Abstract
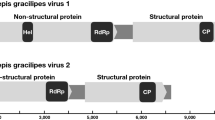
A putative new member of the family Totiviridae was identified in arboreal ants (Camponotus nipponicus). The viral dsRNA consisted of 5,713 nt with two overlapping open reading frames (ORFs). ORF1 encodes a putative capsid protein. ORF2 encodes a viral RNA-dependent RNA polymerase (RdRp). ORF2 could be translated as a fusion protein with the ORF1 product through a -1 frameshift in the overlapping ORF1. Phylogenetic analysis based on the RdRp revealed that the virus from C. nipponicus is closely related to Camponotus yamaokai virus, a member of the family Totiviridae, from another ant species. The name Camponotus nipponicus virus (CNV) is proposed for the new virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The family Totiviridae includes viruses that form small, non-enveloped, isometric particles with a double-stranded RNA (dsRNA) genome of 4.6–7.0 kbp in length [6]. The previously identified members of this family were detected in either parasitic protozoa or fungi. However, an unclassified member of the family Totiviridae, named penaeid shrimp infectious myonecrosis virus (IMNV) was found in the Pacific white shrimp [3]. Following the identification of IMNV, totiviruses have been reported in flies and mosquitoes, and all of those viruses were closely related to each other [2]. A totivirus that is closely related to Giardia lamblia virus was also found in salmon [1].
A virus belonging to the family Totiviridae, Camponotus yamaokai virus (CYV), was isolated from the arboreal ant Camponotus yamaokai [7]. CYV is the first totivirus found in ants. It shares the general characteristics of a totivirus in both genome composition and virus particle structure, but the genome sequence suggests that CYV is phylogenetically distinct from other arthropod totiviruses. In this study, we determined the complete genome sequence of a totivirus from another arboreal ant species, C. nipponicus.
C. nipponicus ants were collected from Shimoda, Shizuoka, Japan, in 2014. Total nucleic acids were extracted from worker ants using the phenol-chloroform method. Then, the dsRNA of the RNA virus was purified from the total nucleic acids and subjected to the fragmented and loop-primer ligated dsRNA sequencing (FLDS) method according to Urayama et al. [5].
A sequencing library was generated and analyzed by using IonPGM (Life Technologies), and the sequence reads were assembled using CLC workbench. As a result, the complete sequence of the dsRNA, 5,713 nt in length (DDBJ accession no. LC101918) was obtained. The sequence was predicted to contain two large ORFs estimated by Genetyx software. These two ORFs overlapped by 442 nt. The amino acid sequence of ORF2 had significant similarity to the RdRp of totiviruses and exhibited eight RdRP motifs similar to those of CYV (Supplementary Figure 1).
The presence of an overlapping region suggested that ORF2 is translated as a fusion protein with a product of ORF1 through a -1 ribosomal frameshift. Similar genomic arrangements have been described for several totiviruses [6]. Sequence analysis revealed the presence of a potential slippery heptamer AAAAAAC at nt position 2912–2918, that could facilitate a ribosomal -1 frameshift (Supplementary Figure 2). The heptamer is followed by a short “spacer” region and then a class 3 pseudoknot of 36 nt (at nt position 3054 – 3089).
The pairwise nucleotide and amino acid sequence identities of the novel RNA viral sequence and CYV are summarized in Table 1. A phylogenetic tree was constructed using the maximum-likelihood method with 1000 bootstrap replicates, based on the amino acid sequences of the RdRp domains (287–658 aa of ORF2) as described by Koyama et al. [7]. The phylogenetic analysis suggested that the viral sequence was closely related to, but distinct from, CYV (Fig. 1).
Phylogenic tree of members of the family Totiviridae. Sequences used in this analysis are shown in reference [7]
The structure of the reconstructed genome sequence suggests that the virus is a member of the family Totiviridae. Phylogenetic analysis revealed that the sequence was closely related to CYV, a totivirus from another ant species. Accordingly, we conclude that the dsRNA in C. nipponicus is derived from a new member of the family Totiviridae that is closely related to CYV. Therefore, the virus was named “Camponotus nipponicus virus” (CNV).
The significant similarity between CNV and CYV suggests two possibilities about the evolutionary relationship between the totiviruses infecting C. nipponicus and C. yamaokai, which belong to the subgenera Colobopsis and Myrmamblys, respectively [4]. First, because C. nipponicus and C. yamaokai share parts of their habitats [4], the totivirus might be transferred horizontally between host ants. Although totiviruses rarely undergo horizontal transmission [6], transmission between two ant species might have occurred over an evolutionary timescale. Second, an ancestral virus might have existed prior to the divergence of the two host ant species, with CNV and CYV later diverging and co-evolving with their hosts. In this case, the totiviruses in ants would have existed before the divergence of these subgenera. Some ant species must have lost their totiviruses, because no totiviruses have been found in ants of the subgenus Myrmamblys, such as C. nawai [7]. Future studies of the phylogeography of the host ants and that of the totiviruses will provide insight into the co-evolutionary relationship between the host insects and the viruses.
References
Haugland O, Mikalsen AB, Nilsen P, Lindmo K, Thu BJ, Eliassen TM, Roos N, Rode M, Evensen O (2011) Cardiomyopathy syndrome of atlantic salmon (Salmo salar L.) is caused by a double-stranded RNA virus of the Totiviridae family. J Virol 85:5275–5286
Isawa H, Kuwata R, Hoshino K, Tsuda Y, Sakai K, Watanabe S, Nishimura M, Satho T, Kataoka M, Nagata N, Hasegawa H, Bando H, Yano K, Sasaki T, Kobayashi M, Mizutani T, Sawabe K (2011) Identification and molecular characterization of a new nonsegmented double-stranded RNA virus isolated from Culex mosquitoes in Japan. Virus Res 155:147–155
Poulos B, Lightner D (2006) Detection of infectious myonecrosis virus (IMNV) of penaeid shrimp by reverse-transcriptase polymerase chain reaction (RT-PCR). Dis Aquat Org 73:69–72
Terayama M, Kubota S, Egushi K (2014) Encyclopedia of Japanese ants. Asakura Publishing Co. Ltd, Tokyo
Urayama S, Takaki Y, Nunoura T (2016) FLDS: a comprehensive dsRNA sequencing method for intracellular RNA virus surveillance. Microbes Environ 31:33–40
Wickner R, Wang C, Patterson J (2012) Family Totiviridae. In: Fauquet C, Mayo M, Maniloff J, Desselberger U, Ball L (eds) Virus taxonomy ninth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, pp 571–580
Koyama S, Urayama S, Ohmatsu T, Sassa Y, Sakai C, Takata M, Hayashi S, Nagai M, Furuya T, Moriyama H, Satoh T, Ono S, Mizutani T (2015) Identification, characterization and full-length sequence analysis of a novel dsRNA virus isolated from the arboreal ant Camponotus yamaokai. J Gen Virol 96:1930–1937
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All applicable guidelines for the care and use of animals were followed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koyama, S., Sakai, C., Thomas, C.E. et al. A new member of the family Totiviridae associated with arboreal ants (Camponotus nipponicus). Arch Virol 161, 2043–2045 (2016). https://doi.org/10.1007/s00705-016-2876-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2876-x