to 2017. Euro Surveill 22(19):30533
Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J (2005) The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A 102(51):18590–18595
Mok CK, Lee HH, Lestra M, Nicholls JM, Chan MC, Sia SF, Zhu H, Poon LL, Guan Y, Peiris JS (2014) Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol 88(6):3568–3576
Marjuki H, Mishin VP, Chesnokov AP, De La Cruz JA, Davis CT, Villanueva JM, Fry AM, Gubareva LV (2015) Neuraminidase mutations conferring resistance to oseltamivir in influenza A(H7N9) viruses. J Virol 89(10):5419–5426
Marjuki H, Mishin VP, Chesnokov AP, Jones J, De La Cruz JA, Sleeman K, Tamura D, Nguyen HT, Wu HS, Chang FY et al (2015) Characterization of drug-resistant influenza A(H7N9) variants isolated from an oseltamivir-treated patient in Taiwan. J Infect Dis 211(2):249–257
Yang L, Zhu W, Li X, Chen M, Wu J, Yu P, Qi S, Huang Y, Shi W, Dong J et al (2017) Genesis and spread of newly emerged highly pathogenic H7N9 avian viruses in mainland China. J Virol 91:e01277
Chen H, Smith GJ, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT et al (2006) Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc Natl Acad Sci USA 103(8):2845–2850
Huang K, Zhu H, Fan X, Wang J, Cheung CL, Duan L, Hong W, Liu Y, Li L, Smith DK et al (2012) Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J Virol 86(11):6075–6083
Ma C, Lam TT, Chai Y, Wang J, Fan X, Hong W, Zhang Y, Li L, Liu Y, Smith DK et al (2015) Emergence and evolution of H10 subtype influenza viruses in poultry in China. J Virol 89(7):3534–3541
Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C et al (2015) Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci USA 112(2):548–553
Jin Y, Ren H, Teng Y, Hu M, Peng X, Yue J, Liang L (2017) Novel reassortment of avian influenza A(H7N9) virus with subtype H6N6 and H5N6 viruses circulating in Guangdong Province China. J Infect 75(2):179–182
Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B et al (2013) Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ 347:f4752
Zhu Y, Qi X, Cui L, Zhou M, Wang H (2013) Human co-infection with novel avian influenza A H7N9 and influenza A H3N2 viruses in Jiangsu province, China. Lancet 381(9883):2134
Funding
This study was supported by Jiangsu Provincial Medical Talent Project (No. ZDRCA2016031 and ZDRC A2016032), Science & Technology Demonstration Project for Major Emerging Infectious Diseases Control and Prevention (No. BE2015714, BE2017749), Key Medical Discipline of Epidemiology (No. ZDXK A2016008) and the ‘333’ Project in Jiangsu Province.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that we have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of Jiangsu Provincial Center for Disease Control and Prevention.
Informed consent
Informed consent was obtained from all individual participants included in this study.
Additional information
Handling Editor: Ayato Takada.
Rights and permissions
About this article
Cite this article
Qi, X., An, X., Jiao, Y. et al. Co-circulation of multiple genotypes of influenza A (H7N9) viruses in eastern China, 2016-2017. Arch Virol 163, 1779–1793 (2018). https://doi.org/10.1007/s00705-018-3800-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3800-3