Abstract
Endogenous retroviruses (ERVs) are genomic elements that are present in a wide range of vertebrates and have been implicated in a variety of human diseases, including cancer. However, the characteristic expression patterns of ERVs, particularly in virus-induced tumours, is not fully clear. DNA methylation was analysed by bisulfite pyrosequencing, and gene expression was analysed by RT-qPCR. In this study, we first found that the endogenous avian retrovirus ALVE1 was highly expressed in some chicken tissues (including the heart, bursa, thymus, and spleen) at 2 days of age, but its expression was markedly decreased at 35 days of age. In contrast, the CpG methylation level of ALVE1 was significantly lower in heart and bursa at 2 days than at 35 days of age. Moreover, we found that the expression of ALVE1 was significantly inhibited in chicken embryo fibroblast cells (CEFs) and MSB1 cells infected with avian leukosis virus subgroup J (ALVJ) and reticuloendotheliosis virus (REV) at the early stages of infection. In contrast, the expression of the ALVE1 env gene was significantly induced in CEFs and MSB1 cells infected with Marek’s disease virus (MDV). However, the methylation and expression levels of the ALVE1 long terminal repeat (LTR) did not show obvious alterations in response to viral infection. The present study revealed the expression patterns of ALVE1 in a variety of chicken organs and tissues and in chicken cells in response to avian tumour virus infection. These findings may be of significance for understanding the role and function of ERVs that are present in the host genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endogenous retroviruses (ERVs) are genomic elements that are present in a wide range of vertebrates [15, 44]. The expression of endogenous retroviral sequences has been implicated in a variety of human diseases, including cancer [23] and autoimmune diseases [41]. Moreover, characteristic patterns of ERV expression are often observed in various tumours [46], and a cumulative body of evidence from animal models indicates that ERVs may be involved in the process of malignant transformation or the promotion of tumour growth [23, 46]. However, the characteristic expression pattern of ERVs in virus-induced tumours is not fully clear.
Although many studies on ERVs focus on humans and model organisms, the study of ERVs in domestic animals, including chickens, has recently received increasing attention from scientists in this field [6, 10]. Endogenous avian retrovirus ALVEs were the first chicken ERVs identified and are highly homologous to exogenous avian leukosis viruses (ALVs), one of three avian tumour viruses [47]. The endogenous retrovirus ALVE could be involved in host antiviral defence and innate immunity, although the precise cellular mechanism is unknown. ADOL line 0 chickens have been selected to be free of ALVE genes but susceptible to exogenous ALVs (i.e., ALVA, ALVB, ALVC and ALVJ) [2]. As indicated in a previous report, the expression of the endogenous retrovirus ALVE reduces the immunity of line 15I5 chickens, which contain endogenous retrovirus ALVE, to ALVJ infection compared with that of line 0 chickens [33]. Therefore, we speculated that endogenous retrovirus ALVE may be associated with tolerance to the antigens derived from avian tumour viruses and with resistance to infection with exogenous virus.
The aims of this study were to elucidate the expression patterns of endogenous avian retrovirus ALVE1 in various cell lines and tissues and to provide insights into ALVE1 biology. Some ALVEs are actively transcribed from their inherited chromosomal locations, whereas others (e.g., ALVE1) are silent [3]. ALVE1 is located on chromosome 1 and is unable to replicate in chicken lines and cell lines. We first observed the kinetics of ALVE1 expression in chicken tissues and found that ALVE1 is highly expressed in some chicken tissues at 2 days of age but exhibits decreased expression at 35 days of age. We then focused on the expression patterns of endogenous retrovirus ALVE1 in response to infection with three different avian tumour viruses, namely ALV, reticuloendotheliosis virus (REV), and Marek’s disease virus (MDV). We found that ALVE1 expression is significantly induced by MDV infection in chicken embryo fibroblast cells (CEFs) and MSB1 cells and inhibited or interfered with by retrovirus ALVJ and REV at the early stages of infection. These findings reveal the expression patterns of endogenous retrovirus ALVE1, provide insights into the interaction between an endogenous retrovirus and exogenous tumour viruses, and may be of significance for elucidating the function of endogenous retroviruses.
Materials and methods
Viruses
Three avian tumour viruses, namely Marek’s disease virus (MDV), reticuloendotheliosis virus (REV), and avian leukosis viruses (ALV), were used for infection. The RB1B strain of highly virulent MDV, the JS09GY3 strain of ALVJ, and the HA1101 strain of REV were obtained from the Laboratory of Avian Preventive Medicine, Yangzhou University, China.
Cells
Primary chicken embryo fibroblast cells (CEFs) were prepared from 10-day-old specific-pathogen-free (SPF) embryos obtained from Merial Vital Laboratory Animal Technology Co., Ltd. (Beijing, China). The cells were seeded in six-well plates in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies/GIBCO, MD, USA) with 5 % foetal bovine serum (FBS) and incubated at 37 °C with 5 % CO2 and 95 % humidity. The MDV-transformed lymphoblastoid cell line [1] MSB-1 was maintained in RPMI 1640 medium supplemented with 10 % FBS and 2 % chicken serum at 37 °C with 5 % CO2 and 95 % humidity. The chicken HD11 macrophage cell line [4] was maintained in RPMI 1640 medium containing 10 % heat-inactivated newborn calf serum, antibiotics (100 U of penicillin and 100 μg of streptomycin per ml), 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, and 10 mM HEPES at 41 °C with 5 % CO2, and 95 % humidity. HD11 cells were seeded in six-well plates and collected 6, 12 and 24 hours after incubation.
Tissues
All tissues from 2- and 35-day-old chickens were obtained as described previously [17]. Briefly, three chickens were sacrificed, and their tissue organs (including the liver, thymus, spleen, bursa, brain, heart, lung and kidney) were rapidly excised, rinsed with ice-cold phosphate-buffered saline (PBS, pH 7.4) to remove blood contaminants, and immediately stored in liquid nitrogen until analysis.
Virus infection
Virus titration for MDV, ALVJ or REV infection was performed as reported previously [19, 20, 34, 35], and the virus infection levels in cells at different time points were detected by real-time PCR. Briefly, cells were seeded in six-well plates and infected with either the RB1B or JS09GY3 strain at a multiplicity of infection (MOI) of 0.1, 5 or 1 after 24 hours of incubation. The CEF cells were seeded into six-well plates and infected with the RB1B, JS09GY3 or HA1101 strain after 24 hours of incubation. Cells were collected at 6, 12, 24, 48 and 96 hours postinfection (hpi). MSB1 cells were seeded in six-well plates and infected with the RB1B, JS09GY3 or HA1101 strain. HD11 cells were seeded in six-well plates and infected with the JS09GY3 strain. Cells were collected at 6, 12 and 24 hpi.
RNA extraction and real-time PCR
The total RNA from chicken tissues or cell lines was extracted using an AxyPrep™ Multisource Total RNA Miniprep Kit (Axygen, USA), and 1 μg of RNA was used for reverse transcription using a PrimeScript RT reagent kit after gDNA Eraser treatment (Takara, Japan) following the manufacturer’s instructions. The efficiency of DNA destruction was confirmed by RT-PCR without reverse transcriptase.
Primers specific for endogenous retrovirus ALVE1 were designed with Primer Express software (Version 3.0, Applied Biosystems, CA, USA), and primers specific for exogenous viruses (ALVJ, MDV and REV) were described in previous reports [17, 34, 35]. All of the primers used are shown in Table 1 and were synthesized by Invitrogen Company (Shanghai, China). Gene expression was analysed by reverse transcription quantitative PCR (RT-qPCR) using SYBR Green Master Mix (Takara, Japan) with a 7500 Real-Time PCR system (Applied Biosystems, USA), and the gene expression levels were normalized to those of chicken Gapdh and β-actin.
DNA extraction and bisulfite treatment
DNA from cultured MSB1 cells or HD11 cells was extracted at 36 hours using a DNeasy Blood and Tissue Kit (QIAGEN, Germany), and the concentration of DNA was measured using a spectrophotometer. One microgram of DNA from each sample was treated with bisulfite using an EpiTect Fast DNA Bisulfite Kit (QIAGEN, Germany) following the manufacturer’s protocols.
Pyrosequencing methylation analysis
The forward and reverse primers used in PCR and the sequencing primers used in the pyrosequencing methylation assays were designed using PyroMark Assay Design 2.0 (QIAGEN, Germany; see Table 1). A pyrosequencing methylation analysis was conducted using the PyroMark Q24 system (QIAGEN, Germany) according to the manufacturer’s recommended protocol. Briefly, a volume of 5-20 μl of the PCR product was used for each pyrosequencing reaction based on the concentration of the PCR product and immobilized to streptavidin-coated Sepharose beads (QIAGEN, Germany). After the immobilized PCR product was purified, denatured and washed using a PyroMark Q24 Workstation (QIAGEN, Germany), DNA strands were separated and released into a PyroMark Q24 Plate (QIAGEN, Germany). The sequencing primers were then annealed to DNA strands, and pyrosequencing was performed using the PyroMark Q24 system. The DNA methylation level was analysed using PyroMark Q24 Advanced Software (QIAGEN, Germany). Non-CpG cytosine residues were used as controls to verify bisulfite conversion.
Comparative analysis of transcription and long terminal repeat (LTR) methylation of ALVE1 in chicken cells
The relationship between the DNA methylation of the ALVE1 LTR and its expression was explored in two chicken cell lines (MSB1 and HD11) and CEFs infected with two avian viruses (MDV and ALVJ). PCR and pyrosequencing primers were designed to amplify three to six CpG dinucleotide sites in conserved regions of the ALVE1 LTR, which revealed a negative relationship between DNA methylation of the ALVE1 LTR and the RNA level of ALVE1 [56].
To further investigate the role of methylation in ALVE1 LTR expression, MSB1 and HD11 cells were treated with the demethylating agent 5-aza-2′-deoxycytidine. Briefly, MSB-1 and HD11 cells were seeded in six-well plates at 5×105 cells/well, cultured for 24 hours before drug treatment, and treated with 5-aza-2′-deoxycytidine (AZA, Sigma-Aldrich; 5 µM) for 24 hours. The dose of AZA was selected based on preliminary studies as well as previously published studies [7, 40].
Statistical analysis
Statistical analysis was performed with either the Statistical Package for the Social Sciences (version 16.0) or GraphPad Prism (version 5.0) software. The p-values were obtained using paired Student’s t-tests or unpaired tests for normal distributions of at least three independent experiments.
Results
Quantitative evaluation of ALVE1 expression in chicken tissues
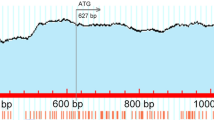
The whole ALVE1 included two CpG islands within 7,525 bp based on BLAT results from the UCSC Genomic Browser (Fig. 1). The first CpG island, which contains 136 CpG sites within 2,176 bp, was selected to evaluate the methylation differences among three different tissues (heart, bursa and liver) from 2- and 35-day-old chickens. The expression of ALVE1 genes (LTR and env regions, Fig. 1) was examined in various chicken tissues, including the heart, liver, lung, kidney, thymus, spleen, bursa, brain and skin, through RT-qPCR. As shown in Fig. 2A and B, ALVE1 was highly expressed in some chicken tissues at 2 days of age, but this level was significantly decreased at 35 days of age in most chicken tissues, including the brain, heart and immune organs (thymus, spleen and bursa). Interestingly, the expression of ALVE1 remained high in the liver from 2 to 35 days of age. In addition, no significant alterations in ALVE1 expression were found in the kidney, skin or lung from 2 to 35 days of age. Furthermore, level of the CpG methylation of the ALVE1 region was significantly lower in the heart and bursa at 2 days of age than at 35 days of age, whereas a significant alteration was detected in the liver (Fig. 2C-K). These findings indicated that the expression of ALVE1 is tissue-specific in chickens and may be associated with tissue development.
Positions of amplified regions of the avian endogenous retrovirus ALVE1 used for pyrosequencing and RT-qPCR. (A) Positions of amplified regions of ALVE1 used for RT-qPCR. (B) Positions of amplified regions of ALVE1 used for pyrosequencing and RT-qPCR for the LTR region. 5′ LTR, 5′ long terminal repeat; U3, U3 region in the LTR; R, R region in the LTR, U5: U5 region in the LTR; gag, gag gene; pol, pol gene; env, env gene; PPT, polypurine tract; 3′ LTR, 3′ long terminal repeat. CpG island 1 contains 136 CpG sites within 2,176 bp, and CpG island 2 contains 24 CpG sites within 304 bp (BLAT results from the UCSC Genome Browser website)
Relative expression and DNA methylation analysis of ALVE1 genes in chicken tissues. (A) Expression of the LTR region in chicken tissues at 2 and 35 days of age. (B) Expression of the env gene in chicken tissues at 2 and 35 days of age. (C-K) Pyrosequencing DNA methylation analysis of the CpG islands of ALVE1 in the chicken heart (C-E), bursa (F-H) and liver (I-K) at 2 and 35 days of age. Representative pyrograms for the CpG islands of ALVE1. The percentage shown at each CpG site is the methylation percentage for this site calculated as mC/(mC+C), where mC is the number of methylated cytosines and C is the number of unmethylated cytosines. The x-axes show the nucleotide dispensation order of the sequencing reactions based on the assayed sequences. The y-axes represent the light emission values obtained as relative light units. *, p < 0.05; **, p < 0.01; n = 3 for each line
Homologous exogenous retrovirus ALVJ inhibits the expression of the endogenous retrovirus ALVE1
The expression levels of ALVE1 genes were significantly increased at 6 hpi (except the LTR), were decreased at 12 hpi, and showed no significant change at 24, 48 and 96 hpi (Fig. 3A-D) in CEFs. In contrast, infection with ALVJ led to a significantly decreased expression of ALVE1 genes at 6 hpi in MSB1 cells, and expression of the gag gene was persistently downregulated from 6 to 24 hpi (Fig. 3F-I). In HD11 cells, however, we found that the env and pol genes were significantly downregulated at 12 hpi and 24 hpi, whereas LTR and gag were downregulated at 12 and 24 hpi, respectively, by ALVJ infection (Fig. 3K-N). Additionally, we observed that exogenous ALVJ proliferated rapidly after 24 hpi in CEFs and HD11 cells, but the proliferation of exogenous ALVJ was significantly inhibited in MSB1 cells after infection.
Expression patterns of ALVE1 in chicken cells infected with ALVJ. (A-D) RT-qPCR analysis of the expression levels of ALVE1 genes in CEFs at 6, 12, 24, 48 and 96 hours after ALVJ infection. (E) RT-qPCR analysis of the virus replication levels in CEFs at 6, 12, 24, 48 and 96 hours after ALVJ infection. (F-N) RT-qPCR analysis of the expression levels of ALVE1 genes in MSB1 cells (F-I) and HD11 cells (K-N) at 6, 12 and 24 hours after ALVJ infection. (J and O) RT-qPCR analysis of the virus replication levels in MSB1 cells (J) and HD11 cells (O) at 6, 12 and 24 hours after ALVJ infection. The data are presented as the means of relative values ± SEMs. *, p < 0.05; **, p < 0.01; n = 3 for each line
Nonhomologous exogenous avian herpesvirus MDV induces the expression of the endogenous retrovirus ALVE1
The expression of ALVE1 displayed different patterns of response to nonhomologous exogenous MDV infection. We found that the expression of the ALVE1 env gene was abnormally higher in infected CEFs than in controls and remained high with persistent MDV replication from 6 to 96 hpi, whereas the gag, pol, and LTR genes of ALVE1 did not show a similar change (Fig. 4A-D). The expression of the ALVE1 env gene was also induced by MDV infection in MSB1 cells from 6 to 24 hpi, similarly to the expression of the env gene in MDV-infected CEFs (Fig. 4F-I). In addition, increased expression of the pol and gag genes was also detected in MDV-infected MSB1 cells only at 12 hpi. In MDV-infected HD11 cells, only the gag gene was significantly upregulated at 12 hpi (Fig. 4K-N). These results suggest that nonhomologous exogenous avian herpesvirus can significantly induce expression of the env gene of the endogenous retrovirus ALVE1 in CEFs and MSB1 cells but not in HD11 cells.
Expression patterns of ALVE1 in chicken cells infected with MDV. (A-D) RT-qPCR analysis of the expression levels of ALVE1 genes in CEFs at 6, 12, 24, 48 and 96 hours after MDV infection. (E) RT-qPCR analysis of virus replication levels in CEFs at 6, 12, 24, 48 and 96 hours after MDV infection. (F-N) RT-qPCR analysis of expression levels of ALVE1 genes in MSB1 cells (F-I) and HD11 cells (K-N) at 6, 12 and 24 hours after MDV infection. (J and O) RT-qPCR analysis of virus replication levels in MSB1 cells (J) and HD11 cells (O) at 6, 12 and 24 hours after MDV infection. The data are presented as the means of relative values ± SEMs. *, p < 0.05; **, p < 0.01; n = 3 for each line
Nonhomologous exogenous avian retrovirus REV interferes with the expression of endogenous retrovirus ALVE1 at early stages of infection
Exogenous avian retrovirus REV significantly interfered with the expression of ALVE1 genes in infected chicken CEFs and MSB1 cells at the early stages of virus infection. Reduced expression of ALVE1 genes was observed in CEFs infected with REV only at 6 hpi, and after this time point, no significant alteration was observed, even though persistent replication of REV was detected from 6 to 96 hpi (Fig. 5A-D). In MSB1 cells, the expression of ALVE1 genes was significantly downregulated at 6 hpi and then clearly upregulated at 12 hpi with persistent replication of REV (Fig. 5F-I). In addition, the expression of only the gag gene was significantly downregulated at 24 hpi in REV-infected HD11 cells (Fig. 5K-N).
Expression patterns of ALVE1 in chicken cells infected with REV. (A-D) RT-qPCR analysis of expression levels of ALVE1 genes in CEFs at 6, 12, 24, 48 and 96 hours after REV infection. (E) RT-qPCR analysis of virus replication levels in CEFs at 6, 12, 24, 48 and 96 hours after REV infection. (F-N) RT-qPCR analysis of expression levels of ALVE1 genes in MSB1 cells (F-I) and HD11 cells (K-N) at 6, 12 and 24 hours after REV infection. (J and 0) RT-qPCR analysis of virus replication levels in MSB1 cells (J) and HD11 cells (O) at 6, 12 and 24 hours after REV infection. The data are presented as the means of relative values ± SEMs. *, p < 0.05; **, p < 0.01, n = 3 for each line
Association of ALVE1 LTR expression with DNA methylation
We found no significant change in the DNA methylation of the ALVE1 LTR in response to viral infection (Fig. 6A-H), which is consistent with its expression in CEFs at 6 and 96 hours after ALVJ or MDV infection. In the MSB1 and HD11 cell lines, however, we found that the expression of the ALVE1 LTR is correlated with the DNA methylation level, and the methylation level of LTR DNA sequences in HD11 cells was significantly higher only at CpG sites 1 and 2, and not at sites 3 and 4,compared with that found in MSB1 cells, whereas the expression of LTR in HD11 cells was significantly lower than that in MSB1 cells (Fig. 6I-6J). To confirm the regulation of the ALVE1 LTR by DNA methylation, MSB1 and HD11 cells were treated with the DNA methyltransferase inhibitor AZA for 24 hours. The RT-qPCR results indicated that ALVE1 LTR expression was significantly activated in both MSB1 and HD11 cells (Fig. 6K). Taken together, these results indicate that the expression level of the ALVE1 LTR is negatively correlated with the DNA methylation level of the LTR.
Quantitative DNA methylation analysis of the LTR region of ALVE1 in chicken cells. (A-G) Representative pyrograms for the LTR region in CEFs at 6 hours (A-C) and 96 hours (E-G) infected with MDV or ALVJ. (D and H) Methylation levels for each CpG site in the LTR region of ALVE1 in CEFs after 6 hours (D) and 96 hours (H) of infection with MDV or ALVJ. (I) Methylation levels for each CpG site in the LTR region of ALVE1 in HD11 and MSB1 cells at 36 hours. (J) Expression levels of the LTR region in HD11 and MSB1 cells. (K) Expression levels of the LTR region in HD11 and MSB1 cells treated with AZA or DMSO for 24 hours. The data are presented as the means of relative values ± SEMs. *, p < 0.05; **, p < 0.01; n = 3 for each line
Discussion
In this study, we first observed distinct expression patterns of avian endogenous retrovirus ALVE1 in various tissues from 2- and 35-day-old chickens and then analysed the variation in the DNA methylation patterns of ALVE1 in the bursa, heart and liver. To better elucidate ALVE1 biology, we further analysed the variations in the expression patterns of the ALVE1 gag, pol and env genes in various chicken cells in response to exogenous avian tumour viruses and the association between ALVE1 LTR expression and DNA methylation in chicken cells.
ALVE1 was found to be ubiquitously expressed in nearly all of the chicken tissues tested, with the lowest levels observed in the thymus and bursa. The expression patterns of the ALVE1 LTR and the env gene in chicken tissues (with the exception of the liver, kidney, lung and skin) were consistent with high levels at 2 days of age and low levels at 35 days of age. However, more-obvious differences were observed in the expression of the ALVE1 env gene in various tissues from 2- and 35-day-old chickens. Notably, the expression of the ALVE1 LTR region and the env gene was increased in the liver at 35 days of age compared with that observed at 2 days of age. In contrast, this phenomenon was not observed in other tissues. Higher expression of the ALVE env gene was significantly correlated with lower body weights in female but not male chickens [22]. In contrast, ERVs are also regulated through transcriptional mechanisms during early development [45]. In this study, we found that the ALVE1 LTR region and the env gene are negatively correlated with the methylation level of ALVE1 in the chicken heart and bursa. A previous study also found that higher methylation levels are linked to lower mRNA levels of ALVE in chicken tissues and indicated that the hypermethylation pattern of ALVE may be relevant for resistance against ALV-induced tumours in chickens [56]. More interestingly, there was substantially higher expression of the ALVE1 env gene in the spleen and heart at 2 days of age and in the lung both at 2 and 35 days of age than in the brain, bursa, thymus, liver, kidney, and skin. The higher expression in the spleen and lung than in other tissues has also been observed for other endogenous retroviruses [9, 28].
Next, the expression pattern of ALVE1 was further studied in vitro to elucidate ALVE1 biology and why the ALVE1 env gene was highly expressed in chicken spleen and lung, which we speculated might be involved in the host immune response. In lung and spleen, macrophages and lymphocytes are the main immune cells involved in immune response. Thus, representative innate and adaptive immune cells (the chicken macrophage cell line HD11 and lymphocyte cell lines MSB1) and non-immune cell (chicken embryo fibroblasts [CEFs]) were chosen in this study to investigate the expression pattern of ALVE1 after viral infection. Three avian tumour viruses (MDV, ALVJ and REV) have long been used in the study of the pathogenesis and immune control of virus-induced tumours in an easily accessible small-animal system [37, 48]. Thus, they are ideal infective agents for understanding the role and function of ERVs that are widely found in the genome. MDV is a DNA virus and an avian herpesvirus, whereas REV and ALVJ are RNA viruses and members of the family Retroviridae and have the gene arrangement LTR-gag-pol-env. Different viruses can elicit different host immune responses, and this could result in different expression levels of ALVE1, which may reflect the specific host immune system or cell innate immune system in which endogenous viruses may play a role, as has been discussed recently [42, 43, 55].
In spleen and thymus, the higher expression of ALVE1 may be associated with T lymphocyte selection and virus infection. Several studies have suggested that ERVs have a strong influence on T lymphocyte selection and improve the sensitivity with which T lymphocytes react to retroviral infection [11, 52, 54]. Moreover, T cells responding to HERV can kill target cells carrying HERV protein [11]. In the chicken T-lymphocyte cell line MSB1, ALVE1 expression was inhibited by the homologous exogenous retrovirus ALVJ at 6 hpi but induced at 12 hpi by the nonhomologous exogenous retrovirus REV. However, the selective induction of the ALVE1 env gene was observed during infection with the exogenous herpesvirus MDV. Our results further revealed that the interaction between exogenous viruses and endogenous retroviruses in T lymphocytes and the expression pattern of ALVE1 after viral infection may be associated with the antiviral immune response.
A higher expression level of ALVE1 env in lung at 2 and 35 days of age might be also associated with the host defense response (such as the macrophage response). Compared with other tissues, the lungs are susceptible to exogenous viral and bacterial infections. Higher expression in lung than in other tissues has also been observed for other endogenous retroviruses [9, 28]. Macrophages are present essentially in all tissues (especially in the lung) and are involved in host defense [38]. Macrophage cell lines (such as the human U-937 cell line) are suitable model systems for the study of endogenous retroviruses biology [25, 26]. It has been reported that the human endogenous retrovirus HERV-W induces a proinflammatory response in macrophage cells through the TLR4 activation pathway [42]. In the chicken macrophage cell line HD11, ALVE1 expression was inhibited by infection with the homologous exogenous retrovirus ALVJ (Fig. 3K-4O). However, no obvious difference in expression (except that of the gag gene) was found in HD11 cells infected with the nonhomologous exogenous retrovirus REV or the herpesvirus MDV (Fig. 4K-O and 5K-O). Inhibition of ALVE1 expression by ALVJ infection in macrophages may be involved in host antiviral defense. Especially, the env gene products derived from ERVs have been shown to act as restriction factors against related exogenous retroviruses in chickens, sheep, mice, and cats [31].
Interestingly, the ALVE1 env gene exhibited selective overexpression during MDV infection, whereas the methylation level and expression of the ALVE1 LTR showed no obvious alterations in MDV-infected cells. Several studies have suggested that the human herpesvirus HSV-1 increases the affinity of several transcription factors for their DNA-binding sites in the ERV LTR by producing the immediate early protein ICP0 [24] or the immediate early protein 1 (IEP1) [29] and thereby transactivates HERVK. Similar results were also observed in other studies [16, 50, 51]. ERVs can also be activated by the innate immune system and associated inflammatory transcription factors [32], and, in turn, ERVs can induce an innate immune response by pattern-recognition receptors (such Toll-like receptors) [5, 55]. Thus, we hypothesize that viral proteins or microRNAs encoded by MDV may have the ability to influence LTR activity or to induce innate immunity and thereby activate ALVE1 expression. It is known that innate immunity (such as TLR immunity) can be activated by MDV infection [14, 20, 39]. In addition, MDV-encoded Meq protein can bind to several factors involved in cell cycle control, including CDK2, p53, and RB [37]. A functional orthologue of miR-155, encoded by MDV called miR-M4, has been shown to activate the oncogene c-Myc and regulate TLR3 expression [8, 18, 57]. However, it remains unclear why MDV infection activates ALVE1 env expression, and the mechanisms remain to be further investigated.
In contrast, ALVE1 expression can be inhibited or interfered with by exogenous retroviruses, and we found reduced expression of the ERV LTR in response to homologous avian exogenous retrovirus (ALVJ) and nonhomologous avian exogenous retrovirus (REV) in CEFs and MSB1 cells. Moreover, ALVE1 expression was inhibited, particularly at the early stages of infection. Nevertheless, in individuals infected with exogenous human retrovirus, such as HIV-1 [13, 21] and HTLV-1 [53], the expression of ERV is often abnormally elevated. These exogenous human retroviruses can transactivate ERVs by producing viral proteins, which increases the affinity of several transcription factors to their DNA-binding sites in the ERV LTR [32]. Thus, this report describes an exception to this phenomenon. This finding reveals a unique interaction between ERV and exogenous retrovirus in chicken cells: ERV is inhibited or interfered with by a homologous or nonhomologous exogenous retrovirus. However, it remains unclear why MDV infection, but not homologous ALVJ, could activate ALVE1 env gene expression, and this finding remains to be further investigated.
The present study also revealed a negative correlation between the methylation level of the ALVE1 LTR and its expression in chicken cells and virus-infected cells, consistent with other reports [27, 36, 49]. Moreover, expression of the ALVE1 LTR could be stimulated by treatment with the demethylating agent 5-aza-2′-deoxycytidine (Fig. 5). The mechanisms involved in ALVE1 LTR regulation after virus infection remain to be investigated, but the methylation of ERV LTRs could be involved in the modulation of their activity [12]. The expression of ERVs usually depends on transcriptional regulatory elements present within retroviral LTRs. These LTRs contain many regulatory sequences, such as promoters, enhancers and factor-binding sites, and are responsive to both viral and cellular transcription factors [32]. Additionally, epigenetic mechanisms, particularly CpG methylation of the LTR, control the basal expression of ERVK in various cell types and tissues, and low levels of LTR methylation have been shown to result in high levels of ERVK expression [30]. However, the induction or inhibition of ALVE1 expression by virus infection does not fully follow the methylation level of LTR, and the methylation level and expression of the ALVE1 LTR do not show any obvious correlation in virus-infected CEFs, suggesting that the ALVE1 LTR might be controlled by other epigenetic mechanisms, but the DNA sequences controlling ALVE1 expression have not yet been identified.
In summary, the present study revealed the expression patterns of ALVE1 in a variety of chicken tissues and in various cell lines in response to avian tumour virus infection and may provide new insights into ALVE1 biology. In addition, we also observed the selective upregulation of the ALVE1 env gene in chicken cells infected with Marek’s disease virus. These findings may be of significance for understanding the role and function of ERVs that are present in the genome. In the future, more work is needed to elucidate the mechanism underlying the functions of ALVE1.
References
Akiyama Y, Kato S (1974) Two cell lines from lymphomas of Marek’s disease. Biken J 17:105–116
Bacon LD, Fulton JE, Kulkarni GB (2004) Methods for evaluating and developing commercial chicken strains free of endogenous subgroup E avian leukosis virus. Avian Pathol J WVPA 33:233–243
Baker B, Robison H, Varmus HE, Bishop JM (1981) Analysis of endogenous avian retrovirus DNA and RNA: viral and cellular determinants of retrovirus gene expression. Virology 114:8–22
Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T (1979) Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390
Blasius AL, Beutler B (2010) Intracellular toll-like receptors. Immunity 32:305–315
Bolisetty M, Blomberg J, Benachenhou F, Sperber G, Beemon K (2012) Unexpected diversity and expression of avian endogenous retroviruses. mBio 3:e00344–e00412
Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB (1999) Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21:103–107
Chi JQ, Teng M, Yu ZH, Xu H, Su JW, Zhao P, Xing GX, Liang HD, Deng RG, Qu LH, Zhang GP, Luo J (2015) Marek’s disease virus-encoded analog of microRNA-155 activates the oncogene c-Myc by targeting LTBP1 and suppressing the TGF-beta signaling pathway. Virology 476:72–84
Dieckhoff B, Kessler B, Jobst D, Kues W, Petersen B, Pfeifer A, Kurth R, Niemann H, Wolf E, Denner J (2009) Distribution and expression of porcine endogenous retroviruses in multi-transgenic pigs generated for xenotransplantation. Xenotransplantation 16:64–73
Garcia-Etxebarria K, Sistiaga-Poveda M, Jugo BM (2014) Endogenous retroviruses in domestic animals. Curr Genom 15:256–265
Garrison KE, Jones RB, Meiklejohn DA, Anwar N, Ndhlovu LC, Chapman JM, Erickson AL, Agrawal A, Spotts G, Hecht FM, Rakoff-Nahoum S, Lenz J, Ostrowski MA, Nixon DF (2007) T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathogens 3:e165
Gimenez J, Montgiraud C, Oriol G, Pichon JP, Ruel K, Tsatsaris V, Gerbaud P, Frendo JL, Evain-Brion D, Mallet F (2009) Comparative methylation of ERVWE1/syncytin-1 and other human endogenous retrovirus LTRs in placenta tissues. DNA Res Int J Rapid Publ Rep Genes Genomes 16:195–211
Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ Jr, Kaplan MH, Markovitz DM (2012) Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol 86:7790–7805
Haq K, Schat KA, Sharif S (2013) Immunity to Marek’s disease: where are we now? Dev Comp Immunol 41:439–446
Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K (2010) Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463:84–87
Hsiao FC, Tai AK, Deglon A, Sutkowski N, Longnecker R, Huber BT (2009) EBV LMP-2A employs a novel mechanism to transactivate the HERV-K18 superantigen through its ITAM. Virology 385:261–266
Hu X, Qin A, Qian K, Shao H, Yu C, Xu W, Miao J (2012) Analysis of protein expression profiles in the thymus of chickens infected with Marek’s disease virus. Virol J 9:256
Hu X, Ye J, Qin A, Zou H, Shao H, Qian K (2015) Both microRNA-155 and virus-encoded MiR-155 ortholog regulate TLR3 expression. PLoS One 10:e0126012
Hu X, Zhu W, Chen S, Liu Y, Sun Z, Geng T, Wang X, Gao B, Song C, Qin A, Cui H (2016) Expression of the env gene from the avian endogenous retrovirus ALVE and regulation by miR-155. Arch Virol 161:1623–1632
Hu X, Zou H, Qin A, Qian K, Shao H, Ye J (2016) Activation of Toll-like receptor 3 inhibits Marek’s disease virus infection in chicken embryo fibroblast cells. Arch Virol 161:521–528
Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, Gyenes G, Sheppard NC, Priumboom-Brees IM, Goodwin DA, Chen L, Rieger M, Muscat-King S, Loudon PT, Stanley C, Holditch SJ, Wong JC, Clayton K, Duan E, Song H, Xu Y, SenGupta D, Tandon R, Sacha JB, Brockman MA, Benko E, Kovacs C, Nixon DF, Ostrowski MA (2012) HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J Clin Investig 122:4473–4489
Ka S, Kerje S, Bornold L, Liljegren U, Siegel PB, Andersson L, Hallbook F (2009) Proviral integrations and expression of endogenous avian leucosis virus during long term selection for high and low body weight in two chicken lines. Retrovirology 6:68
Kassiotis G (2014) Endogenous retroviruses and the development of cancer. J Immunol 192:1343–1349
Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL (2002) Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res 86:93–100
Larsson E, Venables PJ, Andersson AC, Fan W, Rigby S, Botling J, Oberg F, Cohen M, Nilsson K (1996) Expression of the endogenous retrovirus ERV3 (HERV-R) during induced monocytic differentiation in the U-937 cell line. Int J Cancer 67:451–456
Larsson E, Venables P, Andersson AC, Fan W, Rigby S, Botling J, Oberg F, Cohen M, Nilsson K (1997) Tissue and differentiation specific expression on the endogenous retrovirus ERV3 (HERV-R) in normal human tissues and during induced monocytic differentiation in the U-937 cell line. Leukemia 11(Suppl 3):142–144
Lavie L, Kitova M, Maldener E, Meese E, Mayer J (2005) CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J Virol 79:876–883
Lee KH, You RN, Greenhalgh DG, Cho K (2012) Identification of a group of Mus dunni endogenous virus-like endogenous retroviruses from the C57BL/6J mouse genome: proviral genomes, strain distribution, expression characteristics, and genomic integration profile. Chromosome Res Int J Mol Supramol Evol Asp Chromosome Biol 20:859–874
Lee WJ, Kwun HJ, Kim HS, Jang KL (2003) Activation of the human endogenous retrovirus W long terminal repeat by herpes simplex virus type 1 immediate early protein 1. Mol Cells 15:75–80
Maksakova IA, Mager DL, Reiss D (2008) Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci 65:3329–3347
Malfavon-Borja R, Feschotte C (2015) Fighting fire with fire: endogenous retrovirus envelopes as restriction factors. J Virol 89:4047–4050
Manghera M, Douville RN (2013) Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10:16
Mays JK, Bacon LD, Pandiri AR, Fadly AM (2005) Response of white leghorn chickens of various B haplotypes to infection at hatch with subgroup J avian leukosis virus. Avian Dis 49:214–219
Mei M, Ye J, Qin A, Wang L, Hu X, Qian K, Shao H (2015) Identification of novel viral receptors with cell line expressing viral receptor-binding protein. Sci Rep 5:7935
Miao J, Bao Y, Ye J, Shao H, Qian K, Qin A (2015) Transcriptional profiling of host gene expression in chicken embryo fibroblasts infected with reticuloendotheliosis virus strain HA1101. PLoS One 10:e0126992
Ogasawara H, Okada M, Kaneko H, Hishikawa T, Sekigawa I, Hashimoto H (2003) Possible role of DNA hypomethylation in the induction of SLE: relationship to the transcription of human endogenous retroviruses. Clin Exp Rheumatol 21:733–738
Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S (2006) Marek’s disease virus: from miasma to model. Nat Rev Microbiol 4:283–294
Ovchinnikov DA (2008) Macrophages in the embryo and beyond: much more than just giant phagocytes. Genesis 46:447–462
Parvizi P, Abdul-Careem MF, Haq K, Thanthrige-Don N, Schat KA, Sharif S (2010) Immune responses against Marek’s disease virus. Anim Health Res Rev 11:123–134
Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grutzmann R, Aust D, Rummele P, Knosel T, Herd C, Stemple DL, Kettleborough R, Brosnan JA, Li A, Morgan R, Knight S, Yu J, Stegeman S, Collier LS, ten Hoeve JJ, de Ridder J, Klein AP, Goggins M, Hruban RH, Chang DK, Biankin AV, Grimmond SM, Australian Pancreatic Cancer Genome I, Wessels LF, Wood SA, Iacobuzio-Donahue CA, Pilarsky C, Largaespada DA, Adams DJ, Tuveson DA (2012) The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486:266–270
Perl A (2003) Role of endogenous retroviruses in autoimmune diseases. Rheum Dis Clin N Am 29:123–143, vii
Perron H, Dougier-Reynaud HL, Lomparski C, Popa I, Firouzi R, Bertrand JB, Marusic S, Portoukalian J, Jouvin-Marche E, Villiers CL, Touraine JL, Marche PN (2013) Human endogenous retrovirus protein activates innate immunity and promotes experimental allergic encephalomyelitis in mice. PLoS One 8:e80128
Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN (2006) The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol 176:7636–7644
Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240
Rowe HM, Trono D (2011) Dynamic control of endogenous retroviruses during development. Virology 411:273–287
Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N (2008) Endogenous retroviruses and cancer. Cell Mol Life Sci 65:3366–3382
Sacco MA, Nair VK (2014) Prototype endogenous avian retroviruses of the genus Gallus. J Gen Virol 95:2060–2070
Schat KA, Erb HN (2014) Lack of evidence that avian oncogenic viruses are infectious for humans: a review. Avian Dis 58:345–358
Stengel S, Fiebig U, Kurth R, Denner J (2010) Regulation of human endogenous retrovirus-K expression in melanomas by CpG methylation. Genes Chromosomes Cancer 49:401–411
Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT (2001) Epstein–Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 15:579–589
Sutkowski N, Chen G, Calderon G, Huber BT (2004) Epstein–Barr virus latent membrane protein LMP-2A is sufficient for transactivation of the human endogenous retrovirus HERV-K18 superantigen. J Virol 78:7852–7860
Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, Hanada K, Mena O, Kurlander R, Tawab A, Srinivasan R, Lundqvist A, Malinzak E, Geller N, Lerman MI, Childs RW (2008) Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Investig 118:1099–1109
Toufaily C, Landry S, Leib-Mosch C, Rassart E, Barbeau B (2011) Activation of LTRs from different human endogenous retrovirus (HERV) families by the HTLV-1 tax protein and T-cell activators. Viruses 3:2146–2159
Young GR, Ploquin MJ, Eksmond U, Wadwa M, Stoye JP, Kassiotis G (2012) Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathogens 8:e1002709
Yu P, Lubben W, Slomka H, Gebler J, Konert M, Cai C, Neubrandt L, Prazeres da Costa O, Paul S, Dehnert S, Dohne K, Thanisch M, Storsberg S, Wiegand L, Kaufmann A, Nain M, Quintanilla-Martinez L, Bettio S, Schnierle B, Kolesnikova L, Becker S, Schnare M, Bauer S (2012) Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 37:867–879
Yu Y, Zhang H, Tian F, Bacon L, Zhang Y, Zhang W, Song J (2008) Quantitative evaluation of DNA methylation patterns for ALVE and TVB genes in a neoplastic disease susceptible and resistant chicken model. PloS One 3:e1731
Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V (2011) Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek’s disease lymphomas. PLoS Pathogens 7:e1001305
Acknowledgments
This research was supported by the National Key Basic Research Program of China (973 Program, 2012CB517605) and the National Natural Science Foundation of China (81171965, 81372237 and 91540117) to HC, the China Postdoctoral Science Foundation (Grant No. 2015M571828) to HX, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Animal Science and Veterinary Medicine).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This study was performed in strict accordance with the recommendations provided in the Guide for the Care and Use of Laboratory Animals of Yangzhou University. The protocol was approved by the Committee on the Ethics of Animal Experiments of Yangzhou University (License Number: 06R015). This article does not contain any studies with human participants performed by any of the authors.
Additional information
X. Hu, W. Zhu and S. Chen contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hu, X., Zhu, W., Chen, S. et al. Expression patterns of endogenous avian retrovirus ALVE1 and its response to infection with exogenous avian tumour viruses. Arch Virol 162, 89–101 (2017). https://doi.org/10.1007/s00705-016-3086-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3086-2