Abstract
Cotton (Gossypium spp.) is the world’s most important natural textile fiber crop, and Gossypium hirsutum L. has been responsible for 90 % of the annual cotton crop in the world. Few studies are conducted about the population structure and genetic diversity of elite cotton (G. hirsutum) germplasm based on a worldwide collection. In this study, we analyzed population structure and genetic diversity of 157 elite G. hirsutum cultivar accessions collected from China, American, Africa, Former Soviet Union, and Australia using 146 SSR markers randomly distributed on the 26 chromosomes of cotton. The average allele number and PIC were 2.26 and 0.2857, respectively, indicating a relatively low degree of genetic diversity. By predefining the total cotton panel as three groups according to their geographic origin, we proved that the predefined American-origin group had higher average genetic diversity than China-origin group, but the China-origin group had the widest range of genetic variation, and the highest genetic distance happened between the varieties from America and from China, implying a direction for genetic improvement. Population structure analysis revealed two groups existed in the whole cotton panel, but the two groups inferred from structure were not consistent with the predefined three groups, indicating that the exchange and domestication of germplasm all over the world. Further independent STRUCTURE run on the two inferred groups indicated that they could be subdivided into three and two subgroups. Analysis of molecular variance showed that the American-origin group had higher population substructure than China-origin group, and only a little variation between groups was explained by population structure, suggesting that our population was suitable for association mapping. Finally, the evolutionary implication of cotton population structure was discussed. These results have provided valuable information for the association mapping of important agronomic traits, as well as for the breeding and exploit of new cotton germplasms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Gossypium includes 46 diploid (2n = 2x = 26) and five allotetraploid (2n = 2x = 52) species, which distributed in Central, South, and North America (18 species), Africa and Asia (14 species), and Australia (17 species) (Wendel and Cronn 2003). Among the 50 species, only four are cultivated, including two diploids (2n = 2x = 26): G. arboreum L. (A2A2) and G. herbaceum L. (A1A1), which originated from Africa–Asia, and two tetraploids (2n = 4x = 52): G. hirsutum (AD1AD1) and G. barbadense (AD2AD2), which originated from America. Currently, G. hirsutum and G. barbadense have been responsible for 90 and 8 % of the annual cotton crop production in the world, respectively. Because of its economic importance, such as high-yield and environmental suitability, G. hirsutum has attracted considerable scientific interest for plant breeders and agricultural scientists and been planted widespreadly. However, other cultivated species possess many favorable traits for cotton production which the G. hirsutum cultivars lack. For example, G. herbaceum possesses the traits of earliness and drought resistance (Stewart 1992), G. arboreum of resistance to diseases and pests (Kantartzi et al. 2009), and G. barbadense of good fiber quality (Wendel and Cronn 2003). So, Gossypium-cultivated species are important germplasm for cotton improvement.
Though G. hirsutum was planted in more than 50 countries, most of these countries were not cotton’s original home (Wendel and Cronn 2003). The domestication and breeding history of cultivated cotton after its introduction from Mesoamerica to these countries have led to low level of genetic polymorphism (Rungis et al. 2005; Chen and Du 2006). Any crop with a narrow genetic base is more prone to natural disasters, such as the outbreak of a disease. Epidemics of cotton Verticillium wilt disease in China since 1993 is a typical example, and none of the upland cotton varieties was found to be resistant (Zhai and Luo 1994). So, it is crucial to explore novel germplasm resources for potential natural genetic diversity and develop innovative genomics tools to efficiently mobilize these useful genetic variations to breeding germplasm, which should help to overcome existing and potential problems of cotton production associated with narrow genetic base of the cultivar germplasm.
Several studies showed that new genetic variation could be produced by interspecific hybridization and induced mutagenesis and thus enriched the germplasm resources in cotton (Ahoton et al. 2003; Hussain et al. 2002). Recently, some new elite cotton (G. hirsutum) germplasm with the following one or more characters of high yield, good fiber quality, earliness, disease, and pest resistance was created by interspecific hybridization combining induced mutagenesis, polymerization backcross, marker-assisted selection, and introgression of exogenous genes in China (Sun et al. 2004). For cotton geneticists and breeders, the precise evaluation of the genetic diversity of excellent germplasm of G. hirsutum will provide a guide for choosing parents and predicting the degree of inheritance, variation, and level of heterosis, which are essential for realizing the breeding goal. SSR loci are particularly useful for the study of genetic diversity and population structure of domesticated species because their high level of allelic diversity facilitates the detection of the fine structure of diversity more efficiently than an equal number of RFLP, AFLP, or SNP loci (Akagi et al. 1997). Using SSRs, the cotton source germplasm in China was divided into five groups on the basis of the average similarity coefficient (0.610) among the source germplasm (Chen and Du 2006), and 334 G. hirsutum variety accessions from Uzbek cotton germplasm collection performed the analysis of genetic diversity and population structure (Abdurakhmonov et al. 2009).
In this study, we selected a large number of elite G. hirsutum cultivar accessions from the China cotton germplasm collection. These elite cultivar accessions originated from either different cotton-growing areas in China or outside of China, among which some innovative germplasms from interspecific hybridization or induced mutagenesis among G. hirsutum and either G. arboreum or G. barbadense were included. The genotypes of these accessions were analyzed to study the extent and distribution of diversity, population structure, and kinship by using SSR markers. The specific goals of this study are to characterize genetic diversity and population structure within elite G. hirsutum cultivar accessions, to examine the differences between and relationships among genetically defined groups. The resulting framework will be used to pose questions about the origin and diversity of gene pools that exist within world cultivated G. hirsutum cultivars and to lay the foundation for characterizing the genes that distinguish them.
Materials and methods
Plant material
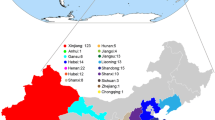
We sampled 157 cotton accessions (Supplementary Table 1) representing the geographic range of elite G. hirsutum cultivar accessions from the China cotton germplasm collection. The sampled accessions were collected from different cotton growing areas in China (106) as well as from America (41), Africa (3), Former Soviet Union (4), France (1), Pakistan (1), and Australia (1).
Genomic DNA extraction and SSR genotyping
Genomic DNA of all materials was isolated from pooled young leaves of ten seedlings following Paterson et al. (1993). SSRs distributed on the 26 chromosomes on the AD-genome wide Reference Map (http://www.cottondb.org/cgi-bin/cmap/viewer) were screened for their polymorphism, and 146 pairs of SSR markers (an average of five on each of the 26 chromosomes, Supplementary Table 2) showing polymorphism among the 157 cotton accessions were retained for genotyping. The chromosome locations of these SSR markers and positions of each locus were obtained from the AD-genome wide Reference Map (http://www.cottondb.org/cgi-bin/cmap/viewer). PCR amplification for SSRs was performed in 67 mM of Tris–HCl (pH 8.8), 16 mM of (NH4)2SO4, 2.5 mM of MgCl2, 0.2 mM of dNTPs, 0.6 μM of primers, with 0.5 units of Sangon (Sangon, Shanghai, China) Taqase, and 25 ng of genomic DNA per 10 μl using a Thermal Cycler 9600 (Perkin-Elmer). PCR runs were performed 35 cycles of 45 s at 94 °C, at the annealing temperature for 45 s and 72 °C for 90 s, and a final extension step at 72 °C for 10 min. PCR products were run on 10 % polyacrylamide gels, using a DYCZ-30 vertical electrophoresis gel apparatus (Produced by China). The gel was run for about 50 min after loading the sample. After electrophoresis, the gel was separated from the plates and treated for 10 min in fixation solution (10 % v/v ethanol and 0.5 % v/v acetic acid) with gentle shaking. After incubating for 12–15 min in staining solution (0.2 % w/v silver nitrate), the gel was washed 2 times with distilled water for 2 min, and with 0.0002 % w/v sodium thiosulfate for about 2 min, and then, the gel was transferred to developing solution (1.5 % w/v sodium hydroxide, 0.4 % formaldehyde) to develop the silver-staining DNA bands. The stop and stored solution was 0.75 % sodium carbonate.
Molecular genetic diversity and phylogenetic analyses
Since the cotton germplasm used in this study was strictly self-pollinated during the past decades for germplasm renewing, we genotyped our cotton accessions according to the methods reported by Abdurakhmonov et al. (2009), in which the SSR data were scored like a dominant marker type. Genetic diversity was assessed using the program POWERMARKER V3.25 (http://www.powermarker.net), measured by number of alleles per locus, gene diversity, and polymorphism information content (PIC). Pairwise genetic distances were calculated, and phylogeny was analyzed using the software Powermarker 3.25 under the Nei 1983 model (Liu and Muse 2005). Genetic variation within and among predefined groups, the entire population, and the inferred groups and pairwise FST genetic distances was measured by analysis of molecular variance (AMOVA) using ARLEQUIN2.0 (Schneider et al. 2000).
Population structure and kinship analysis
The model-based (Bayesian) cluster software STRUCTURE 2.2 (Pritchard et al. 2000) was chosen to estimate the population structure of the predefined groups and the entire population with the 146 unlinked markers which distributed across all cotton chromosomes. We ran STRUCTURE under the ‘admixture model’ with a burn-in period of 10,000 followed by 100,000 replications of Markov Chain Monte Carlo. Five independent runs each were performed with the number of clusters (K) varying from 1 to 10. An ad hoc measure ∆K based on the relative rate of change in the likelihood of the data between successive K values was used to determine the optimal number of clusters (Evanno et al. 2005). That run with the maximum likelihood was adopted to subdivide the cotton accessions into different subgroups with the membership probabilities threshold of 0.50 or the maximum membership probability among subgroups. No a priori population information was used. Relative kinship matrix was constructed using the software SPAGeDi, and negative values between two individuals were changed to 0 (Hardy and Vekemans 2002).
Results
Inference of genetic structure of the predefined groups and the 157 cotton accessions
According to the geographical origins, we predefined the total cotton panel as three groups: American-origin group (P1), China-origin group (P2), and other-origin group (P3). In order to explore the genetic structure of the predefined groups, we performed population structure inference for American-origin group, China-origin group, and other-origin group, respectively. At the same time, we considered the whole cotton panel as one population with potential substructures and its population structure was inferred. The inferred groups for the whole cotton panel were compared with the predefined groups to judge the conformities between the cultivars’ origination and their genetic structure.
Population structure inference showed that the LnP(D) value of the predefined American-origin group, China-origin group, and the whole cotton panel constantly increased with K from 1 to 10, and the highest △K value was observed at K = 2 for American-origin group, K = 3 for China-origin group, and K = 2 for the whole cotton panel, respectively (Figure S1). This suggested that American-origin group could be assigned into two subgroups (P1a, P1b), China-origin group into three subgroups (P2a, P2b, P2c), and the whole cotton panel into two groups (Figs. 1, 2). While for the other-origin group, which consisted of cultivars from Africa, Former Soviet Union, French, Pakistan, and Australia, the LnP(D) value showed that this group could be assigned into seven subgroups (Figure S1, Fig. 1). The two groups inferred from the whole cotton panel were named G1 and G2, respectively. Using a probability of membership threshold of 0.50 or the maximum membership probability among subgroups, 61 lines were assigned to G1 and 96 lines to G2 (Supplementary Table 3). G1 group consisted of 21 American cultivar accessions, 34 Chinese cultivar accessions, 3 Former Soviet Union cultivar accessions, 2 Africa cultivar accessions, and 1 Australia cultivar accession. G2 group consisted of 21 American cultivar accessions, 71 Chinese cultivar accessions, 1 Pakistan cultivar accession, 1 Former Soviet Union cultivar accession, 1 Africa cultivar accession, and 1 French cultivar accession. The two groups inferred from structure were not consistent with the predefined three groups, reflecting the probable extensive exchange of parental lines by breeders worldwide.
For the whole cotton panel, due to the gradual increase of LnP(D) from k = 1 to k = 10 and there were small peaks of ∆K after k = 2 (Figure S1), we performed an independent STRUCTURE run for each of the two groups. Their ∆K indicated there were three and two subgroups in the group G1 and G2, respectively (Figure S1, Fig. 2). The G1 group was classified into three subgroups. Subgroups were named as G1a, G1b, and G1c. G1a contained 9 lines, which were representative of five lines collected from north early-maturity cotton area in China; G1b contained 19 lines and consisted of 8 American cultivars, 9 Chinese cultivars, 1 African cultivar, and 1 Former Soviet Union cultivar, which were representative of PD6186, a breeding line from America; G1c contained 33 lines derived from Huanghe River valley area in China(8), Yangtze River valley area in China (3), northwest inland area in China (3), America (13), Former Soviet Union (2), Australia (1), and Africa (1), respectively.
The G2 group was classified into two subgroups. The subgroups included G2a, containing 90 lines, which originated from USA (20), Africa (1), Pakistan (1), Former Soviet Union (1), France (1), and China (66); G2b, containing 6 lines, which were representative of Arcot-1, a breeding line from America, and its four innovative lines created by atomic energy mutation (Supplement Table 1, Supplement Table 4).
The pairwise kinship estimates based on 146 informative SSR markers showed that the majority of the pairs of cotton accessions (53.75 %) had zero estimated kinship values, while 17.55 % of the pairwise kinship estimates ranged from 0 to 0.05, 13.43 % of the pairs ranging from 0.05 to 0.1, and 11.70 % of the pairs ranged from 0.1 to 0.20. The remaining estimates (3.57 %) had >0.25 kinship values, with a continuously decreasing number of pairs falling in higher estimate categories, and these high kinship values implied some common parental genotypes were utilized in the breeding history of these germplasm groups (Fig. 3). These results indicated that most lines in the panel have no or very weak kinship, which might be attributed to the broad range collection of genotypes and the exclusion of similar genotypes before analysis.
Genetic diversity
A total of 146 SSR loci, randomly distributed across the genome with an average of five on each of the 26 chromosomes, were used to evaluate the genetic diversity of the elite cotton germplasm. These SSR loci were polymorphic across the 157 variety accessions, and a total of 330 alleles were detected (Table 1). The number of alleles per locus varied among marker loci, ranging from 2 to 5 with an average of 2.26. The frequency of major alleles (the alleles have highest frequency per locus) varied from 0.3774 to 0.9936. The average gene diversity of predefined American-origin group (P1), China-origin group (P2), and other-origin group (P3) was 0.3527, 0.3434, and 0.3234, respectively, while the average gene diversity of the entire population, the inferred G1 group, and G2 group (see below) was 0.3502, 0.3695, and 0.3148, respectively. In addition, the average PIC value of predefined American-origin group (P1), China-origin group (P2), and other-origin group (P3) was 0.28809, 0.2798, and 0.2639, respectively, while the average PIC value of the entire population, the inferred G1 group, and G2 group (see below) was 0.2857, 0.3003, and 0.2572, respectively (Table 1).
Phylogenetic analyses revealed genetic distance (GD) of all elite G. hirsutum cultivar accessions ranged from 0.0182 to 0.5651 with an average of 0.3457, demonstrating significant genetic diversity ranges. The average GD within the G. hirsutum cultivar accessions of predefined groups (P1, P2, and P3) was very close and ranged from 0.3398 to 0.3599. Among predefined groups, the American-origin group had the highest average genetic distance and the China-origin group had the lowest, but the latter had the widest range of genetic variation (0.0182–0.5556, Table 1). While among the inferred groups, the average GD within G1 group (0.37) was higher than that in G2 group (0.3122). The highest GD (0.5651) among all elite G. hirsutum cultivar accessions was observed between the cultivar Nashangqudahua (from China) and Mei8123 (from America) (Table 1).
The NJ trees of American-origin group, China-origin group, other-origin group, and the whole cotton panel were created to judge the consistency between clusters based on the phylogenic tree and groups based on the STRUCTURE running (Figs. 1, 2). For American-origin group, the cultivars from the same subgroup in STRUCTURE running were assembled into the same cluster in phylogenic tree (Fig. 1a). For China-origin group, the cultivars from each of the three subgroups in STRUCTURE running were apt to disperse into different clusters in phylogenic tree (Fig. 1b). For the whole cotton panel, the cultivars from the same group or subgroup in STRUCTURE running were also apt to disperse into different clusters in phylogenic tree (Fig. 2). The cultivars in the dispersion state were often assigned to intermediates if we use a probability of membership threshold of 70 %, instead of membership threshold of 0.50 or the maximum membership probability among subgroups (Table S4). This showed that compared to American-origin group, the more exchange of parental lines happened in China-origin group or the whole cotton panel.
Analysis of molecular variance (AMOVA)
The genetic diversity within and among predefined groups was estimated by AMOVA test conducted using predefined P1 and P2 as groups (P3 was not included because of only a few samples) and their corresponding subgroups (P1a, P1b, P2a, P2b, and P2c) as populations. For the whole cotton panel, the same AMOVA test was conducted using inferred groups (G1, G2) as groups, and inferred subgroups (G1a, G1b, G1c, G2a, and G2b) within group as populations.
Analysis of molecular variance results indicated that 14.56 % of the total molecular variation for the inferred groups and 8.97 % of the total molecular variation for the predefined groups could be attributed to the differentiation among corresponding subgroups (Tables 2, 4). But the variation among both the inferred groups and the predefined groups was not significant, implying the low differentiation between American-origin group and China-origin group, or between G1 group and G2 group. About 86.49 % of total genetic variance for the inferred groups and 92.18 of total genetic variance for the predefined groups were attributed to the difference within subgroups (Tables 2, 4). Pairwise Fst showed that differentiation between subgroups from American-origin group (P1) was higher (Fst = 0.12237, P < 0.0001) than that from China-origin group (P2), with Fst ranging from 0.07593 to 0.11158 (Table 5). Similarly, the differentiation between subgroups (G2a and G2b) from the G2 group was higher than that from the G1 group (Table 3). Interestingly, each Fst value in Table 3 was larger than that in Table 5. It seemed that differentiation between subgroups could be greatly improved by STRUCTURE running.
Discussion
Genetic diversity of elite cotton germplasm
We genotyped a total of 157 elite cotton germplasm accessions using 146 genomic SSR markers covering all 26 chromosomes of cotton. Our results demonstrated that the level of detected diversity was relatively low, with an average number of alleles per locus of 2.26 (between 2 and 5 alleles/locus), a gene diversity of 0.3502, and a PIC of 0.2857 (Table 1). These values were very similar to those reported by Fang et al. (2013) (an average of 2.64 alleles per primer pair and an average PIC of 0.2869 detected in 193 upland cotton cultivars collected from 26 countries), and similar to those reported by Rungis et al. (2005) (an average of 2.4 alleles per primer pair and an average PIC of 0.37 detected in 9 cotton cultivars consisted of 8 G. hirsutum and 1 G. barbadense) and by Bertini et al. (2006) (an average of 2.13 alleles per microsatellite locus and an average PIC of 0.40 detected in 53 G. hirsutum cotton cultivars), but lower than those reported by Lacape et al. (2007) (an average of 5.6 alleles per locus and an average PIC of 0.55 detected in 47 accessions including 38 G. hirsutum, 2 G. darwinii, 2 G. tomentosum, and 5 G. barbadense) and by Moiana et al. (2012) (an average of 6.9 alleles per locus and a mean PIC of 0.646 detected in 35 cultivars and eight inbred lines of G. hirsutum from Africa, USA, and Brazil). Although the number of alleles in this study was lower than that reported by Liu et al. (2000) (an average of 5 alleles per locus detected in 97 cultivars and primitive species of G. hirsutum derived from various wild race stocks), the PIC values in the two studies were very similar (0.2857 in our study and 0.31 in Liu’s study). These differences in genetic diversity values might be attributed to the types of germplasm used. As expected, the level of polymorphism among races and wild species of Gossypium was significantly higher than that within cultivated G.hirsutum. What’s more, cultivars domesticated directly in a native cotton growing area usually reserved their higher level of polymorphism than those in a non-native cotton-growing area. It was reported that the G. hirsutum cultivated around the world is derived from the USA, which were exported to other countries in the nineteenth and early twentieth century, with most upland cotton used in early cotton breeding in the world coming from this source (Iqbal et al. 2001). So, American-origin cultivars usually reserved their higher level of polymorphism than those in other countries. Our study also proved that the predefined American-origin group had higher average genetic diversity than China-origin group. Chen and Du (2006) reported that most cotton varieties planted in China were derived from a few sources of germplasm, such as DPL, Stoneville, King, Uganda, Foster, and Trice, all of which were introduced from abroad. Therefore, in terms of allelic richness of our study, the pool of elite cotton germplasm in China owned only a small share of the species variability. However, we detected that the highest GD happened between the varieties from America and from China, which implies a direction for genetic improvement.
Population structure and differentiation of elite cotton germplasm
The elite cotton germplasm lines based on the introduction and domestication of exotic germplasms often have complex genetic background; therefore, understanding population structure and relationships among the elite germplasm lines is of significant importance for cotton improvement and association analysis. In the present study, we firstly predefined the whole cotton panel as American-origin group (P1), China-origin group (P2), and other-origin group (P3), according to their geographic origin. Because the other-origin group had only a few samples, we mainly analyzed American-origin group and China-origin group. At the same time, we considered the whole cotton panel as one population with potential substructures. By selecting ≧0.5 membership as the group subdivision criterion, the analysis showed that when K = 2, the ad hoc measure of △K showed the highest value, which indicated dividing the whole cotton panel into two groups was the most biologically meaningful population structure. It was interesting to note that in each of the two groups, there were germplasm lines from several origins (China, America, Africa, and former Soviet Union), and what’s more, the two groups inferred from structure were not consistent with the predefined three groups, indicating that the exchange and domestication of germplasm between different origins.
Several studies had showed that the pedigree relationships among the cultivars of G. hirsutum strictly relied on the areas where the cultivars originated. For example, the cluster analysis of the genetic distance data for nine cotton cultivars grouped the Australian cultivars separately from most of American cultivars (Rungis et al. 2005), and applying different methods, 35 cultivars and eight inbred lines of G. hirsutum L. were identified as four groups that consisted of American cultivars and inbred lines, African and Brazilian cultivars, BRS Brazilian cultivars, and FM Brazilian cultivars (Moiana et al. 2012). China is not a native cotton growing area, and its cotton breeding and production were based on the introduced germplasms (Chen and Du 2006), which usually led to the close relationships between Chinese cultivars or improved accessions and the introduced germplasms. This viewpoint was further proved by the AMOVA results that the variation among both the inferred groups and the predefined groups was not significant (Tables 2, 4), implying the low differentiation between American-origin group and China-origin group, or between G1 group and G2 group. In Genome-Wide Association Mapping (GWAS) studies, the power of structure-based association studies to detect the effects of single genes would be reduced if a large fraction of variation was explained by population structure (Flint-Garcia et al. 2005). In our population, only a little variation between groups was explained by population structure, suggesting that our population was suitable for association mapping.
From an evolutionary point of view, the low differentiation between groups in our population reflected the evolutionary and domestication history of cultivated upland cotton. Unlike rice, in which the domesticated species Oryza sativa had been domesticated into Japonica varieties and indica varieties over thousands of years (Zhang et al. 2009), the upland cotton only had a domestication history of several hundreds years and had no separate domestication events happened, leading to no special local variety or ecotype. In fact, by analyzing the genetic diversity of upland cotton from different countries using more than one hundred and eighty thousand SNP markers, it was showed that the average genetic similarity coefficient of upland cotton from different countries was close to 0.9 (personal communication). So, we thought all the cultivated upland cotton in the world should be considered as one big population, and different clusters of upland cotton obtained by different researchers should be thought as subgroups from the whole upland cotton population.
Population structure is an indicator of genetic differentiation among groups and subgroups. The results showed that for the predefined American-origin group, the model-based subpopulations corresponded well with distance-based modeled clusters (Fig. 1a), confirming that higher differentiation among subgroups in American-origin group than that in China-origin group (Table 5). So, we thought the American-origin group had higher population substructure than China-origin group.
Since the G. hirsutum cultivated around the world is derived from the USA (Iqbal et al. 2001), the structure of this cotton panel might be a reflection of population structure in the US Upland Cotton. By performing independent structure inference for each group, we had assigned the whole cotton panel into five subgroups. Among the five subgroups, we only detected one subgroup (G1a), in which all the lines were originated from China, but other four subgroups consisted of accessions derived from both China and USA (Supplementary Table 1). This result was very similar to those reported by Tyagi et al. (2014), who divided the US upland cotton accessions into four differentiated subpopulations corresponding to major cotton-growing regions: western, southwestern, midsouth, and eastern. Therefore, we deduced that the genetic structure of upland cotton accessions in the world might be mediated by those in USA.
Through the apparent lack of diversity in cultivated G. hirsutum, Van-Esbroeck and Bownam (1998) had argued that there is enough allelic variation, mutation, or recombination in crosses between closely related individuals to allow improvement in agronomic performance and/or that the coefficient of parentage may not reflect the real genetic distance. In our study, we observed that all the accessions in the subgroup G2b had good fiber qualities with a fiber length of >30 mm and a fiber strength of >30 cN/tex, and most accessions in the subgroup G1a had fine Verticillium wilt resistance (unpublished), which would effectively improve the fiber quality and disease resistance by justifying crosses between these accessions and other related individuals in cotton cultivar breeding programs.
References
Abdurakhmonov IY, Saha S, Jenkins JN, Buriev ZT, Shermatov SE, Scheffler BE, Pepper AE, Yu JZ, Kohel RJ, Abdukarimov A (2009) Linkage disequilibrium based association mapping of fiber quality traits in G. hirsutum L. variety germplasm. Genetica 136:401–417
Ahoton L, Lacape JM, Baudoin JP, Mergeai G (2003) Introduction of Australian diploid cotton genetic variation into upland cotton. Crop Sci 43:1999–2005
Akagi H, Yokozeki Y, Nagaki A, Fujimura T (1997) Highly polymorphic microsatellites of rice consist of AT repeats and a classification of closely related cultivars with these microsatellite loci. Theor Appl Genet 94:61–67
Bertini CHCM, Schuster I, Sediyama T, Barros EG, Moreira MA (2006) Characterization and genetic diversity analysis of cotton cultivars using microsatellites. Genet Mol Biol 2:321–329
Chen G, Du XM (2006) Genetic diversity of source germplasm of upland cotton in China as determined by SSR marker analysis. Acta Genet Sin 33(8):733–745
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fang DD, Hinze LL, Percy RG, Li P, Deng D, Thyssen G (2013) A microsatellite-based genome-wide analysis of genetic diversity and linkage disequilibrium in Upland cotton (Gossypium hirsutum L.) cultivars from major cotton-growing countries. Euphytica 191:391–401
Flint-Garcia SA, Thuillet AC, Yu JM, Pressoir G, Romero SM, Mitchell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES (2005) Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J 44:1054–1064
Hardy OJ, Vekemans X (2002) SpaGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hussain M, Khan MSI, Khanl AI, Akhtar KP, Gong WK (2002) Induced mutagenesis as a genetic improvement tool of cotton (Gossypium hirsutum L.) in conformance with needs in Pakistan. Cotton Sci 14(6):374–377
Iqbal J, Ouk Reddy, Km El-Zik, Pepper AEA (2001) Genetic bottleneck in the evolution under domestication of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor Appl Genet 103:547–554
Kantartzi SK, Ulloa M, Sacks E, Stewart JM (2009) Assessing genetic diversity in Gossypium arboreum L. cultivars using genomic and EST-derived microsatellites. Genetica 136:141–147
Lacape JM, Dessauw D, Rajab M, Noyer JL, Hau B (2007) Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol Breed 19:45–58
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu S, Cantrell RG, Mcarty JCJ, Stewart JM (2000) Simple sequence repeat-based assessment of genetic diversity in cotton race stock accessions. Crop Sci 40:1459–1469
Moiana LD, Filho PSV, Goncalves-Vidigal MC, Lacanallo GF et al (2012) Genetic diversity and population structure of cotton (Gossypium hirsutum L. race latifolium H.) using microsatellite markers. Afr J Biotechnol 11(54):11640–11647
Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Report 11:122–127
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multi-locus genotype data. Genetics 155:945–959
Rungis D, Llewellyn D, Dennis ES, Lyon BR (2005) Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Aust J Agric Res 56:301–307
Schneider S, Roessll D, Excoffier L (2000) ARLEQUIN: a software for population genetics data analysis, version 2.0. Genetics and Biometry Laboratory, Department of Anthropology, Geneva, Switzerland, University of Geneva
Stewart JM (1992) Germplasm resources and enhancement strategies for disease. Proc Beltwide Cotton Conf 3:1323–1325
Sun JL, Du XM, Zhou ZL, Pan ZE, Pang BY (2004) Innovation of elite cotton germplasm. CHINA COTTON 31(4) (in Chinese)
Tyagi P, Gore MA, Bowman DT, Campbell BT, Udall JA, Kuraparthy V (2014) Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.). Theor Appl Genet 127:283–295
Van-Esbroeck G, Bownam DT (1998) Cotton germplasm diversity and its importance to cultivar development. J Cotton Sci 2:121–129
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186
Zhai XJ, Luo GT (1994) The characteristics of cotton Verticillium wilt breaking out and control methods of this disease. Hebei Agricultural Sciences 2:32 (in Chinese)
Zhang DL, Zhang HL, Wang MX, Sun JL, Qi YW, Wang FM, Wei XH, Han LZ, Wang XK, Li ZC (2009) Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor Appl Genet 119:1105–1117
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant No. 31000733) and by the National Hi-Tech Research and Development Program of China (Grant No. 2012AA101108-02-02).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Y., Wang, H., Chen, W. et al. Genetic diversity and population structure of elite cotton (Gossypium hirsutum L.) germplasm revealed by SSR markers. Plant Syst Evol 301, 327–336 (2015). https://doi.org/10.1007/s00606-014-1075-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-014-1075-z