Abstract
Recent advances in somatic cell reprogramming is one of the most important developments in neuroscience in the last decades since it offers for the first time the opportunity to work with disease/patient-specific neurons or other neural cell types. Induced pluripotent stem cells (iPSCs) can be differentiated into all cell types of the body enabling investigations not only on neurons but also on muscle or endothelial cells which are cell types often also of great interest in neurodegenerative diseases. The novel technology of direct lineage conversion of somatic cells into neurons (induced neurons; iNs) or into expandable multipotent neural stem cells (induced neural stem cells; iNSCs) provides interesting alternatives to the iPSC technology. These techniques have the advantage of easier cell culture, but only neurons (iNs) or neuroectodermal cells (iNSCs) can be generated. Although there are several open questions coming along with these new neural cell types, they hold great promises for both cell replacement and cell modelling of neurodegenerative diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Intoduction
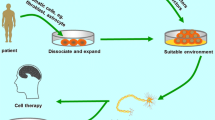
Recent advances in direct lineage conversion of somatic cells into neurons (induced neurons; iNs) or expandable multipotent neural stem cells (induced neural stem cells; iNSCs) provide interesting alternatives to the induced pluripotent stem cell (iPSC) technology to generate neurons or other neural cell populations for both cell replacement and disease modelling for neurodegenerative diseases (see Fig. 1 for schematic presentation of the strategies). Takahashi and Yamanaka (2006) pioneered the field of somatic cell reprogramming (Takahashi et al. 2007). The principle of somatic cell reprogramming was long known by the technique of somatic nuclear transfer, but this technique is highly complicated and always needs an enucleated oocyte as the acceptor cell (for review see Jaenisch and Young 2008). Takahashi and Yamanaka identified four transcription factors which are—when overexpressed in a somatic cell—sufficient to induce a cell harbouring all properties of a pluripotent stem cell (Takahashi et al. 2007; Takahashi and Yamanaka 2006). The so-called induced pluripotent stem cells (iPSCs) are thus able to differentiate into all cell types of the body. This technology is a tremendous breakthrough in medical science since any cell type can now be derived patient-specific with the potential for autologous cell replacement strategies and personalized medicine (Fig. 1). For neuroscientists, a new era begun with the possibility to get access to patient/disease-specific neurons.
Schematic representation of the three strategies to generate patient/disease-specific neural cell types for cell replacement strategies or disease modelling of neurodegenerative diseases. The first strategy is to reprogram somatic cells into pluripotent stem cells, which are subsequently differentiated into the neural cell type of interest (with or without intermediate generation of NSCs [not shown in scheme, grey arrows)]. The second approach is to directly convert somatic cells into neurons (iNs; green arrow). The third strategy is to convert somatic cells into expandable neural stem cells (iNSCs), which are subsequently differentiated into the diseased neural cell type (blue arrows). iNs induced neurons, iNSCs, induced neural stem cells, iPSCs induced pluripotent stem cells
The induction of iPSCs was subsequently shown from many somatic cell sources, e.g. skin fibroblasts (Takahashi et al. 2007; Takahashi and Yamanaka 2006), hair follicle keratinocytes (Linta et al. 2012), liver cells (Kleger et al. 2012) and blood cells (Okita et al. 2013). It is thus believed that they can be generated from any cell source of the body. It was further on successfully demonstrated that iPSCs perform like embryonic stem cells (ESCs) during cell type differentiation, therefore most protocols already established for ES cell differentiation are—in general—applicable for iPSCs. Exemplarily, dopaminergic differentiation from Parkinson’s disease (PD) patient-derived iPSCs was proven in vitro and in vivo by restoration of hemiparkinsonian rats after intrastriatal transplantation (Hargus et al. 2010). This also holds true for sickle cell anemia (Hanna et al. 2007), acute myocardial infarction (Nelson et al. 2009) and diabetes (Alipio et al. 2010). Within this gold rush mood of the recent iPSC years, large parts of the scientific and clinical community neglect some major problems accompanied with the use of pluripotent stem cells such as their tumorigenicity (Miura et al. 2009). Moreover, there are unclear limitations with the use of iPSCs in clinical application such as their genetic and epigenetic instability and heterogeneity (Lister et al. 2011; Gore et al. 2011).
Besides their application in cell replacement strategies, iPSCs are currently believed to be one of the best in vitro cell model systems for various diseases and are hoped to catalyse our understanding in neurodegenerative diseases. However, one major obstacle is whether it will be possible to model neurodegenerative disorders as age-dependent diseases since cellular reprogramming changes the cell to an embryonic stem cell state. Nevertheless, first reports promisingly show neuronal cell degeneration in various iPSC-based disease models, e.g. spinal muscular atrophy (Ebert et al. 2009), PD (Reinhardt et al. 2013), spinocerebellar atrophies (Koch et al. 2011) and Alzheimer’s disease (Koch et al. 2012).
Expression of the reprogramming factors as foreign genes using viral transduction systems leading to genomic integration is of limited suitability for clinical use. Therefore, many attempts have been undertaken in order to overcome this issue such as the use of non-integrating adenoviruses (Stadtfeld et al. 2008), oriP/EBNA1-based episomal vectors (Yu et al. 2009), piggyBac transposition systems (Kaji et al. 2009), transient transfection with reprogramming plasmids (Okita et al. 2008), Cre-recombinase excisable viruses (Soldner et al. 2009), recombinant proteins (Zhou et al. 2009; Kim et al. 2009a) and modified RNA (Warren et al. 2010). In the latter approaches, the need for genomic manipulation and DNA transfection that is associated with viral or other DNA-based reprogramming methods is eliminated. Another attempt has been the reduction of reprogramming factors with or without the combination with small molecules (Kim et al. 2008, 2009b, 2009c). This was accomplished by the use of NSCs as cell origin already expressing three of the four Yamanaka factors. Recent data suggest that there is a dose dependency of the extrinsic factors for optimal reprogramming efficacy potentially explaining the differences between cell types in their suitability for factor reduced reprogramming (Nagamatsu et al. 2012). In addition, somatic cells express their specific set of transcription factors which is important for maintaining the cell identity (Hikichi et al. 2013). Overexpressing these factors was shown to strongly interfere with cell reprogramming and it was shown that these factors differed exemplarily between NSCs and hepatic cells (Hikichi et al. 2013). This means, it is not only important whether the somatic cell already express one of the four reprogramming factors. Additionally, it is also important to overcome the cell type-specific transcription factor network to succeed with iPSCs generation.
Interestingly, even though complete reprogramming has been proven for factor reduced NSC-derived iPSCs they might have a decreased differentiation potential (Lohle et al. 2012). In addition, even though believed to be reprogrammed to a fully immature embryonic stem-like cell, iPSCs seem to retain a distinct epigenetic memory still favouring the subtype differentiation of the respective cell source of origin (Kim et al. 2011b). This is due to incomplete erasure of tissue-specific methylation and aberrant de novo methylation (Kim et al. 2011b).
Directly induced neurons (iNs)
The advances in generating of iPSCs stimulated another approach to generate neurons by direct induction of mature neurons from somatic cells even from other germ layers: Using forced expression of various neuronal key-fate determining factors (mainly transcription factors) was demonstrated to generate fully mature neurons (Vierbuchen et al. 2010; Pfisterer et al. 2011; Pang et al. 2011). The major advantage of this technology is the rapid and rather easy induction of the neuronal phenotype without sophisticated protocols as used in iPSC generation. However, some disadvantages are arising from this technique: The derived cells (postmitotic neurons) are not expandable, thus every experiment starts with a new induction process. Although the induction efficacy is relatively high (~5 %) compared to those for iPSCs/iNSCs generation, the population of the cell type of interest is very low thereby limiting their application. To enhance the efficacy of the conversion process and thus overcome this limitation, Ladewig et al. (2012). recently reported an experimental approach combining two-factor neuronal programming (ASCL1 and NGN2) and small molecule-based inhibition of glycogen synthase kinase-3b and SMAD signalling leading to high yields of functional neurons with high neuronal purity (>80 %) In addition, it seems to be necessary to use different transcription factor combinations for any specific neuronal subtype. This was recently shown for midbrain dopaminergic neurons, where midbrain-specific transcription factors Nurr1 and Lmx1a were used (Caiazzo et al. 2011).
Xue et al. (2013) recently reported the transdifferentiation of fibroblasts to iNs by the use of miRNA-124a. This is in close agreement with our studies in NSC-like cells generated by conversion from bone marrow mesenchymal stem cells (MSCs) without genetic manipulation (Hermann et al. 2004, 2006a): Using genome-wide expression profiling and functional network analysis we detected the miR-124a as an important regulator during the neuroectodermal conversion process (Maisel et al. 2010).
The induction of neurons from adult human cortical perivascular cells was successfully achieved by transduction with only two factors, namely MASH1 and SOX2 (Karow et al. 2012). This suggests that similar to iPSCs also for iNs, the choice of the needed transcription factor combination depends on the cell source (via endogenous transcription factor profiles of different somatic cells). This might favour the use of ectodermal or even better neuroectodermal somatic cells as a cell source for iN induction.
Concerning possible clinical applications, the major disadvantage of iNs is the lack of expandability and thus the low cell number not only limiting the cells amount per se for applications but also for genetic and microbiology testing prior of use. This disadvantage translates into another limitation: iN cultures contain neurons in various purities and non-neural cells such as the initial cell type (e.g. fibroblasts), but not other neural cell types. On the one hand, the contamination with the initial cell type might provoke side effects and, on the other hand, in some cases the non-neuronal cells might support the neurons in cell replacement studies or provide a more physiological environment in cell culture models. In addition, without a defined intermediate immature cell type transplantation approaches are difficult, because detachment from the surface is harmful for mature neurons with established axo-dendritic arborisation. Finally, the transgenes might remain active in the iNs without being completely silenced.
Together, clinical applications of iNs for cell replacement approaches are limited with the current protocols, but iNs might be a very suitable cell source for applications in which only single neuronal entities on the single cell level are of investigational interest. Consistently, a first study showed that iNs can be used for disease modelling of Alzheimer’s disease (Qiang et al. 2011).
Directly induced neural stem cells (iNSCs)
Having some of the limitations of iNs in mind, an alternative approach is to derive induced neural stem cells (iNSCs) from somatic cells as an expandable cell type which can be differentiated into all neuroectodermal lineages. Our group and others established protocols to convert bone marrow-derived MSCs into NSC-like cells without genetic modifications, but by culturing the cells in NSC-promoting conditions (Hermann et al. 2004, 2006a; Lee et al. 2003). Although the generated NSC-like cells express some major properties of expandable NSCs, their functional neuronal differentiation capacity is limited and comparative transcriptome analyses revealed significant differences between these NSC-like cells and brain-derived primary NSCs (Maisel et al. 2010). It is thus now believed that these protocols only lead to partial (and probably transient) neuroectodermal conversion of mesenchymal stem cells. The novel approach using heterologous expression of various key fate-determining transcription factor combinations successfully generated expandable iNSCs from somatic cells expressing all major properties of primary NSCs (Han et al. 2012; Lujan et al. 2012; Ring et al. 2012; Thier et al. 2012) (Table 1). These iNSCs easily differentiated into astrocytes, oligodendrocytes and functional neurons including neuronal subtypes (Han et al. 2012). The first report by Kim et al. (2009a) showed induction of iNSCs from fibroblasts by using the classical Yamanaka-factors as used in iPSC generation. The difference was a shorter time of expression of the foreign genes and the change to a neural medium which seems to convert these intermediate cells into a NSC phenotype. Trilineage differentiation capacity of iNSCs was first described by Lujan et al. (2012) using Brn2, FoxG1 and Sox2 as foreign genes. In both reports, no data were shown concerning expandability, silencing of foreign genes, neuronal subtype specification or whole genome analysis (Kim et al. 2009a; Lujan et al. 2012). Our report together with the studies by Thier and colleagues showed subsequently the generation of stably expandable iNSCs exhibiting a NSC-like transcriptome (Han et al. 2012; Thier et al. 2012). Consistently, both protocols led to silenced transgenes within the iNSCs additionally confirming complete and stable conversion (Han et al. 2012; Thier et al. 2012). Glutamatergic, GABAergic, cholinergic and dopaminergic neuronal subtype specification was only with our protocol (Han et al. 2012). The article by Ring et al. (2012) is of additional note since it reports iNSC induction by the single factor Sox2. However, the use of a very specific feeder cell type for iNSC induction might be the crucial point delivering a yet unknown set of reprogramming factors. Corti and co-workers converted human astrocytes using a single genetic factor (OCT4 or KLF4 or NANOG) into iNSCs with neuronal differentiation capacity (Corti et al. 2012). Sertoli cells were recently shown to be a possible cell source for iNSC generation, but a combination of eight transcription factors were needed for iNSC induction (Sheng et al. 2012).
Another big issue in generating iNSCs is that the definition of the NSC state is by far not that strict as for ESCs and iPSCs. NSCs are defined by expression of some key transcription factors (such as Sox2, Pax6) and their differentiation potential into the main neuroectodermal lineages, namely neurons, astrocytes and oligodendrocytes (Hermann et al. 2006b; Storch and Schwarz 2002). What we learned from both embryonic brain development and ESC/iPSC differentiation is that a regionalization takes place already very early in the developmental/differentiation process to region-specific NSCs (Reinhardt et al. 2013; Pankratz et al. 2007) and it remains enigmatic whether a pan-neural NSC state exists (Pankratz et al. 2007). Thus, neuronal subtype specification might depend on the generation methods of iNSCs.
Conclusions
The novel technology for generating iNSCs from easily accessible cell sources such as skin fibroblasts opens up a new field in personalised medicine for neurodegenerative diseases. Although the available studies convincingly demonstrate the similarities of iNSCs with primary NSCs, there are several open questions such as (1) are the retroviral vectors completely silenced in iNSCs?, (2) does the iNSC conversion process leads to genomic alterations?, and (3) is the conversion into the iNSC state stable and thus leading to a stable change of the epigenome to a NSC-like status? In addition, in contrast to the iPSC technology with readily available differentiation protocols, protocols for neuronal (subtype) differentiation are not available for iNSCs yet. However, expandable iNSCs seem to provide some advantages over iNs and possibly also over iPSCs towards both cell replacement and cell modelling of neurodegenerative diseases.
References
Alipio Z, Liao W, Roemer EJ, Waner M, Fink LM, Ward DC, Ma Y (2010) Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. Proc Natl Acad Sci USA 107(30):13426–13431. doi:10.1073/pnas.1007884107
Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V (2011) Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476(7359):224–227. doi:10.1038/nature10284
Corti S, Nizzardo M, Simone C, Falcone M, Donadoni C, Salani S, Rizzo F, Nardini M, Riboldi G, Magri F, Zanetta C, Faravelli I, Bresolin N, Comi GP (2012) Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp Cell Res 318(13):1528–1541. doi:10.1016/j.yexcr.2012.02.040
Ebert AD, Yu J, Rose FF Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN (2009) Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457(7227):277–280. doi:10.1038/nature07677
Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee JH, Loh YH, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Izpisua Belmonte JC, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein LS, Zhang K (2011) Somatic coding mutations in human induced pluripotent stem cells. Nature 471(7336):63–67. doi:10.1038/nature09805
Han DW, Tapia N, Hermann A, Hemmer K, Hoing S, Arauzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, Greber B, Yang JH, Lee HT, Schwamborn JC, Storch A, Scholer HR (2012) Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10(4):465–472. doi:10.1016/j.stem.2012.02.021
Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R (2007) Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318(5858):1920–1923. doi:10.1126/science.1152092
Hargus G, Cooper O, Deleidi M, Levy A, Lee K, Marlow E, Yow A, Soldner F, Hockemeyer D, Hallett PJ, Osborn T, Jaenisch R, Isacson O (2010) Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc Natl Acad Sci USA 107(36):15921–15926. doi:10.1073/pnas.1010209107
Hermann A, Gastl R, Liebau S, Popa MO, Fiedler J, Boehm BO, Maisel M, Lerche H, Schwarz J, Brenner R, Storch A (2004) Efficient generation of neural stem cell-like cells from adult human bone marrow stromal cells. J Cell Sci 117(Pt 19):4411–4422. doi:10.1242/jcs.01307
Hermann A, Liebau S, Gastl R, Fickert S, Habisch HJ, Fiedler J, Schwarz J, Brenner R, Storch A (2006a) Comparative analysis of neuroectodermal differentiation capacity of human bone marrow stromal cells using various conversion protocols. J Neurosci Res 83(8):1502–1514. doi:10.1002/jnr.20840
Hermann A, Maisel M, Storch A (2006b) Epigenetic conversion of human adult bone mesodermal stromal cells into neuroectodermal cell types for replacement therapy of neurodegenerative disorders. Expert Opin Biol Ther 6(7):653–670. doi:10.1517/14712598.6.7.653
Hikichi T, Matoba R, Ikeda T, Watanabe A, Yamamoto T, Yoshitake S, Tamura-Nakano M, Kimura T, Kamon M, Shimura M, Kawakami K, Okuda A, Okochi H, Inoue T, Suzuki A, Masui S (2013) Transcription factors interfering with dedifferentiation induce cell type-specific transcriptional profiles. Proc Natl Acad Sci U S A. doi:10.1073/pnas.1220200110
Jaenisch R, Young R (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132(4):567–582. doi:10.1016/j.cell.2008.01.015
Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K (2009) Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458(7239):771–775. doi:10.1038/nature07864
Karow M, Sanchez R, Schichor C, Masserdotti G, Ortega F, Heinrich C, Gascon S, Khan MA, Lie DC, Dellavalle A, Cossu G, Goldbrunner R, Gotz M, Berninger B (2012) Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11(4):471–476. doi:10.1016/j.stem.2012.07.007
Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR (2008) Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454(7204):646–650. doi:10.1038/nature07061
Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS (2009a) Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4(6):472–476. doi:10.1016/j.stem.2009.05.005
Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR (2009b) Direct reprogramming of human neural stem cells by OCT4. Nature 461(7264):643–649. doi:10.1038/nature08436
Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR (2009c) Oct4-induced pluripotency in adult neural stem cells. Cell 136(3):411–419. doi:10.1016/j.cell.2009.01.023
Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S (2011a) Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA 108(19):7838–7843. doi:10.1073/pnas.1103113108
Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ (2011b) Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 29(12):1117–1119. doi:10.1038/nbt.2052
Kleger A, Mahaddalkar PU, Katz SF, Lechel A, Joo JY, Loya K, Lin Q, Hartmann D, Liebau S, Kraus JM, Cantz T, Kestler HA, Zaehres H, Scholer H, Rudolph KL (2012) Increased reprogramming capacity of mouse liver progenitor cells, compared with differentiated liver cells, requires the BAF complex. Gastroenterology 142(4):907–917. doi:10.1053/j.gastro.2012.01.004
Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D, Doerr J, Ladewig J, Mertens J, Tuting T, Hoffmann P, Klockgether T, Evert BO, Wullner U, Brustle O (2011) Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature 480(7378):543–546. doi:10.1038/nature10671
Koch P, Tamboli IY, Mertens J, Wunderlich P, Ladewig J, Stuber K, Esselmann H, Wiltfang J, Brustle O, Walter J (2012) Presenilin-1 L166P mutant human pluripotent stem cell-derived neurons exhibit partial loss of gamma-secretase activity in endogenous amyloid-beta generation. Am J Pathol 180(6):2404–2416. doi:10.1016/j.ajpath.2012.02.012
Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kogler G, Muller FJ, Koch P, Brustle O (2012) Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods 9(6):575–578. doi:10.1038/nmeth.1972
Lee J, Elkahloun AG, Messina SA, Ferrari N, Xi D, Smith CL, Cooper R Jr, Albert PS, Fine HA (2003) Cellular and genetic characterization of human adult bone marrow-derived neural stem-like cells: a potential anti glioma cellular vector. Cancer Res 63(24):8877–8889
Linta L, Stockmann M, Kleinhans KN, Bockers A, Storch A, Zaehres H, Lin Q, Barbi G, Bockers TM, Kleger A, Liebau S (2012) Rat embryonic fibroblasts improve reprogramming of human keratinocytes into induced pluripotent stem cells. Stem Cells Dev 21(6):965–976. doi:10.1089/scd.2011.0026
Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471(7336):68–73. doi:10.1038/nature09798
Lohle M, Hermann A, Glass H, Kempe A, Schwarz SC, Kim JB, Poulet C, Ravens U, Schwarz J, Scholer HR, Storch A (2012) Differentiation efficiency of induced pluripotent stem cells depends on the number of reprogramming factors. Stem Cells 30(3):570–579. doi:10.1002/stem.1016
Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M (2012) Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA 109(7):2527–2532. doi:10.1073/pnas.1121003109
Maisel M, Habisch HJ, Royer L, Herr A, Milosevic J, Hermann A, Liebau S, Brenner R, Schwarz J, Schroeder M, Storch A (2010) Genome-wide expression profiling and functional network analysis upon neuroectodermal conversion of human mesenchymal stem cells suggest HIF-1 and miR-124a as important regulators. Exp Cell Res 316(17):2760–2778. doi:10.1016/j.yexcr.2010.06.012
Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, Ogawa D, Ikeda E, Okano H, Yamanaka S (2009) Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol 27(8):743–745. doi:10.1038/nbt.1554
Nagamatsu G, Saito S, Kosaka T, Takubo K, Kinoshita T, Oya M, Horimoto K, Suda T (2012) Optimal ratio of transcription factors for somatic cell reprogramming. J Biol Chem 287(43):36273–36282. doi:10.1074/jbc.M112.380683
Nelson TJ, Martinez-Fernandez A, Yamada S, Perez-Terzic C, Ikeda Y, Terzic A (2009) Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation 120(5):408–416. doi:10.1161/CIRCULATIONAHA.109.865154
Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S (2008) Generation of mouse induced pluripotent stem cells without viral vectors. Science 322(5903):949–953. doi:10.1126/science.1164270
Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, Goshima N, Yamanaka S (2013) An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31(3):458–466. doi:10.1002/stem.1293
Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M (2011) Induction of human neuronal cells by defined transcription factors. Nature 476(7359):220–223. doi:10.1038/nature10202
Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC (2007) Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells 25(6):1511–1520. doi:10.1634/stemcells.2006-0707
Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M (2011) Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA 108(25):10343–10348. doi:10.1073/pnas.1105135108
Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, Moreno H, Abeliovich A (2011) Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell 146(3):359–371. doi:10.1016/j.cell.2011.07.007
Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, Wu G, Muller S, Brockmann K, Kluba T, Maisel M, Kruger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, Klingauf J, Kuhlmann T, Klewin M, Muller H, Gasser T, Scholer HR, Sterneckert J (2013) Genetic correction of a LRRK2 mutation in human iPSCs Links Parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell 12(3):354–367. doi:10.1016/j.stem.2013.01.008
Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y (2012) Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11(1):100–109. doi:10.1016/j.stem.2012.05.018
Sheng C, Zheng Q, Wu J, Xu Z, Wang L, Li W, Zhang H, Zhao XY, Liu L, Wang Z, Guo C, Wu HJ, Liu Z, He S, Wang XJ, Chen Z, Zhou Q (2012) Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res 22(1):208–218. doi:10.1038/cr.2011.175
Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R (2009) Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136(5):964–977. doi:10.1016/j.cell.2009.02.013
Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K (2008) Induced pluripotent stem cells generated without viral integration. Science 322(5903):945–949. doi:10.1126/science.1162494
Storch A, Schwarz J (2002) Neural stem cells and neurodegeneration. Curr Opin Investig Drugs 3(5):774–781
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676. doi:10.1016/j.cell.2006.07.024
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5):861–872. doi:10.1016/j.cell.2007.11.019
Thier M, Worsdorfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nothen MM, Brustle O, Edenhofer F (2012) Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10(4):473–479. doi:10.1016/j.stem.2012.03.003
Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463(7284):1035–1041. doi:10.1038/nature08797
Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ (2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7(5):618–630. doi:10.1016/j.stem.2010.08.012
Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu XD (2013) Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152(1–2):82–96. doi:10.1016/j.cell.2012.11.045
Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324(5928):797–801. doi:10.1126/science.1172482
Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan L, Ding S (2009) Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4(5):381–384. doi:10.1016/j.stem.2009.04.005
Acknowledgments
The research of the authors was supported by the Bundesministerium für Bildung und Forschung, the Deutsche Forschungsgemeinschaft (DFG) through the Collaborative Research Center 655 ‘Cells into tissues: stem cell and progenitor commitment and interactions during tissue formation’ (SFB 655, project A23) and the DFG-Research centre and Cluster of Excellence “Centre for Regenerative Therapies Dresden (CRTD)”, the Thyssen-Stiftung, and the Landesstiftung Baden-Württemberg.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hermann, A., Storch, A. Induced neural stem cells (iNSCs) in neurodegenerative diseases. J Neural Transm 120 (Suppl 1), 19–25 (2013). https://doi.org/10.1007/s00702-013-1042-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00702-013-1042-9