Abstract
Tomato spotted wilt virus (TSWV) infects numerous host plants and has three genome segments, called L, M and S. Here, we report the complete genome sequences of three Korean TSWV isolates (TSWV-1 to -3) infecting tomato and pepper plants. Although the nucleotide sequence of TSWV-1 genome isolated from tomato is very different from those of TSWV-2 and TSWV-3 isolated from pepper, the deduced amino acid sequences of the five TSWV genes are highly conserved among all three TSWV isolates. In phylogenetic analysis, deduced RdRp protein sequences of TSWV-2 and TSWV-3 were clustered together with two previously reported isolates from Japan and Korea, while TSWV-1 grouped together with a Hawaiian isolate. A phylogenetic tree based on N protein sequences, however, revealed four distinct groups of TSWV isolates, and all three Korean isolates belonged to group II, together with many other isolates, mostly from Europe and Asia. Interestingly, most American isolates grouped together as group I. Together, these results suggested that these newly identified TSWV isolates might have originated from an Asian ancestor and undergone divergence upon infecting different host plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato spotted wilt virus (TSWV), a member of the genus Tospovirus, family Bunyaviridae [7], affects a wide range of host plants, including 900 plant species worldwide [3], causing serious agronomic losses and reducing the quality of fruits and vegetables, especially tomatoes [3, 8]. It has been reported that TSWV is transmitted by thrips in a persistent manner [12]. The TSWV virion varies in size from 80 to 120 nm and has a spherical envelope structure [9]. Previous studies have shown that the TSWV genome is composed of three single-stranded linear segments, large (L), medium (M), and small (S), named according to their lengths [9]. The L RNA (~9 kb) contains an RNA-dependent RNA polymerase (RdRp) in a negative-sense orientation [2]. In contrast, the M (~4.8 kb) and S (~3 kb) RNAs each contain two genes, one in the positive- and the other in the negative-sense orientation [4]. The M RNA encodes the NSm protein and the Gn-Gc glycoprotein, while the S RNA encodes the NSs nonstructural and N proteins [1, 6]. In the National Center for Biotechnology Information (NCBI) database, a large number of partial sequences related to TSWV are available. However, only one complete genome sequence of an TSWV isolate (Brazilian TSWV BR-01) maintained in Nicotiana rustica was reported previously [1, 2, 6]. Therefore, it is necessary to get the complete genome sequences of TSWV from various other host plants. We report here the complete genome sequences of three TSWV isolates from tomato and pepper plants. We have performed phylogenetic analysis of these three TSWV isolates together with other TSWV isolates from around the world based on sequences of the RdRp and N proteins [11].
Provenance of the virus material
Three TSWV isolates were identified from tomato and pepper plants in Na-Ju (NJ), Jeon-Nam (JN) province, and Chung-Yang (CY), Chung-Nam (CN) province, respectively, during the period of 2008 to 2009. The collection regions and host plants for each isolate can be found in Supplementary Table S1. To simplify annotation of each TSWV isolate, we designated the three TSWV isolates as TSWV-1, TSWV-2 and TSWV-3. TSWV-1 was isolated from tomato (Na-Ju (NJ) city, Jeon-Nam (JN) province), whereas both TSWV-2 and TSWV-3 were isolated from pepper (Chung-Yang (CY) city, Chung-Nam (CN) province). All three virus isolates were inoculated to local-lesion hosts, reisolated from local lesions at least three times in succession, and propagated in Nicotiana rustica. Total RNAs were extracted from TSWV-infected tomato and pepper leaves using TRI Reagent® (Molecular Research Center, Inc., Cincinnati, USA) following the manufacturer’s instruction. For cDNAs synthesis, total RNAs were incubated at 65°C for 5 min with random primers and dNTPs, followed by incubation at 42°C for 1 h after adding M-MLV reverse transcriptase (RT) and M-MLV 5X buffer (Promega, USA). To amplify PCR fragments from individual cDNAs, a total of 50 μl PCR mixture was prepared that included 2 μl cDNA mixture, 5 μl 10X buffer, 3 μl 2.5 mM dNTPs, 2 μl each primer pair (Supplementary Table 2), 37.5 μl dH2O, and 2.5 units Taq DNA polymerase (Takara, Japan). PCR products were amplified for 30 cycles with following steps: denaturation at 92°C for 20 s, annealing at 53°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. For complete genome sequencing, each primer was designed manually based on the conserved regions of previously known sequences. The sizes of each amplified fragment ranged from 1 to 2 kb, and each fragment overlapped with its neighboring fragments by at least 100 nucleotides. The cDNAs were synthesized using total RNA as a template. The 5’ and 3’ untranslated regions of each TSWV isolate were identified by the rapid amplification of cDNA ends (RACE) method. The amplified PCR product was visualized on a 1% agarose gel by ethidium bromide (EtBr) staining. PCR products were purified using a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and subsequently cloned into pGEM-T Easy Vector (Promega, Madison, USA). All clones were sequenced at least three times in both orientations using universal and specific PCR primers at the National Instrumentation Center for Environmental Management, Seoul National University, Korea. All primers used in this study can be found in Supplementary Table S2.
The sequences that were obtained were first analyzed with the MegAlign program implemented in the DNAStar 5.01 package (DNASTAR, Madison, USA). The BioEdit sequence alignment editor (Version 7.0.9) (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and GeneDoc programs were further used for detailed sequence analysis or conversion of sequence file formats (http://www.nrbsc.org/gfx/genedoc/). In order to examine the phylogenetic relationships of the three TSWV isolates, we retrieved amino acid sequences of other known TSWV isolates from NCBI’s GenBank. Phylogenetic trees were generated by the neighbor-joining method with 1000 bootstrap replications using the MEGA program, version 4.0 [10]. The complete nucleotide sequences of three TSWV isolates were deposited in GenBank (accession numbers HM581934-HM581942; Supplementary Table S1).
Sequence properties
TSWV genomes are composed of three RNA segments, i.e., L, M, and S (Supplementary Table S3), and the genome organization of TSWV-1 is depicted in Fig. 1. The genome sequence of TSWV-1 isolated from tomato is very different from those of TSWV-2 and TSWV-3, which were isolated from pepper. The L RNA segment of TSWV-1 (8,913 nt) is one nucleotide shorter than those of TSWV-2 (8,914 nt) and TSWV-3 (8,914 nt). The sizes of the M and S RNA segments of TSWV-2 and TSWV-3 are identical, with 4,768 and 3,013 nt, respectively, and the M RNA segment of TSWV-1 (4,783 nt) is 15 nt longer than those of TSWV-2 and TSWV-3. Furthermore, the S RNA segment of TSWV-1 (2,968 nt) is 45 nt shorter than those of TSWV-2 and TSWV-3. In contrast to the lengths of the nucleotide sequences, the lengths of the five open reading frames (ORFs) are equal: RdRp (2,880 aa), NSm (254 aa), Gn-Gc (1,136 aa), NSs (468 aa), and N (259 aa) (Supplementary Table S3). The lengths of the 5’ UTR regions of three ORFs, RdRp (33 nt), NSm (247 nt), and NSs (88 nt) are identical in the three TSWV isolates. However, the translation initiation positions for the Gn-Gc and N proteins in TSWV-1 (nucleotide positions 4699 and 2817, respectively) are different from those of TSWV-2 and TSWV-3 (nucleotide positions 4684 and 2862, respectively), which are identical. Additionally, the two ambisense RNA genomic segments have different lengths in the intergenic region (IGR). The IGR lengths for the M and S RNA segments in TSWV-1 are 283 nt and 549 nt, respectively, while those in TSWV-2 and TSWV-3 are 268 nt and 594 nt, respectively. Based on these results, it is likely that the differences in RNA size were mostly caused by insertions and/or deletions of nucleotide sequences within the IGR. We compared the complete genome sequences of three TSWV isolates with that of the BR-01 isolate using Vector NTI, version 7 (Invitrogen, Darmstadt, Germany). The sequence identity ranges of the three TSWV isolates in this study compared to the BR-01 isolate were 93.3%-95.1% for L RNA, 90.7%-90.8% for M RNA and 91.4%-93% for S RNA (Supplementary Table S4). Among the three TSWV isolates in this study, TSWV-1 showed the highest degree of sequence homology with the BR-01 isolate. Moreover, TSWV-2 and 3 are very closely related to each other and most closely related to the Korean isolate from paprika, as indicated in Fig. 2. For example, the nucleotide difference between these two isolates was 14 nt in L RNA, 6 nt in M RNA, and 2 nt in S RNA.
Schematic illustration of the genome organization of TSWV-1, which is composed of three RNA segments, large (L), medium (M), and small (S). The numbers above the diagram indicate the positions of individual ORFs in the RNA based on the nucleotide sequences. The white and gray arrows indicate the direction of protein translation in the sense and antisense strand, respectively. The scale bar indicates 1000 nucleotides
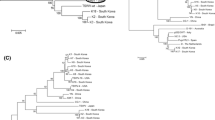
Phylogenetic relationship of the three TSWV isolates from this study to various other TSWV isolates based on the amino acid sequences of the RdRp (a) and N (b) proteins. Phylogenetic trees were constructed in the MEGA version 4.0 program using the neighbor-joining method with 1000 bootstrap replicates. The number at each branch represents the bootstrap value (1000 replicates). The three TSWV isolates from the current study are indicated by gray rectangles. Detailed information for each TSWV isolate, including the region where it was isolated, the name of the host plant, and the protein accession number can be found in the phylogenetic tree
To determine the phylogenetic relationships of these three TSWV isolates to other identified TSWV isolates, we used the amino acid sequences of the RdRp and N proteins, which are the largest and smallest proteins, respectively (Fig. 2). We selected these proteins for phylogenetic analysis due to their presence within the viral envelope and functional importance for TSWV. We retrieved 4 and 24 protein sequences for RdRp and N proteins, respectively, from the protein database at NCBI. On the basis of RdRp protein sequences, the newly isolated TSWV-2 and TSWV-3 clustered together with two known isolates from Japan and Korea (Fig. 2a). The host plants in this group were identified as pepper and paprika, belonging to the species Capsicum annum. The TSWV-1 isolate from tomato was highly homologous to an isolate from Hawaii for which no information about the host plant was given. The isolate from Brazil seemed to be very different from the other TSWV isolates.
A phylogenetic tree based on N protein sequences displayed four different groups of TSWV isolates (Fig. 2b). In group I, all of its members had been isolated in the USA, including North Carolina, Colorado, and California. However, their host plants were diverse, including aster, buttercup, dahlia, chrysanthemum, falso lulo, peanut, and tasselflower. Surprisingly, all three isolates analyzed in this study fell into group II. Except for one isolate from North Carolina, all isolates in group II were from Europe and Asia, including Spain, Bulgaria, Italy, Germany, the Netherlands, Japan, and Korea. The host plant species for this group included tomato, pepper, loosestrife, and dahlia. Group III included three isolates from tobacco plants in Bulgaria as well as two isolates from tomato and pepper in Brazil. Interestingly, group IV consisted of only one isolate from tobacco in Bulgaria, which is different from the other TSWV isolates. The phylogenetic study suggested that three newly identified TSWV isolates might have their origin in Asia. Together, these results suggest that the geographical region plays an important role in distinguishing various TSWV isolates. The complete genome sequences of three TSWV isolates with molecular diversity will provide fundamental information to help elucidate molecular interactions between host plants and TSWV. Furthermore, this information can be applied practically in molecular breeding to develop TSWV-resistant vegetables and crops.
References
de Haan P, Wagemakers L, Peters D, Goldbach R (1990) The S RNA segment of Tomato spotted wilt virus has an ambisense character. J Gen Virol 71:1001–1007
de Haan P, Kormelink R, de Oliveira Resende R, van Poelwijk F, Peters D, Goldbach R (1991) Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J Gen Virol 72:2207–2216
Hanssen IM, Lapidot M, Thomma BP (2010) Emerging viral diseases of tomato crops. Mol Plant Microbe Interact 23:539–548
Heinze C, Letschert B, Hristova D, Yankulova M, Kauadjouor O, Willingmann P, Atanassov A, Adam G (2001) Variability of the N-protein and the intergenic region of the S RNA of Tomato spotted wilt tospovirus (TSWV). New Microbiol 24:175–187
Kim JH, Choi GS, Kim JS, Choi CK (2004) Characterization of Tomato spotted wilt virus from Paprika in Korea. Plant Pathol J 20:297–301
Kormelink R, de Haan P, Meurs C, Peters D, Goldbach R (1992) The nucleotide sequence of the M RNA segment of Tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J Gen Virol 73:2795–2804
Milne RG, Francki RIB (1984) Should Tomato spotted wilt virus be considered as a possible member of the family bunyaviridae? Intervirology 22:72–76
Pappu HR, Jones RA, Jain RK (2009) Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res 141:219–236
Prins M, Goldbach R (1998) The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol 6:31–35
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tsompana M, Abad J, Purugganan M, Moyer JW (2005) The molecular population genetics of the Tomato spotted wilt virus (TSWV) genome. Mol Ecol 14:53–66
Whitfield AE, Ullman DE, German TL (2005) Tospovirus-thrips interactions. Annu Rev Phytopathol 43:459–489
Acknowledgments
We thank Dr. T. L. Sit for reviewing the manuscript. This research was supported in part by grants from the Rural Development Administration (No. PJ006952), the Korea Institute of Planning and Evaluation for Technology of Food, Agriculture, Forestry and Fisheries (No. 710001-03), and the Korea Science and Engineering Foundation grant (No. 20100000126) funded by the Ministry of Education, Science, and Technology (MEST). JSL and WKC were supported by graduate and post-graduate research fellowships, respectively, from the MEST through the Brain Korea 21 Project.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, JS., Cho, W.K., Kim, MK. et al. Complete genome sequences of three tomato spotted wilt virus isolates from tomato and pepper plants in Korea and their phylogenetic relationship to other TSWV isolates. Arch Virol 156, 725–728 (2011). https://doi.org/10.1007/s00705-011-0935-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-011-0935-x