Abstract
Human parechoviruses (HPeVs) are widespread pathogens causing a wide spectrum of diseases. The prevalence and genetic diversity of HPeV in children with acute diarrhea in China is not well known. The purpose of this study was to investigate the epidemiological characteristics of HPeV in Guangzhou, China. A total of 328 stool specimens collected from children under the age of 5 years with acute diarrhea were tested for the presence of HPeV. Of these, 44 (13.4 %, 44/328) were HPeV positive, with the majority of the infected children (97.7 %, 43/44) being younger than two years of age. HPeV was more frequently detected during July and August. The epidemiological profile of co-infections was similar to that observed in a previous study. Six different HPeV genotypes, including HPeV1, -3, -4, -5, -6, and -14, were identified, and of these, HPeV14, a rarely reported genotype, was reported for the first time in children with acute gastroenteritis in China. In summary, this study clearly demonstrated that HPeV circulating in Guangzhou, China, is genetically diverse, including six genotypes, and it provides useful epidemiological data on the features of HPeV infection in this area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human parechoviruses (HPeVs) are members of the large and growing family Picornaviridae [1, 2]. Sixteen HPeV genotypes have been described worldwide [3–10]. Infections with HPeVs are widespread, but different HPeV genotypes have different epidemiological and clinical characteristics.
HPeV1 is believed to be a common pathogen that mainly affects young children with mild gastrointestinal or respiratory symptoms, whereas HPeV2 infections rarely occur and are mostly associated with gastrointestinal symptoms [6, 7, 11, 12]. HPeV3 infection causes more-severe diseases, such as sepsis or sepsis-like illness and meningitis or encephalitis [13–16]. Although HPeV genotypes 4–16 have been identified in a number of different countries [17–23], their clinical role remains to be clarified.
In China, epidemiological surveillance of HPeV infection in children with diarrhea has been reported since 2009 [24]. Little is known about this viral agent causing acute gastroenteritis in China, except for two studies of HPeV detection in acute gastroenteritis in infants and children less than 5 years old [25, 26]. Thus, the role and effect of HPeV infection in acute gastroenteritis is unclear.
The aims of this study were to investigate the prevalence and distribution of HPeV in Guangzhou, China, by direct screening and typing of stool samples from children with acute diarrhea under the age of 5 years during the 2012 to 2013 season, and to characterize the circulating strains by sequence and phylogenetic analysis.
Materials and methods
Sample collection
Three hundred twenty-eight stool specimens were collected from children under 5 years of age with diarrhea who were being treated as outpatients. Stool samples analyzed in this study were collected from June 2012 through May 2013 and stored at −20 °C until further analysis. All collected specimens were also screened for other diarrheal viruses including human group A rotavirus, norovirus (NoV), astrovirus, sapovirus (SaV) and adenovirus [27–30]. Informed consent was obtained from the parents of all children who provided specimens. The study protocol was approved by the ethics committees of the School of Public Health and Tropical Medicine of Southern Medical University.
RNA extraction and parechovirus testing
Total RNA was extracted from supernatants of 10 % (w/v) of stool specimens in PBS using TRIzol (Invitrogen, Carlsbad, CA) as described in the manufacturer’s instructions. Extracted RNA was stored at −80 °C and reverse transcribed into cDNA using a reverse transcription system (Promega, Madison, WI). cDNA was either used directly for PCR or stored at −20 °C.
Testing for HPeV was carried out using highly conserved 5’ untranslated region (5’-UTR) primers [31] under the following reaction conditions: 94 °C for 1 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final incubation at 72 °C for 7 min. Positive samples producing a amplicon with a predicted size of 243 bp were identified by visualization under UV light after 2 % agarose gel electrophoresis and ethidium bromide staining.
Parechovirus genotyping and phylogenetic analysis
For parechovirus genotyping, samples testing positive by PCR of the 5’-UTR were amplified by nested PCR using primers from the VP3/VP1 junction region as described elsewhere [31]. The two rounds of amplification were performed under the following cycling conditions: 40 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s, and a final incubation at 72 °C for 7 min. The final products of 304 bp were identified by agarose gel electrophoresis as described above and were purified by using a QIAquick kit (QIAGEN).
Sequencing was carried out using a Big-Dye Terminator Cycle Sequencing Kit and an ABI 3730 XL DNA Analyzer (Applied Biosystems). The sequences were compared with those of reference strains available in the NCBI GenBank database using the BLAST server (http://www.ncbi.nlm.nih.gov/blast). Sequences were assembled with Sequencher software (version 4.6, Gene Codes Corp., Ann Arbor, MI), and genetic identity was determined by comparing the sequence with standard strains in GenBank. Multiple sequence alignment was conducted using ClustalW [32], and a phylogenetic tree was constructed from partial nucleotide and deduced amino acid sequences of the VP3/VP1 region using MEGA version 5.0 (http://www.megasoftware.net/) [33].
Nucleotide sequence accession numbers
Partial nucleotide sequences of HPeV VP3/VP1 junction region identified in the present study were deposited in the GenBank database. The accession numbers are KF489908 - KF489951.
Results
Study group
The female-to-male gender ratio was 0.84 (150/178). The age of children with acute gastroenteritis ranged from 10 days to 58 months, with a mean age of 11 months, and the difference in the age distributions of the boys and girls was not statistically significant (P = 0.887, α = 0.05).
Epidemiology of HPeV infections
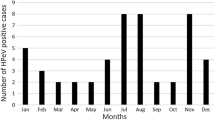
Of the 328 specimens, 44 (13.4 %, 44/328) were HPeV positive. The details of these 44 samples are shown in Table 1. Twenty-four (54.5 %, 24/44) were from boys, and 20 (45.5 %, 20/44) were from girls. Taking into account the gender ratio, the difference in the prevalence of HPeV was not statistically significant (P = 0.968, α= 0.05). The age range of HPeV-positive patients was from 1 month to 48 months (median, 7 months; mean age, 8.97 months); 43.2 % (19/44) were younger than 6 months, 47.7 % (21/44) were 7 to 12 months old, 6.8 % (3/44) were 13 to 24 months old, and 2.3 % (1/44) were 48 months old. HPeV was detected throughout the entire year except February and December, with the peak incidence in July (30.8, 12/39) and August (32.1 %, 9/28) (Fig. 1).
Among of 44 positive samples, co-infection with other enteric viruses was found in 23 (52.3 %) pediatric patients, while monoinfection by HPeV alone was detected in 21 (47.7 %) cases. Coinfection with rotavirus was found most frequently (12 samples, 27.3 %), while another specimens revealed coinfection with norovirus (5 samples, 11.4 %), adenovirus (4 samples, 9.1 %), astrovirus (4 samples, 9.1 %) or sapovirus (1 sample, 2.3 %). HPeV showed an 11.8 % (21/178) positive rate in stool samples that were negative for all of the other five enteric viruses.
Phylogenetic analysis
For genotyping using nucleotide sequences, all HPeV-positive samples were successfully amplified and sequenced. Phylogenetic analysis of the VP1/VP3 region of reference HPeV strains and the isolates studied here showed that the strains could be identified as HPeV1 (32 strains), HPeV3 (4 strains), HPeV4 (2 strains), HPeV6 (4 strains), and HPeV14 (1 strain) (Fig. 2). HPeV1 was the predominant genotype, and more than half of the viruses identified in this study clustered with the prototype Harris strain. Of the 32 HPeV1 isolates, one (GZ-684) was divergent from the rest of the group. The divergence was 10.4 % and 23.4 %, respectively, when compared with the VP3/VP1 junction region of HPeV1A (SH 401, JX441355) and the remaining HPeV1B clade. Since the divergence for a new genotype assignment has to exceed 25 %, the GZ-684 strain was clustered with HPeV1. HPeV1 isolates could be grouped into two clusters of Harris-like clade 1A and clade 1B [34]. Therefore, the isolate GZ-684 was assigned to genotype 1A.
Phylogenetic analysis based on the nucleotide sequence of the VP3/VP1 junction region. The tree was constructed using the neighbor-joining method with the p-distance model. A bootstrap test was replicated 1000 times, and only bootstrap values over 70 are shown. A sample (GZ-990) that could not be assigned to a specific HPeV cluster is indicated by a black circle
Notably, isolate GZ-990 was segregated to a single branch that could not be assigned to a specific HPeV cluster (Fig. 2). GZ-990 had highest nucleotide sequence identity, 77.5 %, to the prototype strain HPeV5 CT86-6760 (AF055846). The second-best match was 73.9 % identity to HPeV4 K251176-02(DQ315670). Importantly, the pairwise distances (0.192–0.298) between isolate GZ-990 and other reference strains was over the threshold of 18 % divergence for nucleotides for the division of intra- and intertype categories [31]. When amino acid sequences were analyzed, isolate GZ-990 was characterized as HPeV type 5 (the pairwise distance between this isolate and HPeV5 CT86-6760 was 0.051, lower than the 8 % divergence threshold) (Fig. 3).
Phylogenetic analysis based on the deduced amino acid sequence of the VP3/VP1 junction region The tree was constructed using the neighbor-joining method with the p-distance model. A bootstrap test was replicated 1000 times, and only bootstrap values over 70 are shown. A sample (GZ-990) that could not be assigned to a specific HPeV cluster is indicated by a black circle
Discussion
Human parechoviruses (HPeVs) have been classified into sixteen serotypes based on neutralization test or molecular identification. Clinical manifestations that have been associated with HPeV infections include gastroenteritis, respiratory diseases, aseptic meningitis, lymphadenopathy, myocarditis, sepsis-like syndromes with necrotizing enterocolitis, and sudden infant death syndrome (SIDS), notably in young children. [15, 17, 19, 35–43]. Epidemiological studies of HPeV are required to further our understanding of disease causation and geographic distribution.
We tested stool specimens collected from children less than 5 years old for HPeV between 2012 and 2013 in Guangzhou, China. The percentage of HPeV-positive specimens (13.4 %) was lower than that found in other studies in China (55 % in Shanghai [25] and 25.3 % in Lanzhou [26]). The discrepancy might be due to whether the samples were negative for other gastroenteritis-associated viruses, the geographical location, the specific year of sample collection, or the use of different PCR methods.
Nearly all of the infections (97.7 %) occurred in the first 2 years of life. This age distribution is consistent with previous studies [7, 8, 44, 45]. The gender-neutral distribution of HPeV infection is also in agreement with studies from Thailand [32], Lanzhou [26] and Shanghai, China [25]. In Guangzhou, HPeV infection had a distinct seasonal distribution that peaked during July and August. Similar seasonal distributions were also found in Shanghai, China [25].
Coinfection with other enteric viruses was found in 23 (52.3%) pediatric patients. A 52.3 % coinfection rate, together with the previously reported 64.4 % from Lanzhou [26], suggests that coinfection is common for HPeV. Rotavirus was the most common partner. By screening fecal samples known to be negative for common enteric viruses, this study provides one more piece of evidence that HPeV infection is associated with acute gastroenteritis. With a detection rate of 11.8 % (21/178) among the samples tested that were negative for common enteric viruses, this study demonstrates that HPeV-related diarrhea among children with acute gastroenteritis is not rare in Guangzhou, China. Comparing our results to those from two other studies, which have shown detection rates of 2.0 [12] and 8.1 % [8], respectively, the difference in detection rate might well be due to study populations and geographical locations.
In this study, six different HPeV genotypes, HPeV1,-3, -4, -5,-6, and -14, were present in infants and children with acute gastroenteritis in the Guangzhou region. The HPeV1 genotype was predominant over other genotypes, which is consistent with the fact that HPeV1 is the major genotype worldwide [14, 46–52]. In our study, most of the HPeV1 isolates (56.25 %, 18/32) clustered with HPeV1A, which has seldom been found in recent years [7, 45, 53]. Changes in the rate of HPeV1A infection in Guangzhou could not be investigated due to the lack of data from previous years; however, this study could confirm that genotype HPeV1A was circulating in Guangzhou, China.
The inconsistent assignment of isolate GZ-990 in phylogenetic trees based on nucleotide and amino acid sequences was due to the fact that most of the differences in the nucleotide sequences between GZ-990 and HPeV5 reference sequence occurred at synonymous sites. In this research, infection with HPeV8, which was detected in other areas of China, was not detected, indicating that HPeV8 is most likely absent in Guangzhou. More studies are needed to confirm the occurrence for HPeV8 in China. It is noteworthy that HPeV14, a rarely reported genotype, was detected unexpectedly in this study. As far as we know, this is the first report of the presence of HPeV14 in China to date.
In conclusion, with the identification of HPeV in fecal specimens, six different genotypes were found. In addition, this is the first report of HPeV14 infection in patients with acute gastroenteritis in China. This study provides useful epidemiological data for future disease control and prevention by documenting the distribution of genotypes, as well as age and seasonal patterns in Guangzhou, China.
References
Stanway G, Joki-Korpela P, Hyypia T (2000) Human parechoviruses–biology and clinical significance. Rev Med Virol 10:57–69
Wigand R, Sabin AB (1961) Properties of ECHO types 22, 23 and 24 viruses. Arch Gesamte Virusforsch 11:224–247
Kim PN, Trinh QD, Takanashi S, Abeysekera C, Abeygunawardene A et al (2010) Novel human parechovirus, Sri Lanka. Emerg Infect Dis 16:130–132
Drexler JF, Grywna K, Stocker A, Almeida PS, Medrado-Ribeiro TC et al (2009) Novel human parechovirus from Brazil. Emerg Infect Dis 15:310–313
Joki-Korpela P, Hyypia T (2001) Parechoviruses, a novel group of human picornaviruses. Ann Med 33:466–471
Pham NT, Takanashi S, Tran DN, Trinh QD, Abeysekera C et al (2011) Human parechovirus infection in children hospitalized with acute gastroenteritis in Sri Lanka. J Clin Microbiol 49:364–366
Ito M, Yamashita T, Tsuzuki H, Kabashima Y, Hasegawa A et al (2010) Detection of human parechoviruses from clinical stool samples in Aichi, Japan. J Clin Microbiol 48:2683–2688
Pham NT, Trinh QD, Khamrin P, Maneekarn N, Shimizu H et al (2010) Diversity of human parechoviruses isolated from stool samples collected from Thai children with acute gastroenteritis. J Clin Microbiol 48:115–119
Harvala H, Simmonds P (2009) Human parechoviruses: Biology, epidemiology and clinical significance. J Clin Virol 45:1–9
Baumgarte S, de Souza LL, Grywna K, Panning M, Drexler JF et al (2008) Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis. J Clin Microbiol 46:242–248
Piralla A, Furione M, Rovida F, Marchi A, Stronati M et al (2012) Human parechovirus infections in patients admitted to hospital in Northern Italy, 2008–2010. J Med Virol 84:686–690
Han TH, Kim CH, Park SH, Chung JY, Hwang ES (2011) Detection of human parechoviruses in children with gastroenteritis in South Korea. Arch Virol 156:1471–1475
Boivin G, Abed Y, Boucher FD (2005) Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11:103–105
Abed Y, Boivin G (2006) Human parechovirus infections in Canada. Emerg Infect Dis 12:969–975
Boivin G, Abed Y, Boucher FD (2005) Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11:103–105
Ito M, Yamashita T, Tsuzuki H, Takeda N, Sakae K (2004) Isolation and identification of a novel human parechovirus. J Gen Virol 85:391–398
Al-Sunaidi M, Williams CH, Hughes PJ, Schnurr DP, Stanway G (2007) Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains. J Virol 81:1013–1021
Pajkrt D, Benschop KS, Westerhuis B, Molenkamp R, Spanjerberg L et al (2009) Clinical characteristics of human parechoviruses 4–6 infections in young children. Pediatr Infect Dis J 28:1008–1010
Watanabe K, Oie M, Higuchi M, Nishikawa M, Fujii M (2007) Isolation and characterization of novel human parechovirus from clinical samples. Emerg Infect Dis 13:889–895
van der Sanden S, de Bruin E, Vennema H, Swanink C, Koopmans M et al (2008) Prevalence of human parechovirus in the Netherlands in 2000 to 2007. J Clin Microbiol 46:2884–2889
Oberste MS, Feeroz MM, Maher K, Nix WA, Engel GA et al (2013) Characterizing the picornavirus landscape among synanthropic nonhuman primates in Bangladesh, 2007 to 2008. J Virol 87:558–571
Williams CH, Panayiotou M, Girling GD, Peard CI, Oikarinen S et al (2009) Evolution and conservation in human parechovirus genomes. J Gen Virol 90:1702–1712
Benschop KS, Williams CH, Wolthers KC, Stanway G, Simmonds P (2008) Widespread recombination within human parechoviruses: analysis of temporal dynamics and constraints. J Gen Virol 89:1030–1035
Shan TL, Guo W, Cui L, Shang XG, Dai XQ et al (2009) The first detection of human parechovirus infections in China. J Clin Virol 45:371–372
Zhong H, Lin Y, Sun J, Su L, Cao L et al (2011) Prevalence and genotypes of human parechovirus in stool samples from hospitalized children in Shanghai, China, 2008 and 2009. J Med Virol 83:1428–1434
Zhang DL, Jin Y, Li DD, Cheng WX, Xu ZQ et al (2011) Prevalence of human parechovirus in Chinese children hospitalized for acute gastroenteritis. Clin Microbiol Infect 17:1563–1569
Ren L, Gonzalez R, Xiao Y, Xu X, Chen L et al (2009) Saffold cardiovirus in children with acute gastroenteritis, Beijing, China. Emerg Infect Dis 15:1509–1511
Gabbay YB, Linhares AC, Oliveira DS, Nakamura LS, Mascarenhas JD et al (2007) First detection of a human astrovirus type 8 in a child with diarrhea in Belem, Brazil: comparison with other strains worldwide and identification of possible three lineages. Mem Inst Oswaldo Cruz 102:531–534
Boxman IL, Tilburg JJ, Te LN, Vennema H, Jonker K et al (2006) Detection of noroviruses in shellfish in the Netherlands. Int J Food Microbiol 108:391–396
Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H (2003) Detection of norovirus (GI, GII), Sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J Virol Methods 114:37–44
Harvala H, Robertson I, McWilliam LE, Benschop K, Wolthers KC et al (2008) Epidemiology and clinical associations of human parechovirus respiratory infections. J Clin Microbiol 46:3446–3453
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Benschop KS, de Vries M, Minaar R et al (2009) Comprehensive full length sequence analyses of human parechoviruses: Diversity and recombination. J Gen Virol 91:145–154
Benschop KS, Schinkel J, Luken ME, van den Broek PJ, Beersma MF et al (2006) Fourth human parechovirus serotype. Emerg Infect Dis 12:1572–1575
Sainato R, Flanagan R, Mahlen S, Fairchok M, Braun L (2011) Severe human parechovirus sepsis beyond the neonatal period. J Clin Virol 51:73–74
Ren L, Xiang Z, Guo L, Wang J (2012) Viral infections of the lower respiratory Tract. Curr Infect Dis Rep 14(3):284–291
Harvala H, McLeish N, Kondracka J, McIntyre CL, McWilliam LE et al (2011) Comparison of human parechovirus and enterovirus detection frequencies in cerebrospinal fluid samples collected over a 5-year period in edinburgh: HPeV type 3 identified as the most common picornavirus type. J Med Virol 83:889–896
Selvarangan R, Nzabi M, Selvaraju SB, Ketter P, Carpenter C et al (2011) Human parechovirus 3 causing sepsis-like illness in children from midwestern United States. Pediatr Infect Dis J 30:238–242
Wakatsuki K, Kawamoto D, Hiwaki H, Watanabe K, Yoshida H (2008) Identification and characterization of two strains of human parechovirus 4 isolated from two clinical cases in Fukuoka City, Japan. J Clin Microbiol 46:3144–3146
Verboon-Maciolek MA, Krediet TG, Gerards LJ, de Vries LS, Groenendaal F et al (2008) Severe neonatal parechovirus infection and similarity with enterovirus infection. Pediatr Infect Dis J 27:241–245
Jokela P, Joki-Korpela P, Maaronen M, Glumoff V, Hyypia T (2005) Detection of human picornaviruses by multiplex reverse transcription-PCR and liquid hybridization. J Clin Microbiol 43:1239–1245
van Zwol AL, Lequin M, Aarts-Tesselaar C, van der Eijk AA, Driessen GA, et al. (2009) Fatal neonatal parechovirus encephalitis. BMJ Case Rep 2009
Chieochansin T, Vichiwattana P, Korkong S, Theamboonlers A, Poovorawan Y (2011) Molecular epidemiology, genome characterization, and recombination event of human parechovirus. Virology 421:159–166
Tapia G, Cinek O, Witso E, Kulich M, Rasmussen T et al (2008) Longitudinal observation of parechovirus in stool samples from Norwegian infants. J Med Virol 80:1835–1842
Chaimongkol N, Khamrin P, Suantai B, Saikhreang W, Thongprachum A et al (2012) A wide variety of diarrhea viruses circulating in pediatric patients in Thailand. Clin Lab 58:117–123
Chen BC, Cheng MF, Huang TS, Liu YC, Tang CW et al (2009) Detection and identification of human parechoviruses from clinical specimens. Diagn Microbiol Infect Dis 65:254–260
Krogerus C, Egger D, Samuilova O, Hyypia T, Bienz K (2003) Replication complex of human parechovirus 1. J Virol 77:8512–8523
Benschop KS, de Vries M, Minnaar RP, Stanway G, van der Hoek L et al (2010) Comprehensive full-length sequence analyses of human parechoviruses: diversity and recombination. J Gen Virol 91:145–154
Benschop K, Minnaar R, Koen G, van Eijk H, Dijkman K et al (2010) Detection of human enterovirus and human parechovirus (HPeV) genotypes from clinical stool samples: polymerase chain reaction and direct molecular typing, culture characteristics, and serotyping. Diagn Microbiol Infect Dis 68:166–173
Williams CH, Panayiotou M, Girling GD, Peard CI, Oikarinen S et al (2009) Evolution and conservation in human parechovirus genomes. J Gen Virol 90:1702–1712
Calvert J, Chieochansin T, Benschop KS, McWilliam LE, Drexler JF et al (2010) Recombination dynamics of human parechoviruses: investigation of type-specific differences in frequency and epidemiological correlates. J Gen Virol 91:1229–1238
Abed Y, Boivin G (2009) Molecular characterization of viruses from clinical respiratory samples producing unidentified cytopathic effects in cell culture. Viruses 1:84–90
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, H., Yao, Y., Liu, X. et al. Molecular detection of human parechovirus in children with acute gastroenteritis in Guangzhou, China. Arch Virol 159, 971–977 (2014). https://doi.org/10.1007/s00705-013-1915-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1915-0