Abstract
A highly pathogenic swine disease designated as ‘porcine high fever disease (PHFD)’ appeared recently in China. Porcine reproductive and respiratory syndrome virus (PRRSV) was identified as an agent associated with PHFD, and two discontiguous sequence deletions were identified as a genetic marker in the Nsp2 region of the viral genome. To examine PHFD in Shandong province, a total of 10 PRRSV isolates were recovered from pig herds that had never been vaccinated for PRRS. Sequence analysis of open reading frame 5 (ORF5) showed that the level of identity among the 10 isolates ranged between 88.2 and 99.2%. For the non-structural protein 2 (Nsp2) gene, three isolates shared high sequence identity with VR-2332, the prototype virus of the North American genotype, while the remaining seven isolates exhibited two discontiguous sequence deletions that were identical to those of PHFD: a one-amino-acid (phenylalanine) deletion at position 482 and a 29-amino-acid deletion at positions 533–561 of Nsp2. Experimental infection of pigs with SD-JN, which was one of the seven isolates containing such deletions, resulted in severe clinical symptoms characterized by red discoloration on the body and hemorrhages in the lungs, kidneys, and inguinal lymph nodes, accompanied by higher mortality and longer duration of viremia. These symptoms were similar to those of PHFD observed in the field. Our results show that VR2332-like PRRSV coexists with PHFD-associated atypical PRRSV in pig herds in the Shandong area, and different PRRSV isolates differ greatly in their pathogenesis and virulence in pigs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine reproductive and respiratory syndrome (PRRS) emerged in Europe and North America almost simultaneously, but independently, in the late 1980s. PRRS is characterized by reproductive failure in pregnant sows and respiratory problems in sucking piglets [13, 14]. Since its emergence, the disease has spread globally to most pig-producing countries and has been the cause of significant economic losses to the pork industry [1, 19]. The causative agent of PRRS is PRRS virus (PRRSV), which is a member of the family Arteriviridae in the order Nidovirales, together with the families Coronaviridae and Roniviridae. PRRSV is a small enveloped virus possessing a single-stranded positive-sense RNA genome of approximately 15 kb in size and contains nine open reading frames (ORFs): ORF1a, ORF1b, and ORFs 2 through 7, in order starting from the 5′-end of the genome [11, 26]. Both ORF1a and ORF1b code for non-structural proteins (Nsp) that are believed to be involved in genome replication and transcription, and ORFs 2–7 code for viral structural proteins GP2a, GP2b, GP3, GP4, GP5, membrane (M) protein, and nucleocapsid (N) protein, respectively [18]. The European-type PRRSV (prototype virus Lelystad) is only about 60% similar to the North American-type PRRSV (prototype virus VR-2332) at the genomic sequence level. Both types form two distinct genotypes, and the isolates in each genotype exhibit significant sequence variation [4, 8]. Among the viral genes, ORF5 and Nsp2 are two of the most variable genes, with sequence variability up to 17 and 30%, respectively, in isolates of the North American genotype [2, 6, 9, 17, 22]. Thus, it appears that the viral genome changes quickly and PRRSV evolves rapidly.
In China, PRRSV was first isolated in 1995 from a fetus of an affected animal [12], and it was shown to belong to the North American genotype [6]. Since then, PRRSV circulating in China has been predominantly of the North American type. In the spring of 2006, a highly pathogenic PRRS broke out in the central region of China and quickly spread throughout the country [23, 24, 29]. Unlike the previous PRRS, the newly appearing PRRS caused high morbidity of 50–100% and a mortality rate of 20–100%. The high mortality in grown pigs is unusual and led to a countrywide epidemic of atypical PRRS. Subsequently, the atypical PRRS that emerged in China was designated as “porcine high fever disease (PHFD)”. The highly virulent atypical PRRS occurred in many herds vaccinated with commercial PRRS vaccines, suggesting that the commercial vaccines were unable to protect pigs from PRRS. Shandong province lies in the east of China and is one of the major pig-producing provinces in China, with an annual production of more than 60 million pigs. Recently, in the Shandong area, increased incidence was observed in pigs with high fever (40.5–42.0°C) accompanied with respiratory disorders and red discoloration of the ears and bodies, which resulted in increased economic losses to the pork industry in the province. Based on serological evidence, we examined PRRS in the affected pigs during 2004–2007 and recovered 10 PRRS virus isolates. The ORF5 and Nsp2 genes were sequenced from these isolates, and pigs were experimentally inoculated with isolated viruses. The clinical signs in the infected pigs were similar to those of PHFD observed in the field, and here, we report the results of the study.
Materials and methods
Cells and virus
MARC-145 cells were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 8% fetal bovine serum (Fisher Scientific, Pittsburgh, PA), 100 units/ml penicillin G and 100 μg/ml streptomycin sulfate, and PRRS was propagated in MARC-145 cells. A total of 10 virus isolates were recovered from the lung and tonsil homogenates of diseased pigs. These pigs had not been vaccinated against PRRS. The virus isolates were plaque-purified in Marc-145 cells and propagated for 10 passages. The 50% tissue culture infective dose (TCID50) was calculated by the Reed–Muench method [21], based on the number of wells showing typical cytopathic effect (CPE) for PRRSV.
Reverse transcription-polymerase chain reaction (RT-PCR) for the ORF5 and Nsp2 genes
Two pairs of primers were designed for use in RT-PCR: primer 1 (5′-TGTGAATTCATGTTGGGGAAATGCTTGACC-3′) and primer 2 (5′-CCACTCGAGCCTTTTGTGGAGCCGTGCTAT-3′) for ORF5; primer 3 (5′-TGGTCCTAACGGTTCGGAAGAAAC-3′) and primer 4 (5′-GTGAGCTGAGTATTTTGGGCGTGT-3′) for the amplification of a portion of Nsp2 gene. Total RNA was extracted from 100 μl of PRRSV using Trizol (Invitrogen) according to the manufacturer’s instructions. One-step RT-PCR was carried out as described previously using reagents purchased from Takara Bio Inc. (Dalian, China) according to the manufacturer’s instructions [5]. The first-strand cDNA was synthesized at 50°C for 30 min, and the reaction was heated at 94°C for 2 min, followed by amplification for 35 cycles of denaturation at 94°C for 1 min, annealing for 50 s at 54°C for ORF5, or 55°C for the Nsp2 gene, and extension at 72°C for 1 min. At the end of the last cycle, the reaction was extended at 72°C for 10 min. PCR products were resolved by 1% agarose gel electrophoresis.
Sequence analysis
PCR products were purified using a purification kit (Invitrogen) and subsequently cloned into a pMD18-T (Takara Bio Inc.) vector for sequencing. To obviate possible base mismatches resulting from PCR, three clones were generated from each cDNA fragment and used for sequencing. Nucleotide sequences were analyzed using sequence analysis software (Hitachi DNAStar, version 7.1, Madison WI). The sequences for Nsp2 and GP5 genes of individual isolates were deposited in the GenBank database with the following accession numbers: Shandong-3 (FJ422121), SD-ZQ (FJ422122), SD-JZ (FJ422123), SD1 (FJ422124), SD2 (FJ422125), SD3 (FJ422126), SD4 (FJ422127), SD5 (FJ422128), SD6 (FJ422129), SD7 (FJ422130).
Animal infection
Fifteen piglets of 6 weeks of age were purchased and randomly allotted to three groups, each group consisting of five pigs. These pigs were prescreened by serology and shown to be free of antibody to PRRS, swine influenza, and mycoplasma. Pigs in each group were inoculated intranasally with 2 ml of PRRSV SD1 isolate (107.1 TCID50/ml), SD-JN isolate (105.0 TCID50/ml) or DMEM. Rectal temperatures and clinical signs were monitored daily, and blood samples were collected at 0, 4, 7, 14, 21, 28 and 35 days post-inoculation (dpi) to determine viremia. The animals were killed at 35 dpi, and autopsy was performed for examination of pathological lesions. Antibody titers were determined using the commercially available PRRSV antibody detection kit (HerdCheck PRRS; IDEXX, Westerbrook, Maine, USA) according to the manufacturer’s instructions. ELISA results were expressed as a ratio of the optical density to the positive control (S/P ratio). According to the assay, an S/P ratio greater than 0.4 is considered positive for the presence of PRRSV antibodies. The animal infection protocol was approved by the Shandong Province Animal Ethics Committee and the study was conducted in the animal isolation facility of the Shandong Provincial Laboratory in accordance with the protocol.
Results
Isolation and characterization of PRRSV associated with PHFD
Clinical specimens were collected from 10 different outbreaks of PRRS in the Shandong area during the period of 2004–2007, and PRRS virus was isolated from each specimen in MARC-145 cells. Typical CPE for PRRSV was observed in MARC-145 cells by 3–5 days post-inoculation as shown by cell conglomeration, dissolution, or desquamation of the monolayer. Cell culture fluids were collected, and virus was purified by plaque assay. The purified virus was amplified in MARC-145 cells and titrated for TCID50. The titers ranged between 104.5 and 107.1 TCID50/ml (Table 1). To confirm the specificity of the virus, the ORF5 sequence of PRRSV was amplified by RT-PCR using the North American type PRRSV-specific primers, and a 700-bp product was specifically amplified. The sequencing confirmed the specificity for PRRSV.
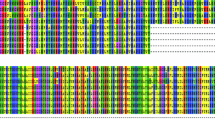
Since ORF5 is one of the most variable genes of PRRSV [15], the full-length ORF5 gene was sequenced for all 10 isolates, and these were compared to each other (Fig. 1). It has been postulated that amino acid residues at positions 13 and 151 of the GP5 protein are virulence-associated for PRRSV [3]. For the Shandong isolates, the amino acid at position 13 was arginine (R) for nine isolates, and only the SD2 isolate had glutamic acid (Q). The amino acid at position 151 was ‘R’ for isolates SD3, SD4, SD5, SD7, Shandong-3, SD-JN and SD-ZQ, whereas that of SD1, SD2 and SD6 was substituted by glycine (G) (Fig. 1). PRRSV SD1, SD2 and SD6 shared 99.3–99.5% nucleotide sequence homology with VR-2332 for ORF5 and shared 64.9% with Lelystad virus. In contrast, the other isolates SD3, SD4, SD5, SD7, Shandong-3, SD-JN, and SD-ZQ exhibited only 87.6–88% and 63.9–64.9% homologies with VR-2332 and Lelystad virus, respectively (Fig. 2). All isolates in the Shandong area were more closely related to the North American genotype than the European genotype based on phylogenetic analysis of ORF5 (Fig. 3). In particular, isolates SD3, SD4, SD5, SD7, Shandong-3, SD-JN and SD-ZQ were similar in their ORF5 sequences to the JXA1, Shanghai, and HEB1 isolates, which were identified to be associated with porcine high fever disease (PHFD) in China [23, 24, 29]. Isolates SD1, SD2, and SD6 belonged to the main branch, closely related to the North American prototype VR2332 (Fig. 3).
Amino acid sequence alignments of the GP5 protein from 10 isolates recovered from PRRSV outbreaks during 2004–2007 in the Shandong area. Boxes represent amino acid positions 13 and 151, respectively. GenBank accession numbers: JXA1 (EF112445), MLV RespPRRS/Repro (AF159149), VR2332 (EF536003), Shandong-3 (FJ422121), SD-ZQ (FJ422122), SD-JZ (FJ422123), SD1 (FJ422124), SD2 (FJ422125), SD3 (FJ422126), SD4 (FJ422127), SD5 (FJ422128), SD6 (FJ422129), SD7 (FJ422130)
The Nsp2 gene is known to be variable in PRRSV [9, 22], and thus the Nsp2 sequences of these isolates were examined for genetic variation. A specific product of 494 bp in size was identified by RT-PCR from three isolates (SD1, SD2 and SD6). For the remaining seven isolates (SD3, SD4, SD5, SD7, Shandong-3, SD-JN and SD-ZQ), a smaller-sized product of 404 bp was amplified. Both the smaller and larger products were sequenced to examine the genetic variation of the isolates. Two separate deletions were identified for isolates SD3, SD4, SD5, SD7, Shandong-3, SD-JN, and SD-ZQ in comparison to SD1, SD2, and SD6 (Fig. 4). These deletions resulted in a single amino acid deletion at position 483 of the Nsp2 protein (position 5 in Fig. 4) and a continuous 29-amino-acid deletion at positions 535–563 of Nsp2 (positions 55–83 in Fig. 4). Both deletions were identical to those of PHFD-associated PRRSV recently reported in China [23, 29, 30]. No deletion was found in isolates SD1, SD2 or SD6. This finding suggests that an isolate containing two discontiguous deletions is likely to be an agent associated with the highly pathogenic PRRS that recently emerged in China.
Amino acid sequence alignments of the partial Nsp2 gene. Amino acid deletions are boxed. The deletion at amino acid position 5 in the figure corresponds to amino acid position 483 of the entire Nsp2 protein, and the 29-amino-acid deletion at positions 55–83 in the figure corresponds to positions 535–563 of the Nsp2 protein
Pathogenesis and virulence of the atypical PRRSV in pigs
To determine the virulence and pathogenesis of the isolated PRRSV, an experimental infection was performed in pigs. For this study, SD1 and SD-JN were chosen as representative viruses: SD1 as a strain without a deletion, and SD-JN as a strain containing the two discontiguous deletions in the Nsp2 gene (Fig. 4). The viruses were plaque-purified once in MARC-145 cells and amplified for preparation of an inoculum. Fifteen 6-week-old pigs were obtained from a PRRS-free herd and randomly divided into three groups. Each group of five pigs was infected with SD1 or SD-JN, or DMEM as placebo, and maintained for 5 weeks. Two pigs in the SD1-infected group showed a moderate level of anorexia and depression at 7 dpi, and the symptoms persisted for 4 days before they disappeared. Other pigs in this group showed no obvious fever, and their rectal temperatures ranged between 39.2 and 39.9°C. None of the pigs in the SD1 group died during the experiment. For pigs in the SD-JN group, high fever of over 41°C was observed at 4–6 dpi, and the fever persisted for 5–10 days thereafter. One pig in this group died at 4 dpi, and two additional pigs died at 9 dpi. Only two pigs remained alive by 35 dpi. The dead pigs all showed red discoloration (Fig. 5c) and multiple lesions in various organs, such as hyperplasia and hemorrhagic spots in the lungs (Fig. 5d), hemorrhagic spots in the kidney (Fig. 5e), and hemorrhages and necrosis in the inguinal lymph node (Fig. 5f). For SD1-infected pigs, inguinal lymph nodes were swollen, and mild lung lesion were observed upon autopsies at 35 dpi, but hemorrhages were absent in the lungs, kidneys, and inguinal lymph nodes. While mild interstitial pneumonia was observed in the pigs infected with SD1 (Fig. 6b), severe interstitial pneumonia characterized by severe atrophy of alveoli and proliferation and soakage of lymphocytes was observed in the SD-JN-infected pigs (Fig. 6c). Consolidation of alveoli and disappearance of septum alveoli were also identified in the SD-JN-infected pigs. Pigs in the control group inoculated with DMEM were normal in appearance, appetite, and rectal temperature during the course of experiment, and no obvious pathological changes were observed in the organs. The mock-infected pigs showed no histopathological changes in the alveoli and alveolar septa (Fig. 6a). To examine the serologic profiles in PRRSV-infected pigs, antibody detection, RT-PCR for viral genome detection, and virus isolation were conducted from weekly serum samples. In pigs infected with either the SD1 or the SD-JN isolate, the N gene was amplifiable from sera taken at 4 dpi (data not shown), and the corresponding virus was recovered from the same sera by cell culture. The viremia caused by the SD1 and SD-JN isolates persisted for 14 and 21 days, respectively, in the infected pigs, (Table 2), and these pigs seroconverted by 14 dpi. In SD1- and SD-JN-infected pigs, specific antibody was detectable by 7 dpi, increased quickly by 14 dpi and thereafter remained high for both groups (Fig. 7). Taken together, the infection study shows that SD-JN virus with two deletions in the Nsp2 gene was indeed an agent associated with PHFD, and in the Shandong area, this agent co-circulates with typical North American-type PRRSV.
Pathological changes in a pig that died at 4 days post-infection from the SD-JN virus infection (a–f) and the plaque morphology of the SD-1 and SD-JN viruses (bottom panels). The pig infected with the SD-1 virus showed normal skin color (a), and no abnormal pathology was observed in the lung (b). In contrast, the pig infected with SD-JN displayed red discoloration of the skin (c), hyperplasia and hemorrhagic spots in the lung (d), hemorrhagic spots in the kidney (e), and hemorrhages and necrosis in the inguinal lymph node (f). The plaque morphology of the SD-1 and SD-JN viruses was similar (bottom panels)
Discussion
In the present study, PRRSV isolates were obtained between 2004 and 2007 from different herds experiencing clinical symptoms of PRRS in the Shandong area. These herds had never been vaccinated against PRRS. By cloning and sequencing the ORF5 and Nsp2 genes, three isolates (SD1, SD2, and SD6) were found to be typical PRRSV, and they shared high sequence homology with VR2332. Of the three isolates, SD1 and SD2 were obtained prior to the 2006 epidemics of PHFD, and SD6 was obtained in 2006 (Table 1). The remaining seven isolates (SD3, SD4, SD5, SD7, Shandong-3, SD-JN, and SD-ZQ) were obtained during the period of 2006–2007, and they contained two discontiguous sequence deletions of 30 amino acids in Nsp2. The Nsp2 gene of the seven isolates showed a high degree of nucleotide sequence identity with PHFD-associated PRRSV. These results indicate that the typical PRRSV and atypical PRRSV have coexisted in pig herds in the Shandong area of China since 2006.
Different types of CPE were observed for different PRRSV isolates that were obtained from the Shandong area [27], and various titers of different PRRSV isolates were also observed in that study, suggesting the co-existence of phenotypically different PRRSV in this area. It is interesting that the SD1 isolate produced a high titer (107.1 TCID50/ml) of virus but showed relatively low pathogenicity in pigs. SD1-infected pigs presented mild clinical signs and shorter duration of viremia during the course of infection. In contrast, SD-JN produced a lower titer (105.0 TCID50/ml) of virus but caused severe clinical signs and pathological changes accompanied by a longer duration of viremia in the pigs. No obvious difference in the plaque morphology was observed between the two viruses (Fig. 5), and the relationship between growth phenotype and virulence is unknown at the present time.
The experimentally infected pigs were serologically negative for porcine parvovirus, pseudorabies, swine influenza, and Mycoplasma hyopneumoniae. The animals were also negative by PCR for porcine circovirus type 2 (PCV2), Pasteurella multocida, and Actinobacillus pleuropneumoniae. These animals however were not screened for swine dysentery, atrophic rhinitis, transmissible gastroenteritis, and other bacterial pathogens of swine, and thus it is possible that unidentified agents may have contributed to the clinical severity during infection by SD-JN, although it is unlikely.
GP5 is one of the major viral envelope proteins. It is encoded by ORF5 and is an important viral component for induction of neutralizing antibodies. Amino acid residues at positions 13 and 151 of GP5 have been postulated to be associated with viral virulence [5, 25]. PHFD-associated PRRSV isolates have been found to contain arginine (R) at positions 13 and 151, while in low-pathogenic PRRSV isolates and in the modified live vaccine virus, R at both positions was replaced with glutamic acid (13Q) and glycine (151G). In the present study, sequence variations were also observed at the signal peptide (positions 1–31) and one (positions 32–36) of the hypervariable regions. Except SD2, all other isolates contained R at position 13, as reported previously [15]. Nsp2 is one of the most variable proteins in PRRSV, and a number of linearized B cell epitopes have been reported [7]. Deletions have been shown in the Nsp2 region of MN184B and SY0608 strains, both of which are highly pathogenic [16]. Similar observations have been made with the atypical PRRSV isolates described in the present study. Based on PepScan analysis, 18 B-cell epitopes were identified in Nsp2, and five of them (431–445, 441–455, 476–490, 496–510, and 536–550) were distributed in the region where the sequence deletions were located in the PHFD-associated PRRSV [7]. The deletion of amino acids may cause partial losses of B-cell epitopes, possibly leading to an impaired immune response and prolonged viremia [20]. It is consistent with our study that SD-JN, which contained deletions in the Nsp2 region, was associated with severe clinical signs with prolonged viremia in infected pigs, whereas SD1, which did not contain the deletions, was associated with a mild clinical outcome. A recent report, however, has shown that the 30-aa deletion in Nsp2 is not associated with the virulence of PRRSV [29]. Thus, the deletions in Nsp2 may instead serve as a genetic marker for atypical PRRSV and for developing genetically modified marker vaccines to differentiate infection from vaccination [10, 28]. In summary, typical PRRSV and highly virulent atypical PRRSV have coexisted in pig herds in the Shandong area since 2006. The virulence of our atypical PRRSV was much higher than that of typical PRRSV. Whether the emergence of atypical PRRSV was due to the evolution from a typical North American PRRSV in this area remains to be determined.
References
Albina E (1997) Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol 55:309–316
Allende R, Lewis TL, Lu Z, Rock DL, Kutish GF, Ali A, Doster AR, Osorio FA (1999) North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J Gen Virol 80:307–315
Andreyev VG, Wesley RD, Mengeling WL, Vorwald AC, Lager KM (1997) Genetic variation and phylogenetic relationship of 22 porcine reproductive and respiratory syndrome virus (PRRSV) field strains based on sequence analysis of open reading frame 5. Arch Virol 1(142):993–1001
Blaha T (2000) The “colourful” epidemiology of PRRS. Vet Res 31:77–83
Chang CC, Yoon KJ, Zimmerman JJ, Harmon KM, Dixon PM, Dvorak CM, Murtaugh MP (2002) Evolution of porcine reproductive and respiratory syndrome virus during sequential passages in pigs. J Virol 76:4750–4763
Chen J, Liu T, Zhu CG, Jin YF, Zhang YZ (2006) Genetic variation of Chinese PRRSV strains based on ORF5 sequence. Biochem Genet 44:421–431
De Lima M, Pattnaik AK, Flores EF, Osorio FA (2006) Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology 353:410–421
Done SH, Patton DJ, White ME (1996) Porcine reproductive and respiratory syndrome (PRRS): a review, with emphasis on pathological, virological and diagnostic aspects. Br Vet J 152:153–174
Fang Y, Kim DY, Ropp S, Steen P, Christopher-Hennings J, Nelson EA, Rowland RR (2004) Heterogeneity in Nsp2 of European-like porcine reproductive and respiratory syndrome viruses isolated in the United States. Virus Res 100:229–235
Fang Y, Rowland RR, Roof M, Lunney JK, Christopher-Hennings J, Nelson EA (2006) A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: expression of green fluorescent protein in the Nsp2 region. J Virol 80:11447–11455
Grebennikova TV, Glouser DF, Vorwald AC, Musienko MI, Mengeling WL, Lager KM, Wesley RD, Biketov SF, Zaberezhny AD, Aliper TI, Nepoklonov EA (2004) Genomic characterization of virulent attenuated, and revertant passages of a North American porcine reproductive and respiratory syndrome virus strain. Virology 321:383–390
Guo B, Chen Z, Liu W, Cui Y (1996) Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from fetuses suspicious of PRRS. Chin J Anim Poult Infect Dis 17:1–4
Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews AA, Rathje JA (1995) Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with of Lelystad virus. Vet Pathol 32:648–660
Johnson W, Roof M, Vaughn E, Christopher-Hennings J, Johnson CR, Murtaugh MP (2004) Pathogenic and humoral immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) are related to viral load in acute infection. Vet Immunol Immunopathol 102:233–247
Key KF, Haqshenas G, Guenette DK, Swenson SK, Toth TE, Meng XJ (2001) Genetic variation and phylogenetic analyses of the ORF5 gene of acute porcine reproductive and respiratory syndrome virus isolates. Vet Microbiol 83:249–263
Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P (2007) Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J 174:577–584
Mateu E, Díaz I, Darwich L, Casal J, Martín M, Pujols J (2006) Evolution of ORF5 of Spanish porcine reproductive and respiratory syndrome virus strains from 1991 to 2005. Virus Res 115:198–206
Meulenberg JJ (2000) PRRSV, the virus. Vet Res 31:11–21
Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ (2005) Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392
Oleksiewicz MB, Bøtner A, Toft P, Normann P, Storgaard T (2001) Epitope mapping porcine reproductive and respiratory syndrome virus by phage display: the nsp2 fragment of the replicase polyprotein contains a cluster of B-cell epitopes. J Virol 75:3277–3290
Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497
Shen S, Kwang J, Liu W, Liu DX (2000) Determination of the complete nucleotide sequence of a vaccine strain of porcine reproductive and respiratory syndrome virus and identification of the Nsp2 gene with a unique insertion. Arch Virol 145:871–883
Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF (2007) Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS one 6:1–10
Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ (2007) Highly pathogenic porcine reproductive and respiratory syndrome in China. Emerg Infect Dis 13:1434–1436
Wesley RD, Mengeling WL, Lager KM, Clouser DF, Landgraf JG, Frey ML (1998) Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF 5. J Vet Diagn Invest 10:140–144
Wootton S, Yoo D, Rogan D (2000) Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch Virol 145:2297–2323
Wu J, Wang J, Liu Y, Wang W, Zhang X, Yoo D (2008) Relationship between herd size and the prevalence of porcine reproductive and respiratory syndrome in China. Vet Rec 163:90–91
Yoo D, Welch SK, Lee C, Calvert JG (2004) Infectious cDNA clones of porcine reproductive and respiratory syndrome virus and their potential as vaccine vectors. Vet Immunol Immunopathol 102:143–154
Zhou L, Zhang J, Zeng J, Yin S, Li Y, Zheng L, Guo X, Ge X, Yang H (2009) The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J Virol 83:5156–5167
Zhou YJ, Hao XF, Tian JZ, Tong GZ, Yoo D, An TQ, Zhou T, Li GX, Qiu HJ, Wei TC, Yuan XF (2008) Highly virulent porcine reproductive and respiratory syndrome virus emerged in China. Transbound Emerg Dis 55:152–164
Acknowledgments
This study was supported by the Shandong Academy of Agricultural Sciences Youth Funds (Grant No. 2005YQ039), Natural Scientific Funds of Shandong Province (Grant No. Z2007-D06), and Main and Special Funds for Science and Technology of Shandong Province (Grant No. 2007-ZHZX11103). DY is a recipient of the grant from USDA National Research Initiative Competitive Grants Program (2008-35204-04634).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, J., Li, J., Tian, F. et al. Genetic variation and pathogenicity of highly virulent porcine reproductive and respiratory syndrome virus emerging in China. Arch Virol 154, 1589–1597 (2009). https://doi.org/10.1007/s00705-009-0478-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0478-6