Abstract
Lily plants exhibiting virus-like symptoms of leaf yellowing, twisting and brownish necrotic spots were collected, and next-generation sequencing of small RNAs was conducted to identify the associated viruses. Cucumber mosaic virus, lily symptomless virus and a hitherto unrecorded potyvirus, tentatively named “lily yellow mosaic virus” (LYMV), were detected. The genomic RNA of LYMV was 9811 nt in length, encoding a large polyprotein of 3,124 amino acids with a predicted Mr of 353.3 kDa. BLAST analysis showed that LYMV shared a high degree of amino acid sequence identity with Thunberg fritillary mosaic virus (55%), bean yellow mosaic virus (52%), clover yellow vein virus (51%), leek yellow stripe virus (51%), and lily mottle virus (52%), and these viruses clustered together in a phylogenetic tree.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Lily plants (species Lilium) are popular ornamentals because of their beautiful flower colors and floral scents, and there are more than 4500 cultivated varieties [1]. Lily plants are frequently infected with viruses, including cucumber mosaic virus (CMV), lily mottle virus (LMoV), lily symptomless virus (LSV), lily virus X (LVX), arabis mosaic virus (ArMV), plantago asiatica mosaic virus (PlAMV), strawberry latent ringspot virus (SLRV), tobacco rattle virus (TRV), and tomato ringspot virus (TRSV) [1,2,3,4]. Once infected by one or more viruses, symptoms of mosaic, chlorosis, streaking and/or leaf deformation often occur [5]. Viruses are transmitted vegetatively and may reduce the value of flowers.
In 2016, lily plants exhibiting virus-like symptoms of leaf yellowing, twisting and brownish necrotic spots were observed in the experimental field of Beijing Agricultural University under natural conditions (Supplemental Fig. 1). To identify the potential viruses involved in the disease, small-RNA deep sequencing was conducted using the Illumina Hiseq2000 sequencing platform. Raw Illumina sRNA reads were trimmed and cleaned by removing sequences shorter than 16 nucleotides (nt) or longer than 30 nucleotides, low-quality tags, and polyA or N tags using an in-house Perl script. A total of 4522 contigs with size ranging from 33-389 nt were assembled de novo using Velvet (k-mer value of 17) [6], and BLAST analysis against the GenBank database showed that most of the contigs were host-derived, while 159 were virus-like. Of these, 42 were of CMV and 61 were of LSV. The remaining virus-like contigs showed a high degree of amino acid (aa) sequence similarity to different proteins of potyviruses in BLASTx analysis, indicating a potential new potyvirus. All three viruses were confirmed by RT-PCR using virus-specific primers (Table S1).
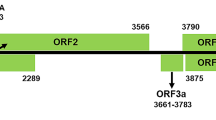
Schematic representation of the genomic organization of LYSV. The 5′ and 3′ nontranslated regions (NTR) are represented by lines, and the large open reading frame (ORF) is depicted by an open box. The numbers below the diagram indicate the starting position (nt) predicted for each gene. The proteinase cleavage sites were also listed
To determine the complete genome sequence of the new potyvirus, conventional RT-PCR with primer pairs targeting different protein regions was carried out, and the 5′ and 3′ ends were determined by RACE PCR (Table S1). This virus was tentatively named “lily yellow mosaic virus” (LYMV), although its contribution to the observed symptoms on the multiply-infected host plant are unknown. The genomic RNA of LYMV-BJ was 9811 nt in length (accession number MF543013), excluding the poly(A) tail at the 3′ end with the 5′ and 3′ untranslated regions of 217 and 219 nt, respectively. Protein sequence analysis revealed nine putative proteinase cleavage sites for ten mature proteins at amino acid positions 364, 821, 1168, 1221, 1856, 1909, 2099, 2342 and 2857 (Fig. 1), which are conserved in potyviruses. The recently identified ORF encoding the PIPO protein [7] was identified from a GA6 motif at position 3140-3146 within the P3 region protein in the +2 reading frame. The conserved domains HX8DX28GXSG and FIMRGR in P1 were located at position 275-316 and 336-341 of the amino acid (aa) sequence, respectively [8]. In HC-Pro, the conserved motifs HXCX27CX2C [9], FRNK [10, 11] and PTK were found at polyprotein positions 388-421, 544-547, and 673-675. The conserved motif KITC was present as 415RITC418 in LYMV-BJ, which has also been found in some potyviruses including pepper vein banding virus and tobacco vein banding mosaic virus [9, 12, 13]. However, the less-conserved motif IGN in the central region of HC-Pro, which is essential for genome amplification [14], was IGK (aa 613-615) in LYMV. In P3, the conserved residues EPY-(X)7-SP-(X)2-L were at aa position 853-867 of the polyprotein [15]. In RNA helicase CI, the conserved motifs GXXGXGKS (aa 1,306-1,313), VLLLEPTRPL (aa 1,274-1,280) [16], DECH (position 1,395-1,398), LKVSATPP (1,422-1,429), LVYV (1,473-1,476), VATNIIENGVTL (1,524-1,535) and GERIQRLGRVGR (1,567-1,578) [15, 17] were all found. In NIa-Pro, the motif H(X)2T(X)3GHCG, which is responsible for the proteinase activity [18], was identified at position 2,244-2,254. In NIb, the conserved residues SIKAEL and ADGSRFD were located at positions 2,514-2,519 and 2,590-2,597, respectively. The conserved amino acids FDSS at position 2,596-2,599 of the polyprotein were located 261 aa upstream of the putative NIb/CP cleavage site [19]. The conserved motif (S/T)G-(X)3-T(X)3-N(S/T)(X)18–37GDD began at position 2,651. In CP, the DAG motif, which interacts with PTK of HC-Pro to regulate potyviruses transmission by aphids [20], was located at position 2,865-2,867. The three consensus motifs found in the CP of potyviruses [19] were also found in LYMV-BJ (MVWCIENGTSP, 2,974–2,984; AFDF, 3057-3060; QMKAAA, 3,077-3,082). Sequence analysis of all of the gene products showed that most of the LYMV-encoded proteins shared high levels of aa sequence identity with Thunberg fritillary mosaic virus, while the CP shared 71%, the highest, with pepper veinal mottle virus, which was below the threshold value used to discriminate between species and strains within the genus Potyvirus [21].
References
Sharma A, Mahinghara BK, Singh AK, Kulshrestha S, Raikhy G, Singh L, Verma N, Hallan V, Ram R, Zaidi AA (2005) Identification, detection and frequency of lily viruses in Northern India. Sci Hortic 106:213–227
Asjes CJ (2000) Control of aphid-borne lily symptomless virus and lily mottle virus in Lilium in the Netherlands. Virus Res 71:23–32
Komatsu K, Yamaji Y, Ozeki J, Hashimoto M, Kagiwada S, Takahashi S, Namba S (2008) Nucleotide sequence analysis of seven Japanese isolates of Plantago asiatica mosaic virus (PlAMV): a unique potexvirus with significantly high genomic and biological variability within the species. Arch Virol 153:193–198
Kwon JY, Ryu KH, Choi SH (2013) Reverse transcription polymerase chain reaction-based system for simultaneous detection of multiple lily-infecting viruses. Plant Pathol J 29(3):338–343
Kim JH, Yoo HN, Bae EH, Jung YT (2012) Development of an indirect ELISA and immunocapture RT-PCR for lily virus detection. J Microbiol Biotechnol 22(12):1776–1781
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Chung BY, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105:5897–5902
Valli A, López-Moya JJ, García JA (2007) Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J Gen Virol 88:1016–1028
Atreya CD, Pirone TP (1993) Mutational analysis of the helper component-proteinase gene of a potyvirus: effects of amino acid substitutions, deletions, and gene replacement on virulence and aphid transmissibility. Proc Natl Acad Sci USA 90(24):11919–11923
Gal-On A (2000) A point mutation in the FRNK motif of the potyvirus Helper Component-Protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology 90(5):467–473
Gal-On Amit (2007) Zucchini yellow mosaic virus: insect transmission and pathogenicity—the tails of two proteins. Mol Plant Pathol 8(2):139–150
Anindya R, Joseph J, Gowri TDS, Savithri HS (2004) Complete genomic sequence of Pepper vein banding virus (PVBV): a distinct member of the genus Potyvirus. Arch Virol 149(3):625–632
Yu XQ, Lan YF, Wang HY, Liu JL, Zhu XP, Valkonen JPT, Li XD (2007) The complete genomic sequence of Tobacco vein banding mosaic virus and its similarities with other potyviruses. Virus Genes 35(3):801–806
Plisson C, Drucker M, Blanc S, German-Retana S, Le Gall O, Thomas D, Bron P (2003) Structural characterization of HC-Pro, a plant virus multifunctional protein. J Biol Chem 278(26):23753–23761
Riechmann JL, Laín S, García JA (1992) Highlights and prospects of potyvirus molecular biology. J Gen Virol 73:1–16
Kadaré G, Haenni AL (1997) Virus-encoded RNA helicases. J Virol 71(4):2583–2590
Adams MJ, Antoniw JF, Beaudoin F (2005) Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol 6:471–487
Dougherty WG, Parks TD, Cary SM, Bazan JF, Fletterick RJ (1989) Characterization of the catalytic residues of the tobacco etch virus 49-kDa proteinase. Virology 172(1):302–310
Dujovny G, Sasaya T, Koganesawa H, Usugi T, Shohara K, Lenardon SL (2000) Molecular characterization of a new potyvirus infecting sunflower. Arch Virol 145(11):2249–2258
Atreya CD, Raccah B, Pirone TP (1990) A point mutation in the coat protein abolishes aphid transmissibility of a potyvirus. Virology 178(1):161–165
Wylie SJ, Adams M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM (2017) ICTV virus taxonomy profile: Potyviridae. J Gen Virol 98:352–354
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Funding
This work was supported by grants from the Beijing Municipal Education Commission (CEFF-PXM2017_014207_000043, KM201610020008) and Da Bei Nong Group, the Project of Construction of Innovative Teams and Teacher Career Development for Universities and Colleges under Beijing Municipality and the National Key Research and Development Program (2016YFF0203203).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that we have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Stephen John Wylie.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Jia, A., Qiao, Y. et al. Virome analysis of lily plants reveals a new potyvirus. Arch Virol 163, 1079–1082 (2018). https://doi.org/10.1007/s00705-017-3690-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3690-9