Abstract
The hemagglutinins (HAs) of H9 influenza viruses isolated from birds and mammals of different species were antigenically and genetically analyzed. Antigenic variants were selected from A/swine/Hong Kong/10/98 (H9N2) and A/duck/Hokkaido/13/00 (H9N2) in the presence of monoclonal antibodies (MAbs). Based on the reactivity patterns of these mutants with a panel of MAbs, at least five non-overlapping antigenic sites were defined using eight MAbs which recognized seven distinct epitopes on the H9 HA molecule. Based on the reactivity patterns with the panel of monoclonal antibodies, 21 H9N2 virus strains isolated from birds and mammals were divided into 7 antigenically distinct groups. The present findings indicate that it is important to monitor the antigenic variation in H9 influenza viruses. The panel of MAbs in the present study, thus, should be useful for detailed antigenic analysis of the H9 HAs for epidemiological studies, the selection of vaccine strains, and diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the 1990s, outbreaks of H9N2 influenza virus infections in poultry have caused great economic losses in many countries in Asia and the Middle East [5–7, 9, 19]. The causal H9N2 virus strains, however, did not reproduce severe disease signs in specific-pathogen-free (SPF) chickens. Co-infection of H9N2 viruses with bacteria such as Staphylococcus aureus and Haemophilus paragallinarum or attenuated coronavirus vaccine strains with H9N2 virus exacerbate the disease [1, 9, 10, 15]. The hemagglutinins (HAs) of H9N2 virus isolates in Asia were antigenically and genetically grouped into three distinct lineages, represented by the viruses A/quail/Hong Kong/G1/97 (H9N2) (G1), A/duck/Hong Kong/Y280/97 (H9N2) (Y280), and A/duck/Hong Kong/Y439/97 (H9N2) (Y439) [5]. Many of the H9N2 viruses isolated from chickens, ducks [6, 7, 17], and pigs in southern China [3, 22, 29] belonged to the Y280 llineage. Antigenic and genetic analyses of H9N2 viruses in poultry revealed that the Y280 lineage designated as the A/chicken/Beijing/1/94-like group was further divided into two subgroups, and triple or quadruple reassotants with gene segments of G1-like, Y280-like, and H5N1-like viruses in Hong Kong in 2001 continued to prevail in southern China [30, 31]. It was also assumed that Y280-like variant viruses originated from those circulating in poultry in Asian countries for over 10 years. These H9N2 influenza viruses had receptor-binding specificity for terminal sialic acid with an α2-6 Gal linkage, which is found on human cells [8, 21]. H9N2 viruses of G1 and Y280 lineage have actually been isolated from humans [18, 23]. Thus, in addition to H5N1 and H7N7, H9N2 viruses are possible candidates with the potential to cause pandemics in humans.
The HA of influenza virus is the major target for immune responses due to its role in mediating attachment to and penetration into host cells. The antigenic structure of HAs of H1, H2, H3, H5, and H9 subtypes of influenza A virus has been investigated by antigenic mapping and sequence analysis [2, 11, 12, 24, 26, 28]. Antigenic mapping of an H1 HA, A/PR/8/34 (H1N1), indicates five immunodominant antigenic sites, designated Sa, Sb, Ca1, Ca2 and Cb [4], which are comparable to those of H3 subtype virus A/Hong Kong/1968 (H3N2), designated A, B, C, D and E [28]. The analysis of H9 antigenic variants defined at least two antigenic sites on the H9 HA molecule [12]. In the present study, antigenic and genetic analyses of the HAs of H9N2 viruses were performed using a panel of monoclonal antibodies (MAbs).
Materials and methods
Viruses
Influenza virus A/swine/Hong Kong/10/98 (H9N2) (Sw/HK/98), A/duck/Hokkaido/13/00 (H9N2) (Dk/Hok/00) and other H9 viruses isolated from birds and mammals of different species were grown in 10-day-old embryonated chicken eggs and were stored at −80°C until used (Table 1). The viruses were purified by differential centrifugation and sedimentation through a sucrose gradient [13].
Monoclonal antibodies
Monoclonal antibodies against Sw/HK/98 (H9N2) and Dk/Hok/00 (H9N2) were prepared as described previously [14]. Briefly, BALB/c mice were immunized with the purified virus. Spleen cells of the mice were fused with myeloma cells, and hybridoma cells secreting specific MAbs were selected. Each of the hybridoma cells was inoculated to mice intraperitoneally, and ascitic fluids containing MAbs were used for assays. Isotypes of MAbs were determined using Mouse Monoclonal Antibody Isotyping Reagents (Sigma, MO, USA).
Serological tests
The hemagglutination-inhibition (HI) test was performed by the standard method [25]. In the neutralization test (NT), titers were determined as the reciprocal of the maximum antibody dilution that prevents the cytopathic effect of 100 TCID50 of virus, using Madin–Darby Canine Kidney (MDCK) cells. Enzyme-linked immunosorbent assay (ELISA) was performed as described previously [14].
Selection of antigenic variants
Antigenic variants were selected and the frequencies of antigenic variants were determined as described previously [14]. Briefly, the virus was incubated with excess antibody for 1 h at room temperature, and the mixture was inoculated into 10-day-old embryonated chicken eggs. The yielded viruses were detected by HA test after 48-h incubation at 35°C and cloned by limiting dilution in embryonated eggs.
Sequence analysis of virus genes
Viral RNAs were isolated from virus-containing allantoic fluid with Trizol LS reagent (Invitrogen, PO, USA). HA genes were sequenced after viral RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR) according to Liu et al. [20]. Other sequence data were assembled and translated to the amino acid sequence using gene analysis software GENETYX-WIN version 6.1.0 (Genetyx Corporation, Tokyo, Japan). The positions of amino acid substitutions on the HA molecule were analyzed on the 3-dimensional structure obtained from the Protein Databank (PDB accession number, 1JSD) with the RasMol 2.7.3 program.
Results
Antigenic mapping of the H9 HA molecule
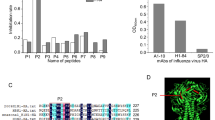
Six anti-HA MAbs against A/swine/Hong Kong/10/98 (H9N2) (Sw/HK/98) and two against A/duck/Hokkaido/13/00 (H9N2) (Dk/Hok/00) were selected as representatives of 50 MAbs obtained in total. Nucleotide sequences of the HA gene of the antigenic variants selected in the presence of each of the MAbs were determined. A single amino acid substitution was found in the deduced amino acid sequence of the HA of each of the antigenic variants (Table 2). A panel of MAbs recognizing eight amino acid positions was established on the basis of the positions of amino acid substitution in the H9 HA molecule. The position of an amino acid substitution at residue 72 (Gly → Glu) on the HA of Sw/L6/2, a mutant selected from Sw/HK/98 in the presence of MAb L6/2, was located in the vicinity of the bottom of the globular head domain of the H9 HA molecule in the proposed antigenic site E in the H3 subtype HA (Fig. 1a). Antigenic variant SwG6/5 had an amino acid substitution at residue 127 (Ser → Asp) at the ‘overlapping site’ of Site I and Site II, that was reported by Kaverin et al. [12]. SwN4/2 had an amino acid substitution at residue 148 (Asn → Asp) at Site I. Substitutions at residues 182 (Thr → Ile) and 183 (Asn → Asp) were located at Site II. An amino acid substitution of SwG12/1 at residue 212 (Leu → His) was found in the vicinity of the trimeric interface of the globular domains of HA1. The Dk/Hok/00 antigenic variants DkD370/4 and DkD272/6 had amino acid substitutions at residues 98 (Leu → Gln) and 131 (Lys → Asn) (137 in H3 HA numbering), respectively.
Schematic representation of monomer structures of the H9 haemagglutinin molecule, showing the locations of amino acid substitutions on HA1. Images were created with RasMol 2.7.3. a Amino acid changes of escape mutants selected with monoclonal antibody against A/swine/Hong Kong/10/98 (H9N2) (red) and A/duck/Hokkaido/13/00 (H9N2) (blue). The positions of the oligosaccharide attachment sites are shown as green lines. Amino acid positions correspond to H9 numbering. b Hypervariable amino acid positions on HAs of H9N2 viruses clustered in the Y280 lineage isolated from 1994 to 2005
The reactivity patterns of antigenic variants with the panel of MAbs showed that the MAbs against Sw/HK/98 and Dk/Hok/00 were divided into seven groups (Table 3). Out of seven groups, five groups of MAbs (groups 1, 2, 5, 6 and 7) recognized independent epitopes, while the other two groups recognized antigenically overlapping epitopes. The mutants SwG6/5 and SwL7/7 did not react with MAb D272/6 in addition to the mutant DkD272/6. The mutants SwG6/5 reacted weakly with MAb N4/2 compared with the parental virus. Therefore, at least five distinct antigenic sites were defined on the H9 HA using the present eight MAbs.
Sequence analysis of the H9 HA molecule
H9N2 influenza viruses have been prevalent in poultry over the last decades. Within this period, many H9N2 viruses were isolated and confirmed to have undergone antigenic variation. Sequence analysis of the H9 HA genes of 133 H9N2 influenza virus strains isolated between 1994 and 2005 showed 28 positions at which amino acid substitutions frequently occurred on the H9 HA molecule (Table 4; Fig. 1b). Twenty-two amino acid substitutions were found on the HA molecule, and two of these (103 and 269) were on the inside of the molecule, whereas the others were found at the HA cleavage site. The positions of the amino acid substitutions in the 3-dimensional structure of the HA molecule revealed that at least four conformational antigenic sites were located on the H9 HA.
Antigenic analysis of the HAs of H9N2 virus isolates using a panel of MAbs
The reactivity of H9N2 influenza viruses isolated from birds and mammals of different species with a panel of MAbs to the HA of Sw/HK/98 and Dk/Hok/00 was analyzed (Table 5). On the basis of the reactivity patterns with the panel of MAbs, 21 H9N2 influenza virus strains were divided into 7 different groups. Viruses belonging to antigenic group I had all seven epitopes, with each MAb recognizing the HA molecule. Viruses belonging to antigenic groups II to VI had several epitopes, and A/shorebird/Delaware/9/96 did not react with any of the MAbs. The HAs of the viruses belonging to the Y280 lineage used in the present study were antigenically conserved and different from those of other lineages.
Discussion
The aim of the present study was to characterize the antigenic structure of the HA molecule of H9N2 influenza viruses recently prevailing in poultry in Asia. The antigenic drift of H9N2 viruses has been detected by reactivity with polyclonal antisera [5, 30, 31]. In the present study, a panel of MAbs to the HA of H9N2 influenza viruses was prepared and used for antigenic comparison of these H9 influenza viruses.
Several HA subtypes have been analyzed using a panel of MAbs [4, 11, 12, 16, 28]. In the present study, the reactivity patterns of antigenic variants with the panel of MAbs showed that the MAbs were divided into seven groups. Of the seven groups, five groups of MAbs recognized independent epitopes. Two antigenic sites on the H9 HA molecules, Site I (amino acid positions 129, 147, and 152) and Site II (amino acid positions 135, 183, and 216), were revealed [20]. According to the positions of amino acid substitutions in the HAs of antigenic variants, MAb G6/5 recognizes Site I, and E2/3 and L7/7 recognizes Site II. MAbs L6/2, G12/1 and D370/4 recognize epitopes in three antigenic sites distinct from sites I and II. The mutants with deduced amino acid changes in Ser127Asn, Lys131Asn and Asn183Asp did not react with MAb D272/6, suggesting that D272/6 recognizes conformational overlapping epitopes. The weak reaction of an antigenic variant selected in the presence of MAb G6/5 with N4/2 may be due to the mutation Ser127Asn, which forms a glycosylation site. The panel of MAbs recognizing seven epitopes in the present study recognized at least five different antigenic sites in the vicinity of the receptor-binding site.
The amino acid comparison of HA molecules of natural isolates belonging to the Y280 lineage of H9N2 viruses showed that 28 amino acid positions were more variable on the HA molecule. These positions are located in the vicinity of the antigenic site known in H1 or H3 subtypes, although there are some differences in 3-dimensional structure. In these positions, six amino acid substitutions were identical to those in the escape mutants selected in the present study and in an earlier publication [12]. It is suggested that at least five antigenic sites were located on the H9 HA molecule and that the antigenic variation occurred in these sites after the long-term prevalence of H9N2 influenza viruses in poultry.
The H9N2 influenza viruses have been divided into Eurasian and North American lineages, and the former lineage has been divided further into Y280, G1 and Y439 sub-lineages [5]. In the present study, it was revealed that reactivity patterns of H9N2 influenza viruses isolated from birds and animals of different species with the panel of MAbs showed that the HA of viruses belonging to the Y280 lineage was antigenically conserved and different from that of other lineages. The present results coincide with the findings that viruses belonging to the Y280 or G1 lineage are antigenically different from other lineages [5].
After long-term prevalence of H9N2 viruses in poultry, it is assumed that the antigenic variants were selected through host immune pressure, and vaccination should also lead to antigenic variation in the viruses. The panel of MAbs prepared in the present study should be useful for detailed antigenic analysis in the epidemiological study of currently circulating H9N2 viruses, especially the viruses belonging to the Y280 lineage. Furthermore, the present panel of MAbs should be useful for the development of an HA subtype-specific diagnosis kit [27].
References
Bano S, Naeem K, Malik SA (2003) Evaluation of pathogenic potential of avian influenza virus serotype H9N2 in chickens. Avian Dis 47:817–822
Caton AJ, Brownlee GG, Yewdell JW, Gerhard W (1982) The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427
Cong YL, Pu J, Liu QF, Wang S, Zhang GZ, Zhang XL, Fan WX, Brown EG, Liu JH (2007) Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J Gen Virol 88:2035–2041
Gerhard W, Yewdell J, Frankel ME, Webster R (1981) Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290:713–717
Guan Y, Shortridge KF, Krauss S, Webster RG (1999) Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA 96:9363–9367
Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M (2000) H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol 74:9372–9380
Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y (2000) Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267:279–288
Ha Y, Stevens DJ, Skehel JJ, Wiley DC (2001) X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci USA 98:11181–11186
Haghighat-Jahromi M, Asasi K, Nili H, Dadras H (2007) Role of infectious bronchitis live vaccine on pathogenicity of H9N2 avian influenza virus. Int J Poult Sci 6:838–841
Haghighat-Jahromi M, Asasi K, Nili H, Dadras H, Shooshtari AH (2008) Coinfection of avian influenza virus (H9N2 subtype) with infectious bronchitis live vaccine. Arch Virol 153:651–655
Kaverin NV, Rudneva IA, Ilyushina NA, Varich NL, Lipatov AS, Smirnov YA, Govorkova EA, Gitelman AK, Lvov DK, Webster RG (2002) Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of antigenic variants. J Gen Virol 83:2497–2505
Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, Webster RG (2004) Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 antigenic variants. J Virol 78:240–249
Kida H, Yanagawa R (1979) Isolation of a new avian paramyxovirus from a rock pigeon (Columba livia). Zentralbl Bakteriol [Orig A] 245:421–428
Kida H, Brown LE, Webster RG (1982) Biological activity of monoclonal antibodies to operationally defined antigenic regions on the hemagglutinin molecule of A/Seal/Massachusetts/1/80 (H7N7) influenza virus. Virology 122:38–47
Kishida N, Sakoda Y, Eto M, Sunaga Y, Kida H (2004) Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch Virol 149:2095–2104
Laver WG, Air GM, Webster RG, Gerhard W, Ward CW, Dopheide TA (1979) Antigenic drift in type A influenza virus: sequence differences in the hemagglutinin of Hong Kong (H3N2) variants selected with monoclonal hybridoma antibodies. Virology 98:226–237
Li KS, Xu KM, Peiris JS, Poon LL, Yu KZ, Yuen KY, Shortridge KF, Webster RG, Guan Y (2003) Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J Virol 77:6988–6994
Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A (2000) Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA 97:9654–9658
Liu J, Okazaki K, Ozaki H, Sakoda Y, Wu Q, Chen F, Kida H (2003) H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathol 32:551–560
Liu JH, Okazaki K, Shi WM, Kida H (2003) Phylogenetic analysis of hemagglutinin and neuraminidase genes of H9N2 viruses isolated from migratory ducks. Virus Genes 27:291–296
Matrosovich MN, Krauss S, Webster RG (2001) H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156–162
Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF (2001) Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 75:9679–9686
Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF (1999) Human infection with influenza H9N2. Lancet 354:916–917
Philpott M, Hioe C, Sheerar M, Hinshaw VS (1990) Hemagglutinin mutations related to attenuation and altered cell tropism of a virulent avian influenza A virus. J Virol 64:2941–2947
Sever JL (1962) Application of a microtechnique to viral serological investigations. J Immunol 88:320–329
Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Li ZN, Nakamura K (2001) Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J Gen Virol 82:2475–2484
Tsuda Y, Sakoda Y, Sakabe S, Mochizuki T, Namba Y, Kida H (2007) Development of an immunochromatographic kit for rapid diagnosis of h5 avian influenza virus infection. Microbiol Immunol 51:903–907
Wiley DC, Wilson IA, Skehel JJ (1981) Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378
Xu C, Fan W, Wei R, Zhao H (2004) Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003(H9N2) virus. Microbes Infect 6:919–925
Xu KM, Li KS, Smith GJ, Li JW, Tai H, Zhang JX, Webster RG, Peiris JS, Chen H, Guan Y (2007) Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J Virol 81:2635–2645
Xu KM, Smith GJ, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang JX, Li KS, Fan XH, Webster RG, Chen H, Peiris JS, Guan Y (2007) The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol 81:10389–10401
Acknowledgments
We thank Liu Jing Hua and Michiko Ushioda for providing materials and excellent technical assistance. We also thank Dr. Kanzhen Yu, Dr. Ian Brown, Dr. Robert G. Webster, Dr. Kennedy F. Shortridge and Dr. Alan Hay for providing H9N2 viruses. This study was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Okamatsu, M., Sakoda, Y., Kishida, N. et al. Antigenic structure of the hemagglutinin of H9N2 influenza viruses. Arch Virol 153, 2189–2195 (2008). https://doi.org/10.1007/s00705-008-0243-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-008-0243-2