Abstract
Vibrio alginolyticus is an opportunistic pathogen of animals and humans; its related strains can also produce tetrodotoxin and hemolysins. A new phage, ϕA318, which lysed its host V. alginolyticus with high efficiency, was characterized. The burst size of ϕA318 in V. alginolyticus was 72 PFU/bacterium at an MOI of 1 at room temperature; the plaque size was as large as 5 mm in diameter. Electron microscopy (EM) of the phage particles revealed a 50- to 55-nm isomorphous icosahedral head with a 12-nm non-contractile tail, similar to the T7-like phages of the family Podoviridae. Phylogenetic analysis based on complete sequences of the DNA-directed RNA polymerase gene revealed that ϕA318 had 28-47% amino acid identity to enterobacteria phages T7 and SP6, and other Vibrio phages, and the phylogenetic distance suggested that ϕA318 could be classified as a new T7-like bacteriophage. Nevertheless, several motifs in the ϕA318 phage RNA polymerase were highly conserved, including DFRGR (T7-421 motif), DG (T7-537 motif), PSEKPQDIYGAVS (T7-563 motif), RSMTKKPVMTL PYGS (T7-627 motif), and HDS (T7-811 motif). Genetic analysis indicated that phage ϕA318 is not a thermostable direct hemolysin producer. The results suggest that the MOI should be higher than 0.1 to prevent the chance of hemolysin production by the bacteria before they are lysed by the phage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aerosols and water from aquaculture are usually open to the environment, resulting in the rapid exchange of transmittable diseases and pollution. The high productivity in aquaculture has concurrently had an ecological impact, including the emergence of a variety of pathogens. Aquatic pathogens such as Vibrio strains are fatal, not only to fish and shellfish, but also to humans, because several virulence factors that they produce—including hemolysin, caseinase, gelatinase, lipase, and phospholipase—can also be detrimental to human health [1]. For instance, an extracellular protease with endopeptidolytic and exopeptidolytic activities from a pathogenic V. pelagius strain is responsible for the lethal effect and vibriosis in turbot [2]. The different virulence factors are produced at different stages of infection. In the example of Vibrio harveyi, quorum sensing negatively regulates phospholipase activity but positively regulates caseinase and gelatinase activity, whereas lipase and hemolysin activity is independent of quorum sensing [3]. Hemolysin-producing bacteria include V. parahaemolyticus, V. alginolyticus, V. cholerae, V. hollisae, Aeromonas veronii, and V. anguillarum (Listonella anguillarum).
Among the strains that are taxonomically related to the harveyi group, Vibrio alginolyticus, a Gram-negative halophilic bacterium that frequently occurs in the normal microbiota of marine environments, is a pathogen of epizootic outbreaks, causing serious mortality in fish and shellfish aquaculture, and it can also be transmitted to humans. The disease caused by V. alginolyticus often has lethal consequences for fish larvae throughout the world, and it has become one of the major limiting factors in aquaculture in developing countries [4, 5]. For instance, related strains have been reported to be a causal agent of vibriosis outbreaks in grouper Epinephelus malabaricus [4] and sea bream Sparus aurata [5]. V. alginolyticus causes several symptoms in sea bream: septicemia, hemorrhaging, dark skin, and ulcers on the skin surface [5]. Some fish accumulate fluid in the peritoneal cavity or have hemorrhagic livers [6]. In several cases of high-mortality outbreaks [6, 7], Vibrio alginolyticus and Vibrio splendidus biovar II were the primary organisms found in moribund clam larvae (Ruditapes decussatus), and these two species processed extracellular products collaboratively at high concentrations to kill the hemocytes after four hours of incubation [7].
The V. alginolyticus ATCC 17749 is also one of the bacteria that have been shown to produce tetrodotoxin, a strong neurotoxin, under laboratory culture conditions [8]. Neurotoxic effects caused by the supernates from diseased strains of Vibrio alginolyticus and Vibrio anguillarum caused several symptoms in trout, including convulsions, wriggling, contortive swimming and respiratory arrest [9]. Injection of the Vibrio strains prompts release of acetylcholine in the motor end plates of muscles in the inoculated fish [9].
To control pathogenic bacteria in aquaculture, chemotherapeutic agents are used widely and indiscriminately due to the high efficiency with which they kill pathogens and cure the illness [see ref. 10 for review]. Antibiotic resistance, however, emerges soon after the antibiotics are applied [11]. For example, in internal organs of diseased gilthead sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax) cultured in fish farms, 91.17% of Vibrio alginolyticus was found to be resistant to nitrofurantoin [11]. As a result of management practices in production cycles, the use of probiotics for prevention of bacterial diseases in aquaculture is becoming a more significant practice [10].
One alternative is to use bacteriophages to control bacterial growth, which is similar to phage therapy in medical care. This idea has been proposed as a cure for coral disease [12]. A variety of bacteriophages that can quickly lyse the pathogens V. alginolyticus, V. carchariae, V. damsela, V. harveyi, V. parahaemolyticus, V. pelagius, or V. vulnificus have high potential for future applications in aquaculture. In this study, we characterized a new phage that can lyse V. alginolyticus ATCC 17749 in a short period of time.
Materials and methods
Bacterial strains and growth conditions
Vibrio strains were bought from the Bioresource Collection and Research Center, Taiwan, including V. alginolyticus ATCC 17749, V. carchariae ATCC 35084, V. damsela ATCC 33536, V. harveyi ATCC 14126, V. parahaemolyticus ATCC 17802, V. pelagius ATCC 25916, and V. vulnificus BCRC15431. The Vibrio strains were maintained in Bacto Brain Heart Infusion medium (BHI; Becton-Dickinson Co, USA) supplemented with 3% NaCl (Panreac Quimica, France). For long-term preservation, bacteria were frozen at −80°C in BHI supplemented with 1% NaCl and 25% glycerol (Nihon Shiyaku, Japan). The strains were streaked onto modified seawater yeast extract (rich MSWYE) agar plates consisting of 23.4 g NaCl, 6.98 g MgSO4·7 H2O, and 0.75 g KCl in 1000 ml distilled water [13]. The pH was adjusted to 7.6 with 1 N NaOH, followed by the addition of 5.0 g of proteose peptone (HIMEDIA Lab, India), 3.0 g of yeast extract (HIMEDIA Lab, India), and 20.0 g of agar per liter.
Isolation and titer of bacteriophages
Water samples were collected from aquaculture waterways in southern Taiwan between 2008 and 2010. The water was centrifuged at 10,000 × g for 30 minutes, and the supernatants were filtered through a 0.45-μm microfilter. After centrifugation and microfiltration, 20% MSWYE medium [13] and 1% overnight-grown Vibrio alginolyticus were added to the filtrate and incubated at 25°C for 4-24 hours to enrich the phages. The bacterial debris was removed by two centrifugations at 10,000 × g for 30 minutes. Using the agar overlay technique, approximate dilutions of the resulting phage suspensions were incubated with actively growing hosts for 5 minutes, mixed with top agar, and plated onto MSWYE agar plates for phage plaque isolation. The plaques were picked into 200 μl MSWYE broth for further titer counting or production amplification. To prepare bacterial cells for determining phage concentrations, the host Vibrio alginolyticus was freshly inoculated as a 1% volume of seed from overnight culture into 10 ml of rich MSWYE broth, and it grew to an OD600 of 0.3-0.4 in about 2 hours. After a series of dilutions, 10 μl of phage was added to 200 μl of bacterial cells, incubated for 5 minutes, mixed with 3 ml of top agar (rich MSWYE with 0.5% agar), and then poured onto the solid surface of a 2% agar plate. The plaques were counted in 3-5 hours; the titer per ml was calculated as 100 × (dilution factor) × (plaque count).
Electron microscopy
Preparation of phage particles for electron microscopy has been described elsewhere [14]. In brief, bacteriophage particles were applied to Parafilm to produce a spherical drop. Carbon-coated nitrocellulose films were fabricated on copper grids, which were placed face down on the sample drop for 1 min to absorb the particles. After being briefly washed twice in 10 μl of 10 mM Tris buffer (pH 8) (Amresco, USA), the samples were then rinsed twice with freshly prepared 2% uranyl acetate (UA; Sigma-Aldrich, USA) in Tris-HCl (pH 8.0) and stained for 60 seconds. Following each step of absorption, washing, and UA staining, the grids were blotted with filter paper until almost dry. The finished grids were dried in vacuo overnight. Images of phage particles were taken at a magnification of 40,000× and a defocus of 3 μm, using a 200-kV electron microscope (JEOL JEM-2010, equipped with a Gatan-832 CCD camera).
Preparation of bacteriophage DNA
For propagation of phage, 10 ml of ϕA318 phage stock was added to 100 ml of V. alginolyticus (3 × 108 CFU ml−1) cultured in MSWYE and incubated in a shaker at 25°C for 3-5 hours until the lysate was clear but still contained some cell debris. The remaining cells and debris were removed by two centrifugation cycles at 10,000 × g for 30 minutes. The supernatant, with a titer of 2 × 1010 PFU ml−1, was stored at 4°C as a phage stock. To concentrate phages using a standard PEG protocol [15], solid NaCl and polyethylene glycol 8000 (Fluka, Germany) were added, and precipitation was performed overnight at 4°C. After centrifugation, the phage particles were resuspended in 2 ml of SM buffer and treated with DNase I and RNase A (Sigma, USA) to remove contaminating nucleic acids from the host. The polyethylene glycol was extracted by adding an equal volume of chloroform (TEDIA, USA) until the interface was clear. The aqueous phase containing the phage was treated with proteinase K (Invitrogen, USA) and sodium dodecyl sulfate (SDS; MD Bio, Inc, USA) at 56°C for 1 h. Phenol extraction was carried out three times at room temperature, and the aqueous phase was further extracted with a 1:1 mixture of equilibrated phenol (Sigma, USA) and chloroform. DNA precipitated by a 2× volume of cold ethanol was redissolved in deionized water.
Adsorption and phage burst size
As described previously [16] for the adsorption of phage on bacteria, 1 ml of broth containing host cells and ϕA318 was taken at the 5th and 10th minutes and centrifuged to remove bacteria and bound phage, and free phage in the supernatants was then measured. Similarly, the burst size was measured as described previously [17]. In brief, V. alginolyticus cells were grown in 20 ml of medium to mid-exponential phase (OD600 = 0.3-0.5) and mixed with 0.1 ml phage ϕA318 solution (MOI = 0.6 and 1). Samples of 1 ml of the mixture were taken at intervals and immediately subjected to centrifugation at 14,000 × g for 3 minutes to remove bacteria. The phage titers in the solutions were determined by the agar overlay technique. Using the growth curve for MOI = 1 according to the one-step criteria, the burst size (Bs) of phage ϕA318 was calculated as Bs = Pt/P0, where Pt is the phage titer at that plateau phase and P0 is the initial infective titer.
Thermal stability of ϕA318
Thermal stability tests have been described elsewhere [18]. Briefly, the bacterium Vibrio alginolyticus was freshly inoculated as 1% volume of seed from overnight culture into 20 ml of rich MSWYE broth. When the cell density reached an OD600 of 0.4-0.5, heat-treated phages from a dilution series were added to infect the host for 5 minutes before they were mixed with top agar and poured onto the solid surface of a 2% agar plate in order to count the plaques in 3-5 hours. Phage particles (2 × 1010 PFU) were treated at 25-80°C, and samples were taken at 15-min intervals. Supernatants obtained by centrifugation at 14,000 × g for 3 minutes were diluted, and the phage titer was determined using the agar overlay technique.
Multiple sequence alignment
To determine the classification status of the newly isolated ϕA318 phage, sequence data for the genes encoding the DNA polymerase, DNA ligase, RNA polymerase, single-strand DNA binding protein, and capsid proteins of enterobacteria phage T7 and Vibrio phages N4, VP4, and ICP3 were employed to find highly homologous regions. Complete genome sequences of three Vibrio T7-like phages and two enterobacteria phages (T7 and SP6) were acquired from NCBI: enterobacteria phages T7 (39,937 bp, GenBank accession no. NC_001604), vibriophage ICP3 (39,162 bp, GenBank accession no. NC_015159), vibriophage N4 (38,497 bp, GenBank accession no. NC_013651), and vibriophage VP4 (39,503 bp, GenBank accession no. NC_007149). The T7 RNA polymerase sequence is also the same as 1QLN (RCSB-PDB ID) and SP6 RNA polymerase is from NC_004831 (GenBank). Sequences of individual genes retrieved from the genome sets were then aligned using ClustalW with default options [19]. Highly conserved 20-mers in both directions were selected for gene sequencing in an Applied Biosystems DNA Analyzer (USA), using as template either total phage DNA from ϕA318 or fragments thereof generated by digestion with restriction enzymes. Sets with mismatched alignment from primers in both directions were discarded. The best alignment of the individual genes was with RNA polymerase; therefore, the preliminary sequence was used for further walking in both directions using the ϕA318 phage DNA as the template. The complete RNA polymerase sequences were analyzed by the neighbor-joining method using the NEIGHBOR program in Phylogeny Inference Package (PHYLIP) [20]. Distances were calculated using the DNADIST program of PHYLIP and displayed in TreeView [21]. ClustalW, PHYLIP, and TreeView were bundled in the BioEdit program version 7.0.5 [22].
Results
Bacteriophage isolation
After enriching the phages in the field samples with the target bacterium Vibrio alginolyticus for 4-16 hours, phage-containing supernatants were incubated with the indicator strain for 5 minutes, suspended in low-percentage agar (top agar), and plated onto solid agar plates so the target strain would form a bacterial lawn with the formation of plaques. The ten largest plaques from each plate were picked and re-amplified three times in bacterium-containing broth using a 5× volume ratio. The plaques of ϕA318 that formed in 4 hours at 25°C were very clear at their center and at the margin in the edge; the size of the plaques was about 3 mm in diameter, which increased to 5 mm overnight. The phage concentration in each plaque increased with gradually increasing amounts of host bacterium, but not higher than a 50× volume ratio.
Morphology study by transmission electron microscopy
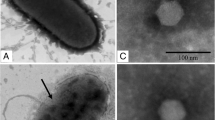
The morphology of phage ϕA318 was observed by transmission electron microscopy, which is the most common method to classify phages. The icosahedral head was almost isomorphous, with a size of 50-55 nm, and the shells on the head capsids sketched out at least three visible rings, although the tail fiber was barely distinguishable from the capsid shell in the micrograph (Fig. 1A). The tail was 12 nm long, elongating from 25 nm wide in the head-neck connector to 5 nm in the nozzle. It was highly similar to the bacteriophages of the family Podoviridae.
In the micrograph shown in Fig. 1B, the prohead carried an internal core of some density, in which the white density was polarized toward the right-hand side in readiness to package the genomic DNA. As Fig. 1C shows, the tail is still attached to the empty head, but no staining core was observed inside the head. The empty head aborted the internal core before the tail attached, presumably not being a precursor of the mature head to package DNAs. Although some broken bubble-like complexes were also observed occasionally (Fig. 1D), chloroform-treated particles maintained the same titer for infection of the bacterium. As Fig. 1E displays, some unknown particles were found occasionally that were one-third smaller in size but with a dense coat.
Host range and phage burst size
The susceptibility of the following Vibrio strains to phage ϕA318 was also investigated by the agar overlay method: V. carchariae, V. damsela, V. harveyi, V. parahaemolyticus, V. pelagius, and V. vulnificus. Among these, V. damsela and V. harveyi, were found to be susceptible to phage ϕA318, while the other four species could not be infected even at an MOI of more than 100. These results indicate that phage ϕA318 has a limited host range within the harveyi group.
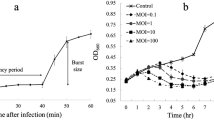
Adsorption is the first step of phage infection of the host bacterium, and the phage then goes immediately through advanced propagation, which is detrimental to host growth for the next round of infection. During experiments to determine the optimal MOI for production of ϕA318 phage, it was found that the higher the MOI, the earlier cell lysis occurred. The optimal production of phage ϕA318 was at an MOI of 0.5-2.0. Infection with an MOI of 0.6 gave an optimal yield of phage progeny (~4 × 1010 PFU ml−1), which was a 143-fold amplification (Fig. 2). When the MOI was too high, the number of viable cells decreased sharply so as to limit phage production, leading to lower magnification folds. When the MOI was too low, phage production was limited by nutrient depletion due to the overgrowth of uninfected bacteria, which used up the nutrients in three hours.
For measurement of the burst size, a bacterial culture in the early exponential growth phase was split into glass tubes with equal numbers of V. alginolyticus cells (~3 × 108 CFU), which were infected with different amounts of phage at the designed MOI of 0.6 and 1.0, and then plaque titers were calculated within 4 hours of incubation. With different MOI values, the time course of phage growth displayed the form of a triphasic curve: absorption phase, growth phase, and plateau phase (Fig. 2). The absorption time was complete in about 10 minutes. According to the one-step method [17], the burst size of phage ϕA318 was 72 PFU per infected cell, which was estimated from the Pt/P0 at an MOI of 1 in which the phages were allowed to propagate in a one-step infection under the assumption that every bacterium was infected by one phage.
Viability of phage ϕA318 in the thermal environment
A thermal stability test was carried out to analyze the heat resistance of phage ϕA318 at pH 7.5-8.0. The phage was incubated at 25, 49.5, 61, 70, and 80°C for one hour. As Fig. 3 shows, the phage titers at different time intervals showed that phage ϕA318 stock solution retained almost 100% infectivity after incubation at temperatures lower than 50°C for one hour. When the temperatures rose above 60°C, the viability of phage ϕA318 declined; less than 10% of phages remained alive after being heated for 60 minutes. At temperatures higher than 70°C, nearly all phages were inactivated after 15 minutes of incubation.
The dose of phage that suppresses hemolysin production
In the early stage of phage amplification from a plaque (normally at low MOI), a small proportion of the cells were arrested by the equivalent amounts of phage. The rest of the cells overgrew by consuming the nutrients in the culture medium, and a small percentage of cells were lysed by phage ϕA318 during the first 5-7 hours. At an MOI lower than 0.1, few phage particles were observed in EM micrographs, while, surprisingly, many four-dot complexes spread over the entire copper grid (Fig. 4). The complex was uniform, with four dense domains holding together tightly, and these were tetramers of thermostable direct hemolysin, formed by four subunits of a 21.6-kDa protein of 189 amino acids. In whole genome analysis, it has been predicted that V. alginolyticus ATCC 17749 and some strains of V. parahaemolyticus can produce it [23]. Using bioinformatics to find sequences with homology to thermostable direct hemolysin (TDH) sequences in V. alginolyticus (GenBank accession no. DQ440529.1) and V. parahaemolyticus (GenBank accession no. BA000032.2), we failed to detect TDH and related genes in the ϕA318 genome (data not shown). Therefore, ϕA318 is not a TDH producer.
Transmission electron micrograph of tetra-dot particles occurring in the V. alginolyticus lysate, even at low-MOI infection with phage ϕA318. After centrifugation at 270,000 × g, the proteins were negatively stained with uranyl acetate. The particles resemble a tetra-dot of TDH. Panel B is a section of panel A with twofold magnification
Sequence similarity among the T7-like phages
The EM morphology of ϕA318 demonstrated that this phage could be related to T7. Comparison of the sequences of the genes for DNA polymerase, DNA ligase, RNA polymerase, single-strand DNA binding protein, and capsid proteins showed that, the enterobacteria phage T7 shares 60-65% homology with Vibrio phages N4, VP4, and ICP3. Using this information on conserved regions, we designed several primers to sequence the ϕA318 genomic DNA. Primers of 21 nucleotides (forward primer CGCTCCTAACTTTGTTCACAG and reverse primer GATTACATCA TGTTCTTCATA) corresponding to regions conserved among known T7-like phages were successful in amplifying a fragment of 197 base pairs in ϕAB, which matched to RNA polymerase. Using the sequence information of the 197 bp, the complete sequence of the ϕAB RNA polymerase gene was determined by walking the sequences in both directions, and the resulting sequence was submitted to GenBank (JQ088079). As Fig. 5 shows, the translated protein sequence of ϕAB RNA polymerase was in alignment with those of other T7-like phages. The similarities of ϕA318 to SP6, T7, and other Vibrio phages were as low as 28-47% (Fig. 6A), while the closest one was enterobacteria phage SP6. Phylogenetic analysis revealed that the tree had a tetrapod shape, with the sequence being an almost equal distance from each other (Fig. 6B). At each vertex were located phages ϕA318, SP6, T7, and the other Vibrio phages (N4/ICP3, VP4).
In comparison with other T7-like RNA polymerases, there were five consensus regions, corresponding to residues 421-425, 537-538, 563-575, 627-641, and 811-813 residues in the T7 RNA polymerase, which were named as motifs T7-421, T7-537, T7-563, T7-627, and T7-811, respectively. As shown in Fig. 5, the motifs were highly conserved among the T7-like phages. In phage ϕA318, the sequences of the motifs were DFRGR (motif T7-421), DG (motif T7-537), PSEKPQDIYGAVS (motif T7-563), RSMTKKPVMTLPYGS (motif T7-627), and HDS (motif T7-811), respectively.
Discussion
Electron microscopy revealed that phage ϕA318 particles were morphologically similar to T7-like phages. ϕA318 can be most likely identified as type C in Bradley’s classification of Podoviridae phage [24], based on its morphological characteristics (Fig. 1). The podovirus family is characterized by phages with an isomorphous icosahedral head and a short non-contractile tail [25, 26]. Furthermore, the lengths of the icosahedral capsid and tail of phage ϕA318 were morphologically indistinguishable from those typically observed for the other members of the T7 phage group.
The protein profiles of known T7 structural proteins contain at least 16 clearly discrete bands in a gradient gel, ranging from approximately 16 to 140 kDa [27, 28]. The predominant polypeptide band of ϕA318 had an apparent size of approximately 42 kDa (data not shown), which can be assigned to major capsid protein gp10A by its size in the T7 phage. There are three other protein bands that can be correlated with T7 structural proteins based on their size: a ~27 kDa band, with be tail protein gp11; a ~25 kDa band, with the inner core protein, and a ~95 kDa band, with the head-tail connector protein gp8 (data not shown). Protein bands of 38 and 40 kDa could be truncated capsid polypeptides. The sixth minor band of 60 kDa could be similar to a hypothetical protein of Thalassomonas phage BA3 [12] and ϕIBB-PF7A [26]. The structural proteins and EM morphology clearly demonstrated that ϕA318 belongs to T7-like phages. Furthermore, based on the complete sequence of the gene for the DNA-directed RNA polymerase, which is a unique viral protein (Figs. 5 and 6), phage ϕA318 is phylogenetically distant from both T7 and other Vibrio phages. Phage ϕA318 is a new bacteriophage of V. alginolyticus.
Similarity and phylogenetic tree of RNA polymerase genes from different T7-like phages. ϕA318 is the phage isolated in this study; SP6 and T7 are enterobacteria phages; Vibrio phages N4, and VP4 are T7-like. (A) The identity of protein sequences between phages. (B) Phylogenetic tree. Phage ICP3 is not shown because its RNA polymerase was identical to that of phage N4
Our TDH micrograph is identical to EM results from Yanagihara et al. [23], who also presented the crystal structure of the TDH tetramer at 1.5-Å resolution. The TDH tetramer forms a central pore with dimensions of 23 Å in diameter and ~50 Å in depth. The central pore was not well defined in our EM micrographs (Fig. 4), which was due to the negative staining, which can only enhance certain strong contrasts in parts, but some image density around the pore was lost. Since there exists high homology of TDH sequences between V. alginolyticus and V. parahaemolyticus, detection of TDH or related genes was carried out to ensure that the ϕA318 genome did not carry the virulent gene (data not shown). Additionally, we found TDH tetramers at low-MOI infection of ϕA318, but few were present at high MOI. TDH production is probably caused by the lysis action of ϕA318 lysin, which releases host TDH protein, while at high MOI, cellular metabolism may be dominated by phage production instead of synthesis of TDH before lysis. Under such conditions, the application of this phage to control V. alginolyticus in aquaculture should be further examined.
In phage therapy for remedying the disease caused by Vibrio alginolyticus, the new phage ϕA318 in this study does not carry a hemolysin-related gene; hence, it is as a promising agent for such applications. We suggest using as high an MOI as possible to achieve the cure, or at least an MOI of 0.1. The optimal yield in the production process for phage preparation is achieved by growing the phage at an MOI of 0.6 in order to reach a yield of 143-fold. For security, the purified phages can be incubated at 50-55°C for 30 minutes to remove contamination from the viable Vibrio host.
Cheetham and Steitz [29] had resolved the structure of T7 RNA polymerase by X-ray diffraction at 2.9 Ǻ. Structural and functional analysis of bacteriophage T7 RNA polymerase 1QLN shows four motifs. In the palm subdomain, DWRGR (motif T7-421) interacts with the base in the DNA template strand for correct initiation [29, 30], while DG (motif T7-537) and HDS (motif T7-H811) are near the elongating end of the nascent RNA chain during the functioning of the enzyme [29, 31]. In the finger subdomain, R–h–K–VMT–YG (motif T7-627) is in the substrate-binding cleft near the active site of the enzyme [29, 31]. Underlines in the motifs represent highly conserved residues that were also found in our ϕA318 phage (Fig. 5). In T7 phage, W422 in motif T7-421 has been shown to be a critical residue interacting to the template [30], while in ϕA318 phage, the tryptophan is replaced by phenylalanine, which is similar to the serine in the enterobacteria phage SP6 carrying a hydroxyl group (Fig. 5). In addition, the 3D structure of T7 RNA polymerase 1QLN (RCSB-PDB ID) shows that the T7-563 loop is located on the exterior surface of the enzyme. Nevertheless, the S564A mutation enhanced polymerization activity by 30%, whereas the mutations at P563T, Q568A, D569A, and Y571F destroyed the activity. In motif T7-563 (Fig. 5), ϕA318 shared S564 with T7 and SP6, while other Vibrio phages contained A564 instead.
References
Baffone W, Citterio B, Vittoria E, Casaroli A, Pianetti A, Campana A, Bruscolini F (2001) Determination of several potential virulence factors in Vibrio spp. isolated from seawater. Food Microbiol 18:479–488
Farto R, Pérez MJ, Briera AF, Nieto TP (2002) Purification and partial characterisation of a fish lethal extracellular protease from Vibrio pelagius. Vet Microbiol 89(2–3):181–194
Natrah FMI, Ruwandeepika HAD, Pawar S, Karunasagar I, Sorgeloos P, Bossier P, Defoirdt T (2011) Regulation of virulence factors by quorum sensing in Vibrio harveyi. Vet Microbiol 154(1–2):124–129
Lee KK (1995) Pathogenesis studies on Vibrio alginolyticus in the grouper, Epinephelus malabaricus Bloch et Schneider. Microb Pathog 19:39–48
Colorni A, Paperna I, Gordin H (1981) Bacterial infections in gilt-head sea bream Sparus aurata cultured at Elat. Aquaculture 23:257–267
Paperna I (1984) Review of diseases affecting cultured Sparus aurata and Dicentrarchus labrax. In: Barnabé G, Billard R (eds) L’aquaculture du bar et des sparides. INRA Publisher, Paris, pp 465–482
Balebona MC, Andreu MJ, Bordas MA, Zorrilla I, Moriñigo MA, Borrego JJ (1998) Pathogenicity of Vibrio alginolyticus for cultured gilt-head sea bream (Sparus aurata L.). Appl Environ Microbiol 64(11):4269–4275
Simidu U, Noguchi T, Hwang DF, Shida Y, Hashimoto K (1987) Marine bacteria which produce tetrodotoxin. Appl Environ Microbol 53(7):1714–1715
Balebona MC, Krovacek K, Moriñigo MA, Mansson I, Faris A, Borrego JJ (1998) Neurotoxic effect on two fish species and a PC12 cell line of the supernate of Vibrio alginolyticus and Vibrio anguillarum. Vet Microbiol 63(1):61–69
Balcázar JL, de Blas I, Ruiz-Zarzuela I, Cunningham D, Vendrell D, Múzquiz JL (2006) The role of probiotics in aquaculture. Vet Microbiol 114(3–4):173–186
Kahla-Nakbi AB, Chaieb K, Besbes A, Zmantar T, Bakhrouf A (2006) Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet Microbiol 117(2–4):321–327
Efrony R, Atad I, Rosenberg E (2009) Phage therapy of coral white plague disease: properties of phage BA3. Curr Microbiol 58(2):139–145
Schwarz JR, Colwell RR (1974) Effect of hydrostatic pressure on growth and viability of Vibrio parahaemolyticus. Appl Microbiol 28:977–981
Lu M-W, Liu W, Lin C-S (2003) Infection competition against grouper nervous necrosis virus by virus-like particles produced in Escherichia coli. J Gen Virol 84:1577–1582
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Shafia F, Thompson TL (1964) Calcium ion requirement for proliferation of bacteriophage Phi Mu-4. J Bacteriol 88:293–296
Adams MH (1959) Bacteriophages. Interscience, New York
Mitra S, Basu S (1968) Some biophysical properties of a vibriophage and its DNA. Biochim Biophys Acta 155(1):143–149
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Yanagihara I, Nakahira K, Yamane T, Kaieda S, Mayanagi K, Hamada D, Fukui T, Ohnishi K, Kajiyama S, Shimizu T, Sato M, Ikegami T, Ikeguchi M, Honda T, Hashimoto H (2010) Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin. J Biol Chem 285(21):16267–16274
Bradley DE (1967) Ultrastructure of bacteriophages and bacteriocins. Bacteriol Rev 31(4):230–314
Stroud RM, Serwer P, Ross MJ (1981) Assembly of bacteriophage T7: dimensions of the bacteriophage and its capsids. Biophys J 36:743–757
Sillankorva S, Neubauer P, Azeredo J (2008) Isolation and characterization of a T7-like lytic phage for Pseudomonas fluorescens. BMC Biotechnol 8:80–91
Issinger OG, Falk H (1976) Comparative studies on the structural proteins of T3 and T7 phages. Arch Virol 52:217–231
Kemp P, Garcia LR, Molineux IJ (2005) Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340:307–317
Cheetham GM, Steitz TA (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science 286(5448):2305–2309
Imburgio D, Anikin M, McAllister WT (2002) Effects of substitutions in a conserved DX2GR sequence motif, found in many DNA-dependent nucleotide polymerases, on transcription by T7 RNA polymerase. J Mol Biol 319:37–51
Tunitskaya VL, Kochetkov SN (2002) Structural–functional analysis of bacteriophage T7 RNA polymerase. Biochemistry (Moscow) 67(10):1124–1135
Acknowledgments
This research fund is partially supported by grants from the National Science Council, Taiwan (NSC96-2313-B-110-002-MY3 and NSC99-2313-B-110-002-MY3), and the Ministry of Education, Taiwan (NSYSU95-99C031701; the second term of Top University Program: NSYSU 00C030205 and NCHU 100-S05-09) under the ATU plan. We thank Professor Long-Huw Lee (National Chung-Hsing University) as the grant organizer of the intercampus ATU plan. Also we appreciate Yu-Tin Liu for helping with gel preparation, and Kenneth B. Lin and Dr. Simon White for comments and editing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lin, YR., Chiu, CW., Chang, FY. et al. Characterization of a new phage, termed ϕA318, which is specific for Vibrio alginolyticus . Arch Virol 157, 917–926 (2012). https://doi.org/10.1007/s00705-012-1244-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1244-8