Abstract
In a previous work, a distinct tymovirus infecting tomato plants in Brazil was reported and tentatively named tomato blistering mosaic virus (ToBMV). In this study, the complete genome sequence of ToBMV was determined and shown to have a size of 6277 nucleotides and three ORFs: ORF 1 encodes the replication-complex polyprotein, ORF 2 the movement protein, and ORF 3 the coat protein. The cleavage sites of the replication-complex polyprotein (GS/LP and VAG/QSP) of ToBMV were predicted by alignment analysis of amino acid sequences of other tymoviruses. In the phylogenetic tree, ToBMV clustered with the tymoviruses that infect solanaceous hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Currently, the genus Tymovirus has 27 recognized species, the majority of which infect dicot plants [1]. In Brazil, a distinct tymovirus infecting tomato plants was isolated from Santa Catarina State and shown to possess a strikingly divergent coat protein (CP) gene sequence [2]. This suggested that this isolate, tentatively named tomato blistering mosaic virus (ToBMV), belongs to a new species. The other tymoviruses reported in Brazil are eggplant mosaic virus (EMV) [3], petunia vein-banding virus (PetVBV) [4], passion fruit yellow mosaic virus (PFYMV) [5], and cassia yellow mosaic-associated virus (CYMaV) [6].
In the previous study, the cp gene sequence of ToBMV showed the highest amino acid (aa) sequence identity (64 %) to chiltepin yellow mosaic virus (ChiYMV) [2]. This parameter satisfied one of the species demarcation criteria according to the Tymoviridae study group of the ICTV [1]. In order to clarify the identification of this virus, the complete genome sequence was determined in this study. Its phylogenetic relationship to other tymoviruses was also analyzed.
Total RNA was extracted using RNA Plant Reagent (Invitrogen, Carlsbad, CA, USA) from tomato plants infected with the ToBMV SC50 isolate. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen) according to a standard protocol with the specific reverse primer ToBMV-5492 Rev (5′-GGA TGT CTC TGG CCT TTT GT-3′), which anneals upstream of the cp gene. A degenerate forward primer at the 5′ end (Tymo 526 For, 5′-ATG CAC GAY GCB CTS ATG TA-3′) was designed based on a genome sequence alignment of tymoviruses infecting solanaceous hosts. After amplification by RT-PCR using LongAmp Taq DNA polymerase (New England Biolabs, NEB, Ipswich, MA, USA), the resulting cDNA fragments (ca. 5 kb) were cloned into the pCR4 TOPO plasmid vector (Invitrogen), and four selected clones were sequenced at Macrogen Inc. (Seoul, Korea). The 5′ end of the genome was determined using the 5′ RACE procedure. The first-strand cDNA was synthesized using the same total RNA described above and the specific reverse primer ToBMV-1283 Rev (5′-GAG CGA AGG TTT GTA GAT TGT C-3′). cDNA products were treated with RNaseH/RNaseA (NEB), and the 3′ end of the cDNA was polycytidylated using terminal transferase (NEB). The subsequent 5′ RACE steps, including nested PCR, were performed using specific reverse primers (GSP1-872-Rev, 5′-GAA TGA GAA GAG AGT GGA CAG G-3′, and GSP2-691-Rev, 5′-AAT CTG TGG GAA GAG AGA CAG A-3′) together with forward primers (Oligo dGI-primer, 5′-AAA CCA CGA CAC CTC CAA GCA AGG GGI IGG GII GGG IIG-3′ and then, the anchor primer, 5′-ACC ACG ACA CCT CCA AGC AAG-3′) and LongAmp Taq DNA polymerase (NEB). The cDNA fragments of the 5′ end were cloned into pGEM-T Easy Vector (Promega, Madison, WI, USA), and four selected clones were sequenced at Macrogen Inc. The complete genome sequence was assembled using the Staden Package [7]. ORFs were predicted using Geneious Software (http://www.geneious.com), and the polyprotein (ORF 1) cleavage sites were determined according to Jakubiec et al. [8] by the alignment of aa sequences of ToBMV, ChiYMV (FN563123), EMV (J04374), turnip yellow mosaic virus (TYMV) (X07441), and kennedya yellow mosaic virus (KYMV) (D00637). The aa domains of the polyprotein were analyzed using the InterPro program (http://www.ebi.ac.uk/interpro). Phylogenetic analysis was performed using the MEGA 6.0 program [9] with the maximum-likelihood method and the general time-reversible model. The complete genome sequence was deposited in GenBank under the accession number KC 840043. The distance plot of ToBMV and other related tymoviruses was drawn using the RDP3 program [10].
The genome organization was inferred using Geneious software. ORF 1 (nt 152–5578) encodes a polyprotein that has a molecular mass of ca. 201 kDa and contains the metyltransferase, protease, NTPase-helicase and RNA-dependent RNA polymerase (RdRp) domains predicted by InterPro. ORF 2 (nt 145–711), which overlaps almost entirely with ORF 1, encodes the putative movement protein (ca. 21 kDa). ORF 3 (nt 5583–6155) encodes the CP (ca. 20 kDa). The 5′ UTR has 144 nt and the 3′ UTR is 121 nt long with a TLS (tRNA-like structure) as described previously [2]. The 5′ UTR sequence of the ToBMV SC50 isolate appears to start with the nucleotide U (T); however, the possibility that it starts with G cannot be excluded due to the method that we used (5′RACE as described above). The genomes of two tymoviruses that infect solanaceous plants have also been reported to start with U—tomato yellow blotch virus (EU779803) and physalis mottle virus (Y16104)—although the genomes of most tymoviruses start with G. The polyprotein cleavage sites were predicted by the alignment of polyprotein aa sequences and identified as described [8]. The cleavage site of protease/NTPase-helicase was predicted to be within the aa sequence GS/LP (aa position 842/843 of the polyprotein), and that of NTPase-helicase/RdRp, between VAG/QSP (aa position 1217/1218).
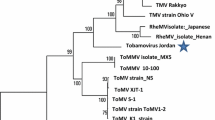
A phylogenetic analysis was performed using the complete genome sequences of all tymoviruses (Fig. 1). ToBMV was closely related to Andean potato latent virus (APLV) isolated from potato plants in Colombia, sharing 72.1 % nt sequence identity. The ToBMV clustered with the tymoviruses infecting solanaceous hosts, such as APLV, Andean potato mild mosaic virus (APMMV), ChiYMV, and EMV. Recently, a near-full-length genome sequence of a tymovirus infecting a tobacco plant was reported and shown to be a strain of ToBMV [11]. The tobacco isolate of ToBMV (KJ940970), which was obtained in 1986 [3], shares 88 % nt sequence identity with this ToBMV tomato isolate, which was obtained in 2010. Various factors, such as the elapsed time, host adaptation, and geographical origin may have contributed to the sequence divergence observed between these two isolates of ToBMV.
Phylogenetic tree of tymoviruses based on the nucleotide sequences of the complete genome. The maximum-likelihood method with the general time-reversible model was used to construct the tree. The branches corresponding to partitions that were reproduced in more than 70 % of bootstrap replicates (500 replications) are shown
A distance plot was drawn using both ToBMV sequences (SC50 from tomato and BR001 from tobacco), the most closely related (Andean potato latent virus—JX508291) and the most distantly related tymovirus (EMV—J04374) that infect solanaceous plants (Fig. 2). Based upon this analysis, the overlapping region of ORF 1 and ORF 2 is highly conserved, whereas the protease domain of the polyprotein is highly variable. The helicase domain and the C-terminal region of RdRp are also conserved, but the CP is moderately variable. Comparing the SC50 tomato isolate and the tobacco isolate of ToBMV, the most divergent regions were in the protease and the RdRp gene regions. These two viruses differ strikingly in their host response in tobacco plants, as the tomato isolate is not able to infect Nicotiana tabacum cv. TNN, in which the tobacco isolate causes a systemic infection. A virus evolution study on the host adaptation of these isolates might be interesting and should include the analysis of more isolates and the use of reverse genetics to identify the determinants of host specificity.
Distance plot of tymoviruses infecting solanaceous hosts. The complete genome of ToBMV tomato isolate SC50 was used as the base line. ToBMV tobacco isolate BR001, one isolate of Andean potato latent virus (JX508291), which represents a closely related virus, and one isolate of EMV as a more distantly related virus (J04374), were used for building this plot. The figures of the genome and the distance plot are to scale. Met, methyltransferase; Pro, protease; Hel, helicase; RdRp, RNA-dependent RNA polymerase; MP, movement protein; CP, coat protein
References
Dreher TW, Edwards MC, Gibbs AJ, Haenni A-L, Hammond RW, Jupin I, Koenig R, Sabanadzovic S, Martelli GP (2012) Family Tymoviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB (eds) Virus taxonomy, 9th report of the International Committee on Taxonomy of Viruses. Elsevier/ Academic Press, San Diego, USA, pp 944–952
Oliveira VC, Nagata T, Guimarães FC, Ferreira FA, Kitajima EW, Nicolini C, Resende RO, Inoue-Nagata AK (2013) Characterization of a novel tymovirus on tomato plants in Brazil. Virus Genes 46:190–194
Ribeiro SG, Kitajima EW, Oliveira CRB, Koenig R (1996) A strain of eggplant mosaic virus isolated from naturally infected tobacco plants in Brazil. Plant Dis 80:446–449
Alexandre MAV, Duarte LML, Rivas EB, Chagas CM, Barradas MM (2000) Petunia vein banding virus: characterization of a new tymovirus from Petunia × hybrida. Plant Dis 84(7):739–742
Crestani OA, Kitajima EW, Lin MT, Marinho VLA (1986) Passion fruit yellow mosaic virus, a new tymovirus found in Brazil. Phytopathology 76:951–955
Nicolini C, Pio-Ribeiro G, Andrade GP, Melo FL, Oliveira VC, Guimarães FC, Resende RO, Kitajima EW, Resende JAM, Nagata T (2012) A distinct tymovirus infecting Cassia hoffmannseggii in Brazil. Virus Genes 45:190–194
Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5:233–241
Jakubiec A, Drugeon G, Camborde L, Jupin I (2007) Proteolytic processing of turnip yellow mosaic virus replication proteins and functional impact on infectivity. J Virol 81(20):11402–11412
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463
Melo FC, Fernandes JEA, Ribeiro BM, Ribeiro SG (2014) Complete genome sequence of a tobacco-infecting tomato blistering mosaic virus. Genome Announc 2:e00701–e00714
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nicolini, C., Inoue-Nagata, A.K. & Nagata, T. Complete genome sequence of a proposed new tymovirus, tomato blistering mosaic virus. Arch Virol 160, 609–612 (2015). https://doi.org/10.1007/s00705-014-2289-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-014-2289-7