Abstract
Most mastreviruses (family Geminiviridae) infect monocotyledonous hosts and are transmitted by leafhopper vectors. Only two mastrevirus species, Tobacco yellow dwarf virus from Australia and Bean yellow dwarf virus (BeYDV) from South Africa, have been identified whose members infect dicotyledonous plants. We have identified two distinct mastreviruses in chickpea stunt disease (CSD)-affected chickpea originating from Pakistan. The first is an isolate of BeYDV, previously only known to occur in South Africa. The second is a member of a new species with the BeYDV isolates as its closest relatives. A PCR-based diagnostic test was developed to differentiate these two virus species. Our results show that BeYDV plays no role in the etiology of CSD in Pakistan, while the second virus occurs widely in chickpea across Pakistan. A genomic clone of the new virus was infectious to chickpea (Cicer arietinum L.) and induced symptoms typical of CSD. We propose the use of the name Chickpea chlorotic dwarf Pakistan virus for the new species. The significance of these findings with respect to our understanding of the evolution, origin and geographic spread of dicot-infecting mastreviruses is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Geminiviruses (family Geminiviridae) are small single-stranded (ss) DNA plant viruses that are subdivided taxonomically into four genera on the basis of genome organization, host range and insect vector [44]. Members of the genus Mastrevirus are transmitted by leafhoppers, and the majority infect monocotyledonous plants. There are currently only two known members of the genus that are adapted to dicotyledonous hosts, namely tobacco yellow dwarf virus (TbYDV), originating from Australia [38], and bean yellow dwarf virus (BeYDV), originating from South Africa [14, 29]. Their widely separated geographical locations and genetic diversity suggest that many more distinct dicot-infecting mastreviruses may await discovery.
The genomes of mastreviruses contain four open reading frames (ORFs) which are conserved in all the viruses characterised (reviewed by [4]). Two ORFs (V1 and V2, encoding the coat protein [CP] and movement protein [MP], respectively) are encoded on the virion-sense strand, and two ORFs (C1 and C2, also known as Rep A and Rep B, respectively) are transcribed from the complementary-sense strand [30, 31]. A characteristic feature of mastrevirus genomes is that the virion- and complementary-sense ORFs are separated by a large intergenic region (LIR) and a small intergenic region (SIR) which contain regulatory elements. The pattern of complementary-sense ORF translation found in mastreviruses is unique among the geminiviruses. The full-length replication-associated protein (Rep) is expressed from a spliced transcript of the C1 and C2 ORFs [49], while Rep A is expressed from a transcript spanning the C1 ORF. Rep is the only virus-encoded product required for viral DNA replication [42] and is involved in nicking and joining of DNA strands at the origin of replication [18]. The Rep A protein activates virion-sense gene expression [8].
The agent causing chickpea stunt disease (CSD) has long been suspected to be a geminivirus [20, 21]. CSD occurs across North Africa, the Middle East and the Indian subcontinent [20, 22, 28, 33–36]. The disease affects several pulse crops including chickpea (Cicer arietinum L.), faba bean (Vicia faba L.) and lentil (Lens culinaris Medik.) and more recently has been shown to affect bean (Phaseolus vulgaris L.) and sugarbeet (Beta vulgaris L.). Although numerous viruses have been shown to cause diseases similar to chickpea stunt [22], the virus causing the disease across North Africa, the Middle East and the Indian subcontinent was shown to be a geminivirus. Horn et al. [20] showed the presence of characteristic “geminate” virus particles in affected chickpea samples originating from India and showed the virus to be transmitted by a leafhopper (Orosius orientalis (Matsumura)). Serological analyses showed the virus, for which the name chickpea chlorotic dwarf virus (CpCDV) was coined, to be unrelated to the only other dicot-infecting, leafhopper-transmitted geminiviruses known at that time; beet curly top virus and TbYDV. Despite these advances in characterising the virus causing CSD, no sequence of CpCDV has been obtained, and the precise nature and relationship of the virus to other geminiviruses remains unclear.
We have surveyed the chickpea-growing areas of Pakistan, screening for the presence of dicot-infecting mastreviruses and have identified viruses of two distinct species associated with plants showing CSD-like symptoms. Isolates of the viruses were cloned and their genomes sequenced. The significance of these results for our understanding of the evolution and present distribution of these viruses are discussed.
Materials and methods
Sample collection and DNA extraction
Samples of chickpea (Cicer arietinum) were collected in Faisalabad in 2004, 2006 and 2007 and from Layyah in 2005 and 2007. Nucleic acids were extracted from frozen leaf samples by the CTAB method [9]. Total nucleic acid extracts were resuspended in sterile distilled water and maintained at −20°C until use.
Amplification and cloning of viruses
A pair of oligonucletide primers (DMF—5′-GTCGACGATGATATTATAGGTGGCG-3′; DMR—5′-GTCGACATCCCCTTCAAGTTCGTCC-3′) was designed based on the aligned sequences of all available dicot-infecting mastreviruses available in the databases. The primers were designed to amplify the whole genome of the virus and contain a SalI restriction endonuclease site for downstream manipulation.
Rolling-circle amplification (RCA [13]) using φ29 polymerase (Fermentas) was used to amplify circular DNA molecules from nucleic acid extracts. Amplification reactions (20 μl) contained 50 μM random hexamer primers, 5 units φ29 polymerase, 1 mM dNTPs and 1 unit pyrophosphate in 1× reaction buffer (provided by the manufacturer). Amplifications were initially heated to 94°C in a PCR machine and slowly cooled to room temperature and the polymerase added before being maintained at 30°C overnight. Reaction products were digested with HindIII and cloned into suitably restricted pTZ57R (Fermentas).
A diagnostic procedure to distinguish the two mastreviruses was developed. This multiplex PCR-based procedure used three primers designed to the sequences of CCD6 and CCD14 (a common reverse primer [5′- GAAGTACACTCGGAAATAACCATTTACATA -3′; spanning nucleotides 1,139–1,110 of CCD6 and 1,136–1,107 of CCD14], a forward primer specific for CCD6 [5′-TAAAAGGCGCACTAATGGGTAGACCGTAGA-3′; spanning nucleotides 102–131] and a forward primer specific for CCD14 [5′-CTATATGAAAGTGTTTAGAGCTCCATTTCA-3′; spanning nucleotides 713–742]). In reactions containing sequences equivalent to CCD6, an approximately 1,000-bp product is produced. In the presence of sequences equivalent to CCD14, reactions yield a product of approximately 400 bp.
Sequencing and sequence analysis
The complete nucleotide sequences of clones were determined by dideoxynucleotide chain-termination sequencing using the PCR-based BIG DYE kit (Perkin-Elmer Cetus) and specific internal primers. Sequencing products were resolved commercially (UCT Sequencing Services and Macrogen, Korea). Sequence information was stored, assembled and analysed using the Lasergene sequence analysis package (DNAStar Inc., Madison, WI, USA) running on an IBM-compatible PC.
Phylogenetic analyses were conducted on matrices of aligned sequences using the neighbour-joining and bootstrap options of Phylip (ver. 3.5c) running on an IBM-compatible personal computer. Sequence alignments were produced using CLUSTAL X [46]. Phylogenetic dendrograms were viewed, manipulated and printed using Treeview [39].
Inoculation and analysis of plants
Clone CHP3, resulting from φ29 polymerase-mediated RCA, was selected to produce a construct for Agrobacterium-mediated inoculation. The clone was digested with HindIII and EcoRI to yield an 850-bp fragment, which was ligated into the binary vector pGreen [16] to yield p0.3CHP3. The full-length HindIII insert of CHP3 was then ligated into the unique HindIII restriction site to yield p1.3CHP3. p1.3CHP3 was finally transferred into Agrobacterium tumefaciens strain GV3101 by electroporation. Agrobacterium cultures were prepared and inoculated to plants as described previously by Hussain et al. [23] for Nicotiana benthamiana Domin. and N. tabacum L., and by Mandala et al. [37] for chickpea.
Results
Two mastreviruses are present in chickpea in Pakistan
Samples of chickpea showing two distinct symptom phenotypes, a generalised chlorosis of the youngest parts of the plant (Fig. 1a) and a reddening of the younger leaves (Fig. 1b), were collected in Faisalabad and Layyah in 2004 and 2005, respectively. These two symptom types have previously been described and are attributed to differing varietal responses [22]. Initial screening of the chickpea samples from Faisalabad by PCR gave positive amplification with the universal dicot-mastrevirus primers, yielding the expected approximately 2,500-bp product (results not shown). These products were cloned, and two clones (CCD6 and CCD14), originating from a single plant, were sequenced and analysed. Analysis of the sequences (as detailed in the following sections) indicated the presence of two distinct viruses. To be able to distinguish these viruses in plants, a diagnostic multiplex-PCR procedure was developed. Use of this procedure showed the presence of only sequences equivalent to CCD6 in all other chickpea samples analysed from both Faisalabad in 2006 and Layyah in 2005 (10 plants positive out of 18 analysed). RCA of samples collected in 2007, followed by multiplex PCR, showed four samples positive for sequences equivalent to CCD6 (none positive for sequences equivalent to CCD14) out of 12 analysed. These findings indicate that the virus equivalent to CCD6 is the prevalent pathogen of chickpeas in the areas of Pakistan investigated. The virus equivalent to CCD14 was only encountered in a single chickpea plant originating from Faisalabad in 2004.
Typical symptoms of chickpea stunt disease (CSD) on chickpea in the field. The symptoms are usually a chlorosis of the leaves appearing after infection (b). In some varieties there is a pronounced reddening of the leaf margins (a). Symptoms exhibited by Sesbenia bispinosa growing in the vicinity of chickpea fields affected by CSD, showing either leaf curling (c), or yellowing (e), compared to an asymptomatic plant (d)
Sequence analysis
The complete sequences of two clones (CCD6 and CCD14) obtained by PCR-mediated amplification from DNA extracts of chickpea originating from Faisalabad in 2004, and two clones (CHP3 and CHP8) amplified with φ29 polymerase from DNA extracts of chickpea samples originating from Layyah in 2007, were determined in both orientations with no ambiguities remaining. The features of these sequences and database accession numbers are given in Table 1.
With the exception of clone CHP8, the sequences have an arrangement of predicted ORFs typical of geminiviruses of the genus Mastrevirus [4, 44]. This is illustrated for clone CHP3 in Fig. 2 and consists of two ORFs encoded in the virion sense (V2 and V1) and two encoded in the complementary sense (C1 and C2), diverging from an intergenic region which contains a predicted hairpin structure with the loop sequence TAATATTAC (known as the nonanucleotide sequence). For one of the clones (CCD14), the C2 ORF does not include an initiation codon (ATG). This is not unusual, since the C2 ORF of mastreviruses is characteristically translated from a spliced transcript which joins the C1 and C2 ORFs. The spliced mRNA encodes the replication-associated protein (Rep), the only virus-encoded protein required for viral DNA replication (reviewed in [4, 15]). The sequences of all four clones contain consensus splice donor and acceptor sequences which are predicted to excise an intron of 86 nucleotides, yielding a spliced C1:C2 product that encodes a predicted product (Rep) of 334 amino acids (Table 1).
Circular representation of the genomes of mastreviruses based upon the sequence of CHP3. The positions and orientations of conserved open reading frames are indicated by arrows. The intron that is spliced out to produce the C1–C2 fusion is indicated as a grey box. The position of the conserved hairpin, containing the invariant nonanucleotide sequence (TAATATTAC), is indicated at position zero
Comparisons of the sequences of the four clones indicate that they fall into two distinct groups (Table 2). Clones CCD6, CHP3 and CHP8 show a high degree of nucleotide sequence identity (>98.7%). In contrast, CCD14 shows significantly lower levels of nucleotide sequence identity to the other three clones (<87%). This finding suggests that the four clones represent two distinct mastreviruses.
Sequence comparisons with viruses of the genus Mastrevirus
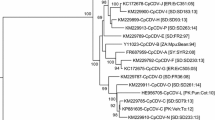
Sequence alignments show CCD14 to be most closely related to isolates of BeYDV (the sequence showed 99.8 and 96.8% nucleotide sequence identity to BeYDV [29] and BeYDV-Mld [14], respectively) and was less related to CCD6/CHP3/CHP8 (<86.5% identity) and TbYDV (67.1% identity). The levels of identity of CCD14 with the monocot-infecting mastrevirus sequences were very low (<50%). In contrast, CCD6/CHP3/CHP8 were less similar to BeYDV (≤86.5 and ≤86.4% nucleotide sequence identity to BeYDV and BeYDV-Mld, respectively) but showed similar levels of identity to TYDV (≤67.1%) and the monocot-infecting mastreviruses (≤51.5%) as CCD14. These findings suggest that the virus represented by CCD14 is an isolate of BeYDV but that CCD6, CHP3 and CHP8 represent a distinct species within the genus Mastrevirus. This is supported by phylogenetic analysis based upon an alignment of the complete nucleotide sequences of the four clones obtained from chickpea with selected other mastreviruses (Fig. 3a). This shows that CCD14 groups closely with the two BeYDV isolates, while the other clones from chickpea form a distinct branch that is approximately equidistant between the BeYDV isolates and TbYDV. Branches separating these groupings have good bootstrap support. The tree additionally shows a distinction between the dicot- and monocot-infecting mastreviruses that has been noted previously [14, 29]. Phylogenetic analysis of alignments of the inferred amino acid sequences of the predicted proteins of the four clones from Pakistan with the homologous products from other mastreviruses show the same topology as that with the full-length nucleotide sequences (results not shown).
Neighbour-joining phylogenetic dendrograms based upon alignments of the complete nucleotide sequences (a), or predicted coat protein amino acid sequences (b), of the four sequences from Pakistan (highlighted with black boxes) and selected other geminiviruses. The full-length nucleotide tree was arbitrarily rooted on the sequence of tomato golden mosaic virus (TGMV; a distantly related geminivirus). The viruses used are bean yellow dwarf virus (BeYDV, Chloris striate mosaic virus (CSMV), Digitaria streak virus (DSV), maize streak virus (MSV), tobacco yellow dwarf virus (TbYDV), and wheat dwarf virus (WDV). The tree based on the coat protein amino acid sequence includes representative viruses from all the Geminiviridae genera. The vector species, where known, is indicated along the right, and vectors that are not leafhoppers are indicated. The viruses used (in addition to those mentioned for panel a) are barley dwarf virus (BDV), beet curly top virus (BCTV), beet severe curly top virus (BSCTV), horseradish curly top virus (HrCTV), oat dwarf virus (ODV), Miscanthus streak virus (MiSV), Panicum streak virus (PanSV), spinach curly top virus (SpCTV), sugarcane streak Egypt virus (SSEV), sugarcane streak Reunion virus (SSREV) and tomato pseudo-curly top virus (TPCTV). The tree was rooted on the whitefly-transmitted Malvastrum yellow vein virus (MaYVV). The standardised isolate descriptors and database accession numbers are shown in each case. The vector of TbYDV, Orosius argentatus, is shown with a question mark, since this may be a synonym of Orosius orientalis. For CHP3, CHP8 and CCD6, the presumed vector Orosius orientalis is shown with a question mark, since the progeny virus of the clones has yet to be shown to be transmissible by this leafhopper species. DSV and its vector Nesoclutha declivata are highlighted since, although the virus groups with the African streak viruses, it is not transmitted by the vector of the African streak viruses (various Cicadulina species). Numbers at nodes equal percentage bootstrap scores (1,000 replicates). Vertical distances are arbitrary; horizontal distance are equal to calculated mutation distances
A phylogenetic analysis based upon an alignment of the predicted amino acid sequences of the coat proteins of the viruses isolated from Pakistan with other selected viruses in shown in Fig. 3b. The viruses include representatives from all four genera with distinct arthropod vectors (whitefly, treehopper, and leafhopper). This shows the coat proteins of CCD6/CHP3/CHP8 to be closely related to those of BeYDV/CCD14. This possibly indicates that the two distinct groups are transmitted by the same vector species. The coat proteins of these viruses are much more closely related than those of Panicum streak virus, sugarcane streak Egypt virus, sugarcane streak Reunion virus and maize streak virus, which are nevertheless transmitted by the same Cicadulina vectors [1, 6, 40]. The only peculiarity in this group is Digitaria streak virus, a virus related to the African streak viruses but transmitted by Nesoclutha declivata (Linnavuori) [25]. The evolutionary origins of this virus and its relationship with N. declivata remain a mystery. The coat proteins of viruses transmitted by Neoalithurus tenellus (Baker) and Pasmmotettix alienus (Dhalb.) also show levels of variation comparable to those between CCD6/CHP3/CHP8 and BeYDV/CCD14. However, based on this analysis one might conclude that TbYDV is transmitted by a vector species distinct from CCD6/CHP3/CHP8 and BeYDV/CCD14.
Comparisons of the predicted amino acid sequences of the Rep proteins of the four chickpea clones with those of BeYDV shows the majority of sequence changes to occur in two clusters between coordinates 211–221 and 310–346 of the alignment (Fig. 4). The region between amino acids 211–221 is immediately downstream of the predicted retinoblastoma binding motif [32] and surrounds the sequences of the splice junction. Possibly, Rep is more tolerant of sequence changes in this region, with only the need, at the nucleotide level, to maintain motifs required for splicing. No function has been identified for the amino acid sequences at the C-terminus of Rep, and it is possible that the sequences between 310 and 346 are structural and thus more tolerant of amino acid changes.
Alignment of the predicted amino acid sequences of the Rep proteins of the viruses isolated from chickpea to those of BeYD and TbYDV. Gaps (-) are introduced to optimise the alignment. Only amino acids at each position differing from CCD14 are shown, otherwise they are indicated with a dot (.). Highlighted are the iteron-related domain (IRD), the retinoblastoma-like protein binding sequence (RBR), three sequence motifs conserved between initiator proteins of rolling-circle DNA replication (Motif I-II; [24, 27]) and the NTP-binding site (Walker A and B; [47]). The position of the splice is indicated by an arrow. Sequences before this position are encoded by the C1 ORF, whereas those after it are encoded by C2
The Rep alignment additionally shows the iteron-related domain (IRD) of CCD14 to be the same as that of BeYDV and TYDV (FRLQ), whereas the IRDs of CCD6, CHP3 and CHP8 are unique (FRFQ), not shared with any other geminivirus. The IRD sequences of Rep are predicted to be the sequences that interact with the viral DNA to initiate rolling-circle replication of the genome [2]. Geminivirus Rep proteins are sequence-specific DNA-binding proteins that recognise repeated motifs (known as “iterons”) adjacent to the nonanucleotide-containing hairpin structure that together form the origin of virion-strand DNA replication. The predicted iteron of CCD14 and BeYDV is ‘TGGAGGCA’ with the core sequence ‘GGAG’ [2]. CCD6, CHP3 and CHP8 share the same ‘GGAG’ core sequence but have a distinct iteron sequence, ‘TGGAGACA’ (CCD6 having a T at position three in the 3′ repeat of the motif). Whether these sequence changes will preclude the trans-replication of the genome of one by the Rep from the other is unclear, but it does show the distinct nature of the two groups of viruses.
Infectivity analysis
The infectivity of clone CHP3 was assessed by Agrobacterium-mediated inoculation of a partial repeat construct to plants. The cloned virus was infectious to N. benthamiana, with symptoms typically appearing within 20 days of inoculation. Nineteen out of 20 plants inoculated were PCR positive with diagnostic primers and showed intense yellowing and downward leaf curling symptoms similar to those described for BeYDV [29]. Inoculation to N. tabacum (two plants infected out of ten inoculated) resulted in plants with greatly reduced apical leaves, which were dark green and with some downward leaf curling (Fig. 5). These symptoms typically appeared within 30–40 days of inoculation, at which point plants ceased growing and failed to flower. Chickpea plants inoculated with p1.3CHP3 showed foliar yellowing and reduced leaf size within approximately 25–30 days of inoculation (three plants infected out of 30 inoculated). Plants were also stunted in comparison to non-inoculated controls.
Identification of alternate hosts
A number of plants species that are common in the vicinity of chickpea fields and frequently show virus-like symptoms were assessed for infection by mastreviruses. Lentils (Lens culinaris), sesame (Sesamum indicum L.; nine plants analysed) and the weed Fumaria parviflora Lam. (eight plants analysed) were uniformly negative when screened with the diagnostic primers by PCR. The leguminous weed Sesbania bispinosa (Jacq.) W. Wight, sometimes grown as a fodder crop, was positive by diagnostic PCR. Of seven plants analysed, one showed the presence of the virus equivalent to CCD6/CHP3/CHP8 and 2 showed the presence of both viruses. Sesbania plants that were shown to be infected exhibited either a generalised chlorosis or leaf curling of the youngest leaves (Fig. 1).
Discussion
Only two dicot-infecting mastreviruses, originating from geographically diverse regions, have thus far been identified and characterised. The virus historically called chickpea chlorotic dwarf virus (CpCDV) is one of a number of viruses that cause CSD. The causative agent has been presumed to be a mastrevirus from the presence of geminate particles in affected plants and its transmission by a leafhopper vector, as well as its serological cross-reactivity to TbYDV. However, the virus was not previously characterised, and is listed as a tentative member of the genus Mastrevirus [11, 44].
The species demarcation criteria for mastreviruses presently include the following: a 75% nucleotide sequence identity level cut-off, above which two viruses should be considered strains of a single species; no trans-replication of genomic components of a second virus; and differences in host range or pathogenicity [10]. Lack of trans-replication, however, is not a reliable character, since, for example, the genome of the African mastrevirus Panicum streak virus is trans-replicated by viruses in the distinct species Maize streak virus [48]. The sequences of CCD6/CHP3/CHP8 show approximately 86% identity to BeYDV, thus being significantly different but still above the threshold value, possibly indicating that CCD6/CHP3/CHP8 are simply divergent strains of BeYDV. However, we demonstrate that BeYDV is not commonly found in chickpea, indicating a distinct host range of these two viruses. In addition, the presence of distinct iteron-related domains as well as distinct interon sequences would suggest that Reps and iterons of the two viruses are incompatible. Based on these findings, we propose that CCD6, CHP3 and CHP8 be considered isolates of a distinct species for which we propose a derivation of the name which has been in common use for some time, Chickpea chlorotic dwarf Pakistan virus (CpCDPV). This is necessary, since a related, but distinct, virus causing CSD has been identified in Sudan (B. Gronenborn, unpublished results). Similar arguments have been made for the proposed mastrevirus species now known as Oat dwarf virus and Barley dwarf virus. These viruses show greater than 75% nucleotide sequence identity to Wheat dwarf virus but are nevertheless considered members of distinct species based on biological properties [26, 43]. This highlights the need to revisit the species demarcation criteria for the genus Mastrevirus, given that they were formulated solely on the properties of the very different African grass-infecting viruses.
The virus now designated CpCDPV has been known to occur in Pakistan since the 1990s, and chickpea is an important pulse crop grown across large areas of Pakistan [34]. To determine the precise aetiology of CSD across Pakistan, we set out to clone and sequence the virus. In the course of doing so, however, we encountered two distinct viruses. The first is an isolate of BeYDV, a virus not previously identified outside of southern Africa. By far the most abundant virus in chickpea is the second virus, which is closely related to but distinct from BeYDV. It is this virus that, at least in Pakistan, is the causative agent of CSD and for which we propose the use of the name CpCDPV. It remains to be seen whether, in the large area where CpCDV has been reported previously based on serological and biological identification, the disease is caused by isolates of the virus we report here, or whether further distinct mastreviruses will be shown to be involved.
The arthropod vector of TbYDV is reported to be the leafhopper Orosius argentatus (Evans), which was previously known as Thamnotettix argentata (Evans), and is a synonym of the species Orosius orientalis (Matsumura) [17, 19, 45]. The vector of BeYDV has yet to be identified. However, for CpCDV (and thus probably for CpCDPV), the vector is reported to be Orosius orientalis [20, 21] suggesting that BeYDV might also be transmissible by this leafhopper species. Geminiviruses encode only a single structural protein, the CP, which mediates interactions with the vector and thus controls vector specificity [5]. Previous studies have shown a weak serological reaction between all three viruses; TbYDV, BeYDV and CpCDV [29]. Comparisons of the predicted CP amino acid sequences of CpCDPV and BeYDV shows them to have above 88% identity, further indicating that they may be vectored by the same species of leafhopper. In contrast, the TbYDV CP shows only 57.5% amino acid sequence identity or less to the BeYDV isolates and CpCDPV. Despite this, TbYDV and CpCDPV are most probably transmitted by the same leafhopper species or possibly closely related races of the same species. It is thus possible that all three virus species are transmitted by distinct biotypes/races of Orosius orientalis, with the viruses and possibly the insect diverging due to geographic separation.
Available evidence suggests that CpCDV occurs across a wide geographical area, from India to northern Africa. However, BeYDV has previously only been reported from South Africa [14, 29]. This raises the question as to whether the presence of BeYDV in South Africa and the subcontinent represent the natural geographical range of the virus or whether some other factor has played a part in its present geographic distribution. The relatively close sequence relatedness between BeYDV and CpCDPV would suggest that these two viruses have a fairly recent common origin, and it is possible that the geographical range of their most recent common ancestor included North Africa, the Middle East or southern Asia. If this is the case, the presence of BeYDV in southern Africa can possibly be explained by the migration of people from the sub-continent to southern Africa, as many southern and East African countries have sizeable expatriate south Asian communities. However, since geminiviruses are not known to be seed transmitted, it is difficult to see how the virus might have been transported. TbYDV is also related to both BeYDV and CpCDPV, albeit distantly (approximately 65% nucleotide identity), meaning that they too have a common origin, but one that is probably as distant as the origin of all begomoviruses. With the Middle East/Indian subcontinent probably being the centre of dicot-infecting mastrevirus diversity, it is not unreasonable to suggest that this region is also their centre of origin. This would suggest that TbYDV was introduced to Australia, possibly upon colonisation of the continent in the 18th Century or even prior to this, by trade with Asia and/or human migration. The relatedness of the viruses, the likely origin of O. argentatus in southern Asia, the fact that BeYDV (in South Africa) and TbYDV affect mainly introduced plant species and the lack of dicot-infecting mastrevirus diversity in southern Africa and Australia would seem to support this hypothesis.
Recently, Ha et al. [12] have shown the presence of a New World-like begomovirus, Corchorus yellow vein virus (CYVV), in Vietnam. Begomoviruses occurring in the New World are a monophyletic group within a larger grouping of Old World viruses, and it was generally accepted that the divergence of these two populations occurred due to an isolated introduction of an Old World virus into the Americas [41]. The identification of CYVV significantly strengthens this assumption. Ha et al. [12] suggested that this introduction could have been made by early Chinese traders. The evidence is far stronger showing that beet curly top virus (species now known as Beet severe curly top virus [BSCTV]) and its close relatives occurring in North America and likely also the rest of the New World, originate in the Mediterranean/Middle East [3, 7]. Neither BSCTV nor its leafhopper vector Neoalithurus tennelus are native to the New World and were probably introduced by early settlers. It is thus clear that mankind has had a major impact on the geographic spread of geminiviruses, and it is not beyond possibility that this has also been the case for the dicot-infecting mastreviruses. However, far more effort will need to be put into the identification and characterisation of viruses in this distinct group of mastreviruses before the validity of our “out of Asia” hypothesis can be examined in detail.
Our screening of various leguminous weed and crop species was able to identify BeYDV in only a single chickpea plant. This leads us to believe that we have yet to identify the (major) natural host of this virus. In South Africa the virus causes problems in beans (Phaseolus vulgaris), but so far no alternate weed hosts have been identified. Beans are not a widely grown crop in central Pakistan, and it is thus possible that, in this region, BeYDV is confined to weeds such as Sesbania, the only other plant species in which we could identify the virus.
The identification of these two mastrevirus species in Pakistan suggests that further species in this genus could possibly occur in this region. Our present efforts are aimed at investigating the diversity of dicot-infecting mastreviruses, examining the availability and mechanism of natural resistance to these viruses in chickpea as well as establishing pathogen-derived resistance by RNA interference.
References
Ammar ED, Kira MT, Abul-Ata AE (1982) Natural occurrence of streak and mosaic diseases on sugar-cane cultivars in upper Egypt, and transmission of sugar-cane streak by Cicadulina bipunctelia zeae (China). Annales de l’Institut Pasteur Virologie 133:183–185
Argüello-Astorga GR, Ruiz-Medrano R (2001) An iteron-related domain is associated to motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Arch Virol 146:1465–1485
Bennett CW, Tanrisever A (1957) Sugar beet curly top disease in Turkey. Plant Dis Rep 41:721–725
Boulton MI (2002) Functions and interactions of mastrevirus gene products. Physiol Mol Plant Pathol 60:243–255
Briddon RW, Pinner MS, Stanley J, Markham PG (1990) Geminivirus coat protein replacement alters insect specificity. Virology 177:85–94
Briddon RW, Lunness P, Chamberlin LCL, Pinner MS, Brundish H, Markham PG (1992) The nucleotide sequence of an infectious insect-transmissible clone of the geminivirus Panicum streak virus. J Gen Virol 73:1041–1047
Briddon RW, Stenger DC, Bedford ID, Stanley J, Izadpanah K, Markham PG (1998) Comparison of a beet curly top virus isolate from the old world with those from the new world. Eur J Plant Pathol 104:77–84
Collin S, Fernández-Lobato M, Gooding PS, Mullineaux PM, Fenoll C (1996) The two nonstructural proteins from wheat dwarf virus involved in viral gene expression and replication are retinoblastoma-binding proteins. Virology 219:324–329
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fauquet CM, Bisaro DM, Briddon RW, Brown JK, Harrison BD, Rybicki EP, Stenger DC, Stanley J (2003) Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch Virol 148:405–421
Fauquet CM, Stanley J (2005) Revising the way we conceive and name viruses below the species level: a review of geminivirus taxonomy calls for new standardized isolate descriptors. Arch Virol 150:2151–2179
Ha C, Coombs S, Revill P, Harding R, Vu M, Dale J (2006) Corchorus yellow vein virus, a New World geminivirus from the Old World. J Gen Virol 87:997–1003
Haible D, Kober S, Jeske H (2006) Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J Virol Methods 135:9–16
Halley-Stott RP, Tanzer F, Martin DP, Rybicki EP (2007) The complete nucleotide sequence of a mild strain of Bean yellow dwarf virus. Arch Virol 152:1237–1240
Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D (1999) Geminviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci 18:71–106
Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42:819–832
Helson GAH (1942) The leaf hopper Thamnotettix argentata Evans, a vector of tobacco yellow dwarf. J Coun Sci Ind Res (Austr) 15:175–184
Heyraud-Nitschke F, Schumacher S, Laufs J, Schaefer S, Schell J, Gronenborn B (1995) Determination of the origin cleavage and joining domain of geminivirus Rep proteins. Nucleic Acids Res 23:910–916
Hill AV (1941) Yellow dwarf of tobacco in Australia. II Transmission by the Jassid Thamnotettix argentata (Evans). J Coun Sci Ind Res (Austr) 14:181–186
Horn NM, Reddy SV, Roberts IM, Reddy DVR (1993) Chickpea chlorotic dwarf virus, a new leafhopper-transmitted geminivirus of chickpea in India. Ann Appl Biol 122:467–479
Horn NM, Reddy SV, Reddy DV (1994) Virus-vector relationships of chickpea chlorotic dwarf geminivirus and the leafhopper Orosius orientalis (Hemiptera: Cicadellidae). Ann Appl Biol 124:441–450
Horn NM, Reddy SV (1996) Survey of chickpea (Cicer arietinum) for chickpea stunt disease associated viruses in India and Pakistan. Plant Dis 80:286–290
Hussain M, Mansoor S, Iram S, Zafar Y, Briddon RW (2007) The hypersensitive response to tomato leaf curl New Delhi virus nuclear shuttle protein is inhibited by transcriptional activator protein. Mol Plant-Microbe Interact 20:1581–1588
Ilyina TV, Koonin EV (1992) Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20:3279–3285
Julia JF, Dollet M (1989) Nesoclutha declivata Homoptera Cicadellidae, vector of Digitaria streak virus (Geminivirus) in Vanuatu. J Phytopathol 127:42–48
Koklu G, Ramsell JNE, Kvarnheden A (2007) The complete genome sequence for a Turkish isolate of Wheat dwarf virus (WDV) from barley confirms the presence of two distinct WDV strains. Virus Genes 34:359–366
Koonin EV, Ilyina TV (1992) Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J Gen Virol 73:2763–2766
Kumari SG, Makkouk KM, Attar N, Ghulam W, Lesemann DE (2004) First report of chickpea chlorotic dwarf virus infecting spring chickpea in Syria. Plant Dis 88:424
Liu L, van Tonder T, Pietersen G, Davies JW, Stanley J (1997) Molecular characterization of a subgroup I geminivirus from a legume in South Africa. J Gen Virol 78:2113–2117
Liu L, Davies JW, Stanley J (1998) Mutational analysis of bean yellow dwarf virus, a geminivirus of the genus Mastrevirus that is adapted to dicotyledonous plants. J Gen Virol 79:2265–2274
Liu L, Pinner MS, Davies JW, Stanley J (1999) Adaptation of the geminivirus bean yellow dwarf virus to dicotyledonous hosts involves both virion-sense and complementary-sense genes. J Gen Virol 80:501–506
Liu L, Saunders K, Thomas CL, Davies JW, Stanley J (1999) Bean yellow dwarf virus RepA, but not Rep, binds to maize retinoblastoma protein, and the virus tolerates mutations in the consensus binding motif. Virology 256:270–279
Makkouk KM, Dafalla G, Hussein M, Kumari SG (1995) The natural occurrence of chickpea chlorotic dwarf geminivirus in chickpea and faba bean in Sudan. J Phytopathol 143:465–466
Makkouk KM, Bashir M, Jones RAC, Kumari SG (2001) Survey for viruses of lentil and chickpea crops in Pakistan. J Plant Dis Protect 108:258–268
Makkouk KM, Fazlali Y, Kumari SG, Farzadfar S (2002) First record of Beet western yellows virus, Chickpea chlorotic dwarf virus, Faba bean necrotic yellows virus and Soybean dwarf virus infecting chickpea and lentil crops in Iran. Plant Pathol 51:387
Makkouk KM, Rizkallah L, Kumari SG, Zaki M, Enein RA (2003) First record of Chickpea chlorotic dwarf virus (CpCDV) affecting faba bean (Vicia faba) crops in Egypt. Plant Pathol 52:413
Mandala B, Varma A, Malathi VG (1997) Systemic infection of Vigna mungo using the cloned DNAs of the blackgram isolate of mungbean yellow mosaic geminivirus through agroinoculation and transmission of the progeny virus by whiteflies. J Phytopathol 145:505–510
Morris BAM, Richardson KA, Haley A, Zhan X, Thomas JE (1992) The nucleotide sequence of the infectious cloned DNA component of tobacco yellow dwarf virus reveals features of geminiviruses infecting monocotyledonous plants. Virology 187:633–642
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Rybicki EP (1988) Maize streak virus: an African pathogen come home. S Afr J Sci 84:30–32
Rybicki EP (1994) A phylogenetic and evolutionary justification for three genera of Geminiviridae. Arch Virol 139:49–77
Schalk HJ, Matzeit V, Schiller B, Schell J, Gronenborn B (1989) Wheat dwarf virus, a geminivirus of graminaceous plants needs splicing for replication. EMBO J 8:359–364
Schubert J, Habekuss A, Kazmaier K, Jeske H (2007) Surveying cereal-infecting geminiviruses in Germany-diagnostics and direct sequencing using rolling circle amplification. Virus Res 127:61–70
Stanley J, Bisaro DM, Briddon RW, Brown JK, Fauquet CM, Harrison BD, Rybicki EP, Stenger DC (2005) Geminiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus Taxonomy. VIIIth Report of the ICTV. Elsevier/Academic Press, London, pp 301–326
Thomas JE, Bowyer JW (1984) Tobacco yellow dwarf virus. CMI/AAB descriptions of plant viruses
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface; flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha and beta subunits of ATPase synthase, myosin, kinases and the the ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951
Willment JA, Martin DP, Palmer KE, Schnippenkoetter WH, Shepherd DN, Rybicki EP (2007) Identification of long intergenic region sequences involved in maize streak virus replication. J Gen Virol 88:1831–1841
Wright EA, Heckel T, Groenendijk J, Davies JW, Boulton MI (1997) Splicing features in maize streak virus virion- and complementary-sense gene expression. Plant J 12:1285–1297
Acknowledgments
N.N. is supported by a Ph.D. fellowship from the Higher Education Commission (HEC), Government of Pakistan. R.W.B. is supported by the HEC under the “Foreign Faculty Hiring Program”. The authors are grateful for the support of NIBGE in conducting this study and to Mr. Muhammad Ilyas for his assistance in inoculating chickpea. Funding for this project came in part from the Ministry of Science and Technology, Pakistan, and from the National Research Foundation, South Africa, and the University of Cape Town Research Committee. The authors are grateful to Darrin Martin and Bruno Gronenborn for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nahid, N., Amin, I., Mansoor, S. et al. Two dicot-infecting mastreviruses (family Geminiviridae) occur in Pakistan. Arch Virol 153, 1441–1451 (2008). https://doi.org/10.1007/s00705-008-0133-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-008-0133-7