Abstract
Tobacco rattle virus from a Hosta hybrid contained one RNA1 (Ho-1) and two RNA2 species (Ho-2a, Ho-2b). Whereas Ho-1 resembles TRV Al RNA1 from Alstroemerias, Ho-2a and Ho-2b resemble TRV TpO1 RNA2 from a potato field. Ho-2a has a complete RNA2-specific sequence, whereas that of Ho2-b carries a large deletion. The short RNA1-related 3’ end of Ho-2a is distinct from that of Ho-1, whereas the longer one of Ho-2b is identical to that of Ho-1. TRV RNA2 molecules may apparently become associated with different TRV RNA1 molecules, from which they can acquire 3’ends of various lengths while often losing large portions of their RNA2-specific sequences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Tobacco rattle virus (TRV) is the type species of the genus Tobravirus [1]. These viruses have bipartite genomes, but due to deletions, recombinations and reassortments, more than one RNA2 species may be present in infected plants [7, 8, 12]. TRV RNA1 contains two 5’ overlapping genes for 134- and 194-kDa replication-associated proteins and two smaller genes for a 29-kDa movement protein and a 16-kDa silencing suppressor [1]. Tobravirus RNA2 molecules consist of a 5’ RNA2-specific region and a 3’ RNA1-related region [2, 7]. Both regions may differ considerably in size and in sequence in different isolates. The 5’ RNA2-specific region contains the coat protein gene and up to three additional ORFs further downstream. The gene product of ORF 2b and with some tobraviruses those of all four ORFs are required for nematode transmission [1]. An unusual RNA2 composition has recently been described for the TRV SYM strain [3]. The 3’ RNA1-related region of tobravirus RNA2 molecules may be short and consist of only c. 250 nucleotides (nt) [4, 8], but it may also be much longer and contain, in addition to the 3’ untranslated region, partial or complete 3’ coding sequences of a tobravirus RNA1 [8, 9]. In the present paper, we describe the genome properties of a recombinant TRV strain (TRV Ho) found in the vegetatively propagated Hosta hybrid ‘Green Fountain’.
In the infected Hosta leaves, which showed necrotic spots, leaf distortions and vein banding, one TRV RNA1 species (Ho-1) and two TRV RNA2 species (Ho-2a and Ho-2b) were found, using the same methods as described previously [8]. Ho-1 is closely related to TRV Al RNA1 from infected Alstroemerias [8]. The complete sequences of Ho-1 and TRV Al RNA1 differ at only 0.6 % of their nucleotide positions, whereas the differences to other previously described TRV RNA1 molecules range from 6.8 to 9.1 % (Table 1). The somewhat deviating type of TRV RNA1 represented by Ho-1 and TRV Al RNA1 may be more widespread than presently recognized. We have identified short sequence portions that shared 99.9 to 100 % sequence identity with Ho-1 and TRV Al RNA1 also in TRV-infected garlic and potatoes (R. Koenig, unpublished). The percentages of nt sequence differences found in individual ORFs mostly resembled those found with the total RNAs. Differences between the various ORFs were observed, however, in the deduced amino sequences (Table 1). In the coding regions for the replicase and movement proteins, the differences between virus isolates were much larger at the nt level than at the amino acid sequence level, because many of the nt differences occurred at codon positions where they did not lead to amino acid differences. This suggests that a strong selective pressure preserves the protein structure. The coding regions for the 16 K silencing suppressor and a potential 13-kDa protein of unknown function, however, contained an increased number of nt substitutions in codon positions where they did lead to amino acid changes. This indicates a higher flexibility of these proteins.
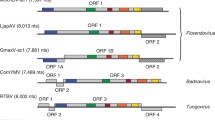
Ho-2a is closely related to TRV TpO1 RNA2 isolated from Trichodorus primitivus in a potato field in England [10]. The two RNA2 molecules have the same size and the same arrangement of four RNA2-specific genes (Fig. 1a), and their RNA 2-specific regions share 99.1 % sequence identity. The much shorter RNA2-specific region of Ho-2b shares 100 % sequence identity with the corresponding regions in Ho-2a and 98.8 % identity with the RNA2-specific region of TRV Ros RNA2 isolated from potatoes in Germany [6]. Ho-2b and TRV Ros RNA2 appear to be deletion mutants that have lost large portions of their coding sequences downstream of their coat protein genes (Fig. 1a). Only low percentages of sequence identity ranging from 50 to 66 % were observed between the RNA2-specific regions of TRV Ho and those of other TRV strains, e.g., TRV PaY4, Al(=TCM), ORY, Mich, PpK20 and PLB (results not shown).
Comparisons of TRV RNA2 molecules from Hosta and other TRV RNA molecules. a Molecular organization of TRV Ho-2a, Ho-2b and the RNA2 molecules of TRV TpO1 and TRV Ros. Equal shading indicate identical or highly similar sequences, different shading indicate the existence of significant sequence differences. b Alignment of the RNA1-like 3’ ends of TRV Ho-2a and TpO1 RNA2 and of the 3’ ends of TRV SYM and PpK20 RNA1. Only those positions are shown where differences occur. Nts which differ from those in the TRV SYM and PpK20 sequences are highlighted by white letters on a black background. The arrow indicates where the sequence of TRV TPO1 RNA2 stops to be identical to that of Ho-2a and starts to be identical to the sequences of TRV SYM and PpK20 RNA1
Whereas the RNA2-specific regions of all four TRV RNA2 molecules shown in Fig. 1a are-deletions excluded-closely related or identical, their RNA1-related regions are all distinct. Ho-2b and TRV Ros RNA2, which carry large deletions in their RNA2-specific regions, have longer RNA1-related 3’ ends than Ho-2a and TRV TpO1 RNA2, which have complete RNA2-specific regions (Fig. 1a). The sequences of the RNA1-related regions of Ho-2a, Ho-2b and TRV Ros RNA 2 share only c. 91 to 93 % identity with each other. Interestingly, however, the 3’ end of Ho-2b shows 100 % sequence identity with the 3’ end of the supporting Ho-1, whereas that of Ho-2a is clearly distinct. A similar situation was encountered previously in Alstroemeria, where several small TRV Al RNA2 variants with partially deleted RNA2-specific sequences have rather long 3’ ends derived from the supporting TRV Al RNA1. The TRV Al RNA2 variant TC3’PE-a, with its full-length RNA2-specific region, however, has only a short RNA1-related 3’ end that differs greatly from that of the supporting TRV Al RNA1 [8]. The closely related Ho-1 and TRV Al RNA1 (Table 1) have recombined in Hosta and Alstroemeria with very different TRV RNA2 species, i.e., the TPO1-like Ho-2a (Fig. 1a) and TRV Al RNA2 [8], respectively. These two RNAs have a different genetic organization and show only 56 % sequence identity [8, 9]. Interestingly, in other hosts, these two RNA2 species have recombined with different RNA1 species (Fig. 1a and [8]). There is only one previous report that partially deleted forms of the same TRV RNA2 have recombined with the 3’ ends of different TRV RNA1 molecules [2].
In Ho-2a and TRV TpO1 RNA2, the RNA1-related sequences of the first 192 nt downstream of their RNA2-specific regions are identical, but they are quite distinct from the 3’ ends of the RNA1 molecules of TRV SYM, PpK20 and other TRV strains (Fig. 1b). Starting with nt 3082, however, the sequence of TRV TpO1 RNA2 becomes identical to those of TRV SYM and PpK20 RNA1, whereas the sequence of Ho-2a continues to be distinct (Fig. 1b). This suggests that an ancestral form of TRV TpO1 RNA2 may have acquired a small portion of a TRV SYM-like RNA1. Unfortunately, no sequence information is available for TRV TpO1 RNA1, and this isolate is no longer available (S. MacFarlane, personal communication). Whereas with most other TRV RNA2 molecules the acquisition of the 3’ end of another RNA1 species is accompanied by deletions in their RNA2-specific regions, such deletions have obviously not occurred with TpO1 RNA2, and also not with TRV PaY4 RNA2 [9]. The opposite situation was observed with the TRV AL RNA2 molecules TC3’PE-b and -c, where deletions in the RNA2-specific regions were not accompanied by the acquisition of 3’-terminal sequences of the supporting RNA1 [8].
As in the case of the TRV Al RNA 2 recombinants [8], an AU content of more than 50 % was found in the sequences of the 50 nt upstream and downstream of the recombination sites in Ho-2b and TRV Ros RNA 2. In the sequences of 100 nt around the recombination sites, conserved secondary structure elements were not predicted for the recombinants or the parental molecules when using two different folding programs [5, 13].
The ability of tobravirus RNA2 molecules to become associated with different TRV RNA1 molecules, to acquire differently-sized portions of the 3’ ends of these RNA1 molecules, and to discard portions of their own RNA2-specific regions may serve as a basis for the pronounced capability of tobraviruses to adapt themselves to new hosts and, in a given host, also to the varying conditions in different infection stages. In the early stages of an infection when the virus is transmitted to the roots of a plant by a nematode, the virus will probably need the whole set of proteins encoded on a full-length RNA2, but later during systemic infections in vegetatively propagated plants, several RNA2-specific genes, e.g., those for nematode transmission, may become redundant, and their elimination from the genome might be advantageous for the virus. Although it has been shown that the specific recognition of tobravirus RNA2 molecules by the RNA1-encoded replication enzymes is essentially determined by sequences at the 5’ ends and not the 3’ ends of the RNA2 molecules [11], the presence of identical 3’ ends in the RNA1 and RNA2 molecules might be advantageous for the viral replication process. It is difficult to imagine, however, what kind of advantages could be gained for the virus from the acquisition of total or partial RNA1-derived coding sequences by its RNA2 molecules. The replacement of the RNA1-related 3’ ends of tobravirus RNA2 molecules by exactly matching parts from a new supporting RNA1 can probably be achieved only with difficulty in a highly regulated homologous recombination process. Thus, 3’ ends with additional coding sequences that may be acquired more easily from a new supporting RNA1 species might be tolerated in the recombined RNA2 molecules.
References
Adams MJ, Heinze C, Jackson AO, Kreuze J, MacFarlane SA, Torrance L (2012) Tobravirus. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds), Virus Taxonomy, Ninth Report of the ICTV. Elsevier/Academic Press, London, pp 1156–1162
Angenent GC, Posthumus E, Brederode FT, Bol JF (1989) Genome structure of tobacco rattle virus strain PLB: further evidence on the occurrence of RNA recombination among tobraviruses. Virology 171:271–274
Ashfaq M, McGavin W, MacFarlane SA (2011) RNA2 of TRV SYM breaks the rules for tobravirus genome structure. Virus Res 160:435–438
Goulden MG, Lomonossoff GP, Davies JW, Wood KR (1990) The complete nucleotide sequence of PEBV RNA2 reveals the presence of a novel open reading frame and provides insights into the structure of tobraviral subgenomic promoters. Nucleic Acids Res 18:4507–4512
Gultyaev AP, van Batenburg FHD, Pleij CWA (1995) The computer simulation of RNA folding pathways using a genetic algorithm. J Mol Biol 250:37–51
Heinze C, van Bargen S, Sadowska-Rybak M, Willingmann P, Adam G (2000) Sequences of tobacco rattle viruses (TRV) from potato. J Phytopathol 148:547–554
Hernandez C, Carette JE, Brown DJF, Bol JF (1996) Serial passage of tobacco rattle virus under different selection conditions results in deletion of structural and nonstructural genes in RNA 2. J Virol 70:4933–4940
Koenig R, Lesemann DE, Pfeilstetter E, Winter S, Pleij CWA (2011) Deletions and recombinations with the RNA1 3 ‘ends of different tobraviruses have created a multitude of tobacco rattle virus TCM-related RNA2 species in Alstroemeria and tulip. J Gen Virol 92:988–996
MacFarlane SA (1999) Molecular biology of the tobraviruses. J Gen Virol 80:2799–2807
MacFarlane SA, Vassilakos N, Brown DJF (1999) Similarities in the genome organization of tobacco rattle virus and pea early-browning virus isolates that are transmitted by the same vector nematode. J Gen Virol 80:273–276
Mueller AM, Mooney AL, MacFarlane SA (1997) Replication of in vitro tobravirus recombinants shows that the specificity of template recognition is determined by 5’ non-coding but not 3’ non-coding sequences. J Gen Virol 78:2085–2088
Schmidt K, Koenig R (1999) Genetic analysis of large-sized RNA 2 species of a TCM-like tobacco rattle virus source from spinach. Arch Virol 144:503–511
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res 31:3406–3415
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank accession numbers for TRV Ho-1, Ho-2a and Ho-2b are JQ235203, JQ235204 and JQ235205, respectively. The sequences lack ca. 20 primer-derived nucleotides at their 5’ and 3’ ends.
Rights and permissions
About this article
Cite this article
Koenig, R., Lesemann, DE. & Pleij, C.W.A. Tobacco rattle virus genome alterations in the Hosta hybrid ‘Green Fountain’ and other plants: reassortments, recombinations and deletions. Arch Virol 157, 2005–2008 (2012). https://doi.org/10.1007/s00705-012-1365-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1365-0