Abstract
The purpose of our study was to investigate the expression of prostate stem cell antigen (PSCA), piwi-like 1 (PIWIL1) and T-box 2 (TBX2) and its correlation with HPV16 infection in cervical squamous cell carcinoma (CSCC). HPV16 was detected by amplifying the HPV16 E7 gene by the polymerase chain reaction (PCR) method, and the expression of PSCA, PIWIL1, TBX2 and HPV16 E7 in 59 CSCCs and matched adjacent normal cervix (MANC) was examined by the streptavidin-peroxidase (SP) method. Fifty-two CSCCs and MANC specimens that were positive for the E7 gene and the E7 protein were identified as infected with HPV16 and included in present study. The rate of infection with HPV16 in CSCC was 52% (27/52), but that in matched adjacent normal cervix (MANC) samples was 4% (2/52). Infection with HPV16 was found to be statistically more frequent in CSCC (P = 0.000). The expression rates of PSCA, PIWIL1 and TBX2 in MANC were 6% (3/52), 8% (4/52) and 2% (1/52), respectively, but those in CSCC were 62% (32/52), 75% (39/52) and 52% (27/52), respectively. Higher expression rates of PSCA, PIWIL1 and TBX2 were observed in CSCC than in MANC (P = 0.000). HPV16 had a statistical positive correlation with PSCA, PIWIL1 and TBX2 in CSCC (P < 0.05). The increased expression of PSCA, PIWIL1 and TBX2 had no correlation with the patient’s age or histological grade P > 0.05). The elevated expression of PSCA and PIWIL1 was associated with invasion of CSCC (P < 0.05). Up-regulated expression of TBX2 had a positive association with lymph node metastasis (P = 0.014). These findings demonstrate for the first time the expression of PSCA, PIWIL1 and TBX2 in CSCC. Their correlation with HPV16 might provide new basic information for investigating the molecular mechanism of HPV and help us to deepen our understanding of the interaction between HPV16 and host cells the carcinogenesis of CSCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Worldwide, cervical cancer remains a leading cause of mortality from gynecologic malignancies [1]. The link between cervical cancer and persistent infection with HPV has been well established [2]. The oncogenic functions of two HPV early proteins, E6 and E7, are crucial for the role of HPV during carcinogenesis: E6 can bind to p53 by forming a trimeric complex with the cellular ubiquitin ligase E6-associated protein, and p53 is rapidly degraded by proteasomes [3–5]; E7 can bind to hypophosphorylated pRB, which is then rapidly degraded by proteasomes, and thereby pRB constitutively releases transcriptional factor E2F [6–8]. The loss of function of these major tumor suppressor gene products due to infection with HPV is a fundamental cause of carcinogenesis in cervical cancer [9]. Investigation of the molecular mechanisms of the effect of HPV16 infection on host cells is an important topic in viral oncology.
Cancer stem cells can play an important role in tumorigenesis and tumor progression [10]. Prostate stem cell antigen (PSCA) is a 123-amino-acid glycoprotein first identified in the LAPC-4 prostate xenograft model of human prostate cancer [11]. PSCA expression in normal tissues is largely prostate-specific, but PSCA transcripts and protein have been found in the transitional epithelium of the bladder and the stomach [12, 13]. PSCA has also recently been shown to be expressed by a majority of bladder and pancreatic cancers [14]. Subsequently, several other tumors were evaluated for PSCA expression, including pancreatic adenocarcinoma [15–17], transitional cell carcinoma [14, 18], renal cell carcinoma [19] and diffuse-type gastric cancer [20].
TBX2 is a member of a family of genes encoding developmental transcription factors, characterized by a 200-amino-acid DNA-binding domain (T-box) [21]. TBX2 performs its function during embryonic development [22, 23] and tumorigenesis [21, 24–26]. It is often amplified in pancreatic cancer [25] and BRCA-1- and BRCA-2-related breast cancers [26]. TBX2 and the closely related gene TBX3 are highly expressed in a number of breast cancer cell lines [27, 28].
PIWIL1 (Hiwi) belongs to the piwi-domain proteins, which are components of ribonucleoprotein complexes [29]. Hiwi is a member of the piwi protein family that plays an important role in stem cell self-renewal, RNA silencing and translational regulation [30]. Overexpression of Piwi, the drosophila homologue, results in an increase both in the number of germline stem cells and in the rate at which they divide [31]. Elevated expression of Mili, one of three mouse Piwi homologues, inhibits apoptosis and stimulates proliferation [32]. Mili and another mouse homologue, Miwi, can form complexes with Piwi-interacting RNAs. These complexes appear to play a regulatory role during mammalian spermatogenesis through either transcription or translation [32]. Expression of Hiwi has been described in seminomas, gastric carcinomas and soft-tissue sarcoma (STS) [31, 33, 34]. There was an association reported between altered expression of Hiwi mRNA and a poor prognosis for STS patients [35].
Few reports have emerged concerning the expression of stem-cell-associated Hiwi, PSCA and TBX2 in cervical squamous cell carcinoma (CSCC), and HPV16 has a causative role in carcinogenesis of CSCC. Therefore, here, we investigate the expression of these cancer stem-cell-associated gene products in CSCC and their relationship to HPV16 in order to provide some basic information on potential molecular mechanisms of the effect of HPV16 infection on carcinogenesis in CSCC.
Materials and methods
Tissue collection
All of the archived formalin-fixed, paraffin-embedded tissue specimens were collected in the First and Second Hospitals of Xi’an JiaoTong University during 2000–2003. The present study was approved by the Ethics Committee of Shaanxi Cancer Institute, and informed consent was obtained from all recruited subjects. None of the patient received preoperative or adjuvant chemotherapy or radiotherapy. Pathological examination of all specimens was performed by two independent experienced pathologists in the pathology departments of the corresponding hospitals according to the World Health Organization (WHO) standard and International Federation of Gynecology and Obstetrics (FIGO) criteria [36]. The patients’ clinicopathological data were obtained from surgical and pathological records. Fifty-nine cases of CSCCs and matched adjacent normal cervix (MANC) samples were tested in our present investigation. Of the 59 CSCCs, 9 cases were classified as grade I (well differentiated), 21 as grade II (moderately differentiated) and 31 as grade III (poorly differentiated). The mean age of patients was 57 years old, and these patients were in a different clinical stages: T1–T2 (n = 35) and T3–T4 (n = 24).
DNA extraction
Samples on 5 to 10 slides were deparaffinized using xylene, rehydrated with graded alcohol and digested with lysis buffer (300 mmol/l NaCl; 50 mmol/l Tris–HCl, pH 8.0, 0.2% SDS) containing proteinase K (200 mg/l), and then incubated at 55°C overnight. DNA was extracted using phenol/chloroform, precipitated with cold alcohol and dissolved in ion-free water. The concentration and purity of the extracted DNA were determined from its optical density at 260 nm.
Polymerase chain reaction for detection of HPV16 infection
For HPV16 DNA detection, E7 was amplified with the forward primer 5′-CGGAATTCATGCATGGAGATACACCTACAT-3′ and the reverse primer 5′-CGGGAAGCTTATGGTTTCTGAGAACAGATGG-3′. The amplicon was approximately 301 bp long. PCR mixtures contained 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 3.5 mM MgCl2, 0.01% gelatin, 200 pmol each primer, 4U Taq DNA polymerase and 100 ng DNA template. The suitability of the samples for PCR amplification was ascertained by testing the β-actin gene. Successful amplification of the β-actin gene fragments indicated that the DNA sample was adequate for PCR analysis and that no PCR inhibitors were present. The primer sequences for the human β-actin gene were 5′-ATCATGTTTGAGACCTTCAACACCCC-3′ and 5′-CATCTCTTGCTCGAAGTCCAGGGCGA-3′, respectively. These sequences flank a 317-bp region in the genomic DNA [37]. The cycling conditions for E7 and β-actin were 94°C for 45 s, 58°C for 45 s and 72°C for 50 s for 30 cycles. To avoid DNA contamination, we carried out DNA extraction, PCR mixture preparation, amplification, and electrophoresis in separated areas. In addition, the plasmid pBR322/HPV16, containing the whole HPV-16 genome, was used as a positive control, and the plasmid PUC19 was used as a negative control to validate the experiment. The amplicons were detected by electrophoresis in 1% agarose and compared with a DL2000 DNA molecular weight marker.
Immunohistochemistry
Immunoperoxidase staining of formalin-fixed, paraffin-embedded tissue sections was performed using the streptavidin-peroxidase (SP) method. After they were deparaffinized and rehydrated in a descending alcohol dilution series, the sections were heated in an 800 W microwave oven at maximum power for 8 min in 0.01 M citrate buffer (pH 6.0) for antigen retrieval. The activity of endogenous peroxidase in the sections was blocked by 0.3% hydrogen peroxide in methanol for 10 min at RT. After being treated with 5% normal goat serum in PBS for 15 min to block non-specific sites, the sections were incubated with primary rabbit monoclonal anti-HPV16 E7 (1:400, Beijing Biosynthesis Biotechnology Co. Ltd), anti-PSCA (1:500, Beijing Biosynthesis Biotechnology Co. Ltd), anti-PIWIL1 (1:600, Beijing Biosynthesis Biotechnology Co. Ltd) and anti-TBX2 (1:400, Beijing Biosynthesis Biotechnology Co. Ltd) antibodies overnight at 4°C. After that, we used an ultrasensitive SP kit (Maixin Biological, Fuzhou, China) and DAB (diaminobenzidine) as a chromogen. The sections were counterstained with hematoxylin and mounted with neutral balsam, and then examined by light microscopy. The immunoactivity of PSCA, PIWIL1 and TBX2 is located in membrane/cytoplasm, cytoplasm and nuclei/cytoplasm of stained cells. A visible brown staining of the cytoplasm/nuclei/membrane of cervical squamous cells in cervical samples indicated a positive result, and negative cells contained a blue color in their nuclei. The staining was scored on a scale from 0 to IV as follows: 0, less than 5% cells were stained; I, 5–25% cells were stained; II, 25–50% cells were stained; III, 50–75% cells were stained; and IV, more than 75% cells were stained. Scores of I–IV were classified as positive, while a score of 0 was negative [38].
Statistical analysis
Statistical significance was determined by the χ2 and Fisher’s exact tests. The correlation between PSCA, PIWIL1, TBX2 and clinicopathological parameters was determined by Spearman’s rank correlation coefficient. Analysis was done by SPSS 11.5 software for windows. Ninety-five percent confidence intervals were calculated to examine statistical significance.
Results
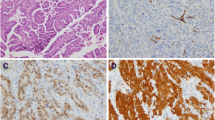
In this study, we examined HPV16 infection by detecting viral oncogene E7 and its protein product in 59 cervix specimens. Both the E7 gene, by PCR detection, and the E7 protein, by immunohistochemical examination, were found to be present in 52 CSCC samples, which were then included in the subsequent analysis. The rate of HPV16 infection in CSCC was 52% (27/52), but that in matched adjacent normal cervix (MANC) samples was 4% (2/52), as shown in Table 1. Statistically, infection with HPV16 was found more frequently in CSCC (P = 0.000) (Fig. 1). The immunoactivity staining of HPV16 E7 was located in the cytoplasm of squamous cells (Fig. 2).
Partial results of PCR detection of HPV16 E7 in CSCC and MANC. M DNA molecular weight standard DL2000 marker, N PCR of PUC19 plasmid as negative control, P PCR of pBR322/HPV16 plasmid as positive control. a Lanes 1 and 3, presence of E7 in CSCC; lane 2, negative PCR results in CSCC. b Lanes 1 and 3, presence of E7 in MANC; lane 2, absence of E7 in MANC
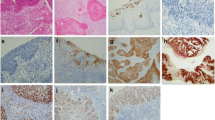
Representative immunohistochemical staining for PSCA, PIWIL1 and TBX2 in CSCC. a Positive staining “3+” for PSCA in CSCC (×200). b Negative staining for PSCA in MANC (×200). c Positive staining “3+” for PIWIL1 in CSCC (×400). d Negative staining for PIWIL1 in MANC (×200). e Positive staining “4+” for TBX2 in CSCC (×200). f Negative staining for TBX2 in MANC (×200). g Positive staining “4+” for HPV16 E7 in CSCC (×200). h Negative staining for HPV16 E7 in MANC (×200)
The expression rates of PSCA, PIWIL1 and TBX2 in MANC were 6% (3/52), 8% (4/52) and 2% (1/52), respectively, but those in CSCC were 62% (32/52), 75% (39/52) and 52% (27/52), respectively. Higher expression rates of PSCA, PIWIL1 and TBX2 were observed in CSCC than in MANC (P = 0.000) (Table 1). The immunoactivity staining of PSCA, PIWIL1 and TBX2 was located in the cytoplasm of cervical squamous cells in CSCC (Fig. 2).
In cervical cancer, 24 (89%) HPV16-positive specimens were scored positive I and above by PSCA staining, while 8 (32%) HPV16-negative specimens were stained to the same degree (P = 0.003). Twenty-six (96%) HPV16-positive specimens were scored positive I and above by PIWIL1 staining, while 13 (52%) HPV16-negative specimens were stained to the same degree (P = 0.020). Twenty-five (93%) HPV16-positive specimens were scored positive I and above by TBX2 staining, while 9 (36%) HPV16-negative specimens were stained to the same degree (P = 0.006). Twenty-one (78%) HPV16 positives and 6 (24%) HPV16 negatives were stained for PSCA as degree II and above (P = 0.000). Nineteen (70%) HPV16 positives and 9 (36%) HPV16 negatives were stained for PIWIL1 as degree II and above (P = 0.012). Twenty-one (78%) HPV16 positives and 5 (20%) HPV16 negatives were stained for PSCA as degree II and above (P = 0.000) (Table 2). HPV16 had a positive correlation with PSCA, PIWIL1 and TBX2 in CSCC (P < 0.05).
In CSCC, the increased expression of PSCA, PIWIL1 and TBX2 had no correlation with the patient’s age or histological grade (P > 0.05). Elevated expression of PSCA and PIWIL1 was associated with invasion of CSCC (P < 0.05), but that of TBX2 was not (P > 0.05). Up-regulated expression of TBX2 had a positive association with lymph node metastasis (P = 0.014), but that of PSCA and PIWIL1 did not (P > 0.05), as indicated in Table 3.
Discussion
It is well recognized that high-risk human papillomavirus type 16 (HPV16) plays an etiological role in the development of cervical cancer [39]. Oncogenic HPV E6 and E7 interrupt host cell functions and therefore make a principal contribution to HPV-induced tumourigenesis. Stem cells and tumour stem cells share several similarities, such as the ability of self-renewal without loss of proliferation capacity [40]. Dysregulation of stem cell self-renewal may be a prerequisite for the development of cancer [41]. In our present study, we examined the variant expression of the stem-cell-associated genes PSCA and PIWIL1 and transcriptional factor TBX2 in CSCC in order to investigate their roles and correlation with HPV16 infection during carcinogenesis of CSCC. Our study results show that elevated expression of PSCA, PIWIL1 and TBX2 might be involved in the onset of CSCC, none of them correlated with histological grade. However, the up-regulated PSCA and PIWIL1 might be candidate markers for clinical prediction of the progression of CSCC, because they were expressed more frequently with increasing clinical stage, while TBX2 might not, due to insignificant differences in its expression in the different clinical stages. Interestingly, TBX2 expression might be correlated with the outcome of CSCC because of its higher expression frequency in CSCC with lymph node metastasis compared to CSCC with no lymph node metastasis.
The subcellular locations of PSCA, PIWIL1 and TBX2 in CSCC were revealed by immunohistochemistry in our present study. PSCA protein was apparently located in the cytoplasm of cervical squamous cells, which is consistent with previous observations in prostate [42] and pancreatic adenocarcinoma [15], but PSCA had been shown to be a cell-surface protein [11].One possible explanation for this discrepancy is that anti-PSCA antibody can recognize PSCA peptide precursors that reside in the cytoplasm. Also, it is possible that the positive staining that appears in the cytoplasm is actually from the overlying cell membrane [14]. Few studies have investigated the subcellular location of PIWIL1, but it was reported by Liu et al. [33] that PIWIL1 was located in the cytoplasm of stained cells in gastric cancer, and our study demonstrated immunoactivity staining of PIWIL1 located in the cytoplasm of squamous cells in CSCC. In previous investigations, TBX2 has been detected at the mRNA level, but there has been no report of the subcellular location of the TBX2 protein. In our present study, TBX2 was observed to be located in the cytoplasm of squamous cells in CSCC.
It has been well documented in cervical cancer that the episomal viral DNA frequently integrates into the host genome as HPV-infected lesions progress to cervical cancer. During viral DNA integration, the E6 and E7 genes always remain together in the host genome [43]. Therefore, the presence of E6 or E7 genes in tumour tissues may better represent the real HPV infection. In some studies, L1 was used as the only indicator, which may have been lost or not expressed in the malignant specimens [39]. Our study examined infection by HPV16 in CSCC by detecting oncogenic E7 in order to avoid false negative results. The immunohistochemical detection of HPV16 E7 protein in CSCC revealed its cytoplasmic location, which was consistent with previous observations [44, 45]. Our study shows that PSCA, PIWIL1 and TBX2 were positively correlated with HPV16, which raised the question whether HPV16 viral oncoprotein might be involved in up-regulating the expression of cancer stem-cell-associated genes and suggests a complex molecular mechanism for the role of HPV16 infection during carcinogenesis of CSCC. Further investigations should be carried out using a cell or animal model to demonstrate a definite role of HPV16 in modulating the expression of PSCA, PIWIL1 and TBX2.
References
Roden R, Wu TC (2006) How will HPV vaccines affect cervical cancer? Nat Rev Cancer 6:753–763
Burd EM (2003) Human papillomavirus and cervical cancer. Clin Microbiol Rev 16:1–17
Scheffner M, Werness BA, Huibregtse JM et al (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136
Huibregtse JM, Scheffner M, Howley PM (1991) A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J 10:4129–4135
Scheffner M, Huibregtse JM, Vierstra RD et al (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495–505
Chellappan S, Kraus VB, Kroger B et al (1992) Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA 89:4549–4553
Boyer SN, Wazer DE, Band V (1996) E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res 56:4620–4624
Jones DL, Thompson DA, Munger K (1997) Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology 239:97–107
Munger K, Baldwin A, Edwards KM et al (2004) Mechanisms of human papillomavirus-induced oncogenesis. J Virol 78:11451–11460
Gangemi R, Paleari L, Orengo AM et al (2009) Cancer stem cells: a new paradigm for understanding tumor growth and progression and drug resistance. Curr Med Chem 16:1688–1703
Reiter RE, Gu Z, Watabe T et al (1998) Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA 95:1735–1740
Gu Z, Thomas G, Yamashiro J et al (2000) Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene 19:1288–1296
Ross S, Spencer SD, Holcomb I et al (2002) Prostate stem cell antigen as therapy target: tissue expression and in vivo efficacy of an immunoconjugate. Cancer Res 62:2546–2553
Amara N, Palapattu GS, Schrage M et al (2001) Prostate stem cell antigen is overexpressed in human transitional cell carcinoma. Cancer Res 61:4660–4665
Argani P, Rosty C, Reiter RE et al (2001) Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res 61:4320–4324
Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL et al (2002) Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 160:1239–1249
Wente MN, Jain A, Kono E et al (2005) Prostate stem cell antigen is a putative target for immunotherapy in pancreatic cancer. Pancreas 31:119–125
Elsamman E, Fukumori T, Kasai T et al (2006) Prostate stem cell antigen predicts tumour recurrence in superficial transitional cell carcinoma of the urinary bladder. BJU Int 97:1202–1207
Elsamman EM, Fukumori T, Tanimoto S et al (2006) The expression of prostate stem cell antigen in human clear cell renal cell carcinoma: a quantitative reverse transcriptase-polymerase chain reaction analysis. BJU Int 98:668–673
Sakamoto H, Yoshimura K, Saeki N et al (2008) Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet 40:730–740
Rowley M, Grothey E, Couch FJ (2004) The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia 9:109–118
Papaioannou VE, Silver LM (1998) The T-box gene family. Bioessays 20:9–19
Harrelson Z, Kelly RG, Goldin SN et al (2004) Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131:5041–5052
Barlund M, Monni O, Kononen J et al (2000) Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res 60:5340–5344
Mahlamaki EH, Barlund M, Tanner M et al (2002) Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer 35:353–358
Sinclair CS, Adem C, Naderi A et al (2002) TBX2 is preferentially amplified in BRCA1- and BRCA2-related breast tumors. Cancer Res 62:3587–3591
Jacobs JJ, Keblusek P, Robanus-Maandag E et al (2000) Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet 26:291–299
Fan W, Huang X, Chen C et al (2004) TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res 64:5132–5139
Grochola LF, Greither T, Taubert H et al (2008) The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer 99:1083–1088
Lingel A, Sattler M (2005) Novel modes of protein-RNA recognition in the RNAi pathway. Curr Opin Struct Biol 15:107–115
Qiao D, Zeeman AM, Deng W et al (2002) Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene 21:3988–3999
Lee JH, Schutte D, Wulf G et al (2006) Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet 15:201–211
Liu X, Sun Y, Guo J et al (2006) Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer 118:1922–1929
Taubert H, Greither T, Kaushal D et al (2007) Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 26:1098–1100
Taubert H, Wurl P, Greither T et al (2007) Stem cell-associated genes are extremely poor prognostic factors for soft-tissue sarcoma patients. Oncogene 26:7170–7174
Hermanek P, Sobin LH, International Union against Cancer (1987) TNM classification of malignant tumours, 4th, fully revised edn. Springer-Verlag, New York
Gall K, Pavicic D, Pavelic J et al (1993) PCR amplification of DNA from stained cytological smears. J Clin Pathol 46:378–379
Zhou Y, Pan Y, Zhang S et al (2007) Increased phosphorylation of p70 S6 kinase is associated with HPV16 infection in cervical cancer and esophageal cancer. Br J Cancer 97:218–222
Bosch FX, Lorincz A, Munoz N et al (2002) The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 55:244–265
Ribacka C, Pesonen S, Hemminki A (2008) Cancer, stem cells, and oncolytic viruses. Ann Med 40:496–505
Soltysova A, Altanerova V, Altaner C (2005) Cancer stem cells. Neoplasma 52:435–440
Zhigang Z, Wenlv S (2004) Prostate stem cell antigen (PSCA) expression in human prostate cancer tissues and its potential role in prostate carcinogenesis and progression of prostate cancer. World J Surg Oncol 2:13
zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2:342–350
Kim KH, Yoon DJ, Moon YA et al (1994) Expression and localization of human papillomavirus type 16 E6 and E7 open reading frame proteins in human epidermal keratinocyte. Yonsei Med J 35:1–9
Smotkin D, Wettstein FO (1987) The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein. J Virol 61:1686–1689
Acknowledgments
This work was supported by the Scientific Research Foundation of the Health Ministry of Shaanxi Province in China (NO. 08D03) and the Scientific Research Foundation of postdoctor in China (NO.20090451383).
Conflict of interest statement
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, WK., Jiang, XY. & Zhang, ZX. Expression of PSCA, PIWIL1 and TBX2 and its correlation with HPV16 infection in formalin-fixed, paraffin-embedded cervical squamous cell carcinoma specimens. Arch Virol 155, 657–663 (2010). https://doi.org/10.1007/s00705-010-0635-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0635-y