Abstract
The large intergenic spacer (IGS) region of rDNA is a valuable DNA sequence because it contains the signals for transcription and processing of rRNAs. In this study, we sequenced and analyzed the complete nucleotide of rDNA IGS region of pomegranate for the first time. This IGS sequence was 3,712 bp in length with GC content of 67.8 %. The sequence analysis indicated that the spacer could be divided into three main regions: a central repetitive region and two flanking unique domains on on both sides of the central area. Central region is composed of two repeat families (A and B), and the third region contained two short direct repeats (C1 and C2). By comparisons with the IGS sequence of other plant species and computer-aided searches, we determined the putative transcription initiation and termination sites, and the large number of methylable sites. The presence of some conserved motifs in this region may play an important role in regulation of rRNA transcription. Taken together, the pomegranate IGS revealed the complex structure. However, its organization and structure are similar to most other rDNA spacers studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In higher eukaryotes, ribosomal RNA genes (rDNA) occur as a multigene family organized in tandem arrays of repeats at the nucleolar organizing region(s) (NOR) of chromosomes (Appels and Honeycutt 1986). Each repeat unit consists of coding regions (18S, 5.8S and 25–28S rRNAs), and three spacer sequences including two internal transcribed spacers (ITS1 and ITS2) and an intergenic spacer (IGS) (Kahl 1988; Poczai and Hyvonen 2010).
The rDNA IGS region is composed of nontranscribed spacer (NTS) and external transcribed spacer (ETS) regions (Baldridge et al. 1992; Kahl 1988; Poczai and Hyvonen 2010). This region separates the 25S rRNA gene from the 18S rRNA gene of the following repeat unit (Poczai and Hyvonen 2010). The IGS is a complex regulatory unit. It contains broadly conserved structural features such as repeating elements, repetitive enhancer elements, promoters, terminators of transcription, and conserved secondary structures (Baldridge et al. 1992; Kahl 1988; Poczai and Hyvonen 2010; Reeder 1989). In plants, there are some evidences that the repeated elements may act as enhancers for transcription (Hemleben and Zentgraf 1994; Maggini et al. 1992; Sardana et al. 1993), facilitating the initiation of the RNA polymerase I transcription (Hemleben and Zentgraf 1994; Schiebel et al. 1989). However, little information is available about the function of numerous repetitive elements (Zentgraf et al. 1990).
The coding regions of the rDNA are strongly conserved amongst organisms, whereas the spacer regions, especially the IGS region, show high variability in length and sequence between and within species and even among individuals (Gerstner et al. 1988; Hemleben et al. 1988; Jorgansen and Cluster 1988; Rogers and Bendich 1987; Schaal and Learn 1988; Weider et al. 2005).
Length of the rDNA IGS varies in plant species group, ranging in size from 1 kb to over 12 kb (Kim and Mabry 1991; Rogers and Bendich 1987). The molecular basis for length heterogeneity is mainly as a result of the presence of one to several tandems or dispersed subrepeat sequences in the IGS region (Bhatia et al. 1996; Lakshmikumaran and Negi 1994; Toloczyki and Feix 1986; Yakura et al. 1984). Variation in the copy number of the subrepeats is probably due to unequal crossing over within the repetitive families (Lakshmikumaran and Negi 1994).
The spacers of ribosomal regions provide a valuable tool for analyzing the evolutionary divergence between species (Bhatia et al. 1996). The IGS sequence plays an important role in the regulation of rDNA transcription and contains the signals for processing pre-rRNAs (Da Rocha and Bertrand 1995; Fernández et al. 2000; Hemleben and Zentgraf 1994). Indeed, studies on the structure and organization of IGS will be useful for future analysis in control of expression of rRNA genes. The data of this region are now available for different families of plants such as Poaceae (Barker et al. 1988; Carvalho et al. 2011; Chang et al. 2010; Toloczyki and Feix 1986), Brassicaceae (Bennett and Smith 1991; Lakshmikumaran and Negi 1994), Cucurbitaceae (King et al. 1993), Solanaceae (Borisjuk and Hemleben 1992) and Fabaceae (Kato et al. 1990; Maggini et al. 1992).
However, based on our knowledge, few, if any, studies have been published on IGS sequence and structure for Lythraceae and Punicaceae families. Here, we present and analyze the nucleotide sequence of the rDNA IGS of pomegranate (Punica granatum L.), which has been traditionally placed in Punicaceae family (Koehne 1881). This is the first complete sequence of IGS region in Punicaceae family and Myrtales order. However, according to the latest available projection (Morris 2007), Punica is a member of the Lythraceae, which is closely allied to Onagraceae family (Conti et al. 1997; Dahlgren and Thorne 1984; Johnson and Briggs 1984). Pomegranate is one of the oldest known valuable trees with high nutritional value and medicinal properties (Seeram et al. 2006; Viuda-Martos et al. 2010). This plant has valuable characteristics such as drought tolerance and can be well adapted to different climates and soils (Levin 1994). However, there is low molecular data for this horticultural crop (Ercisli et al. 2007, 2011; Melgarejo et al. 2009; Wang et al. 2007), thus it seems that more molecular studies must be done. The purpose of this study is to: (1) address structural analysis of rDNA IGS sequence of pomegranate; (2) consider the regulatory elements and functional role of some conserved motifs by comparing them with IGS sequence of other plants.
Materials and methods
Plant materials and DNA extraction
The plant samples of Punica granatum (cultivars: Malas danezard Isfahan, Dare Loshan and Anar Zinati Golsefid) were collected from the pomegranate collection of the Agricultural Research Center of Isfahan, Iran, and the Toryu-shibori cultivar was obtained from the germplasm collection maintained at the USDA National Clonal Germplasm Repository, Davis, California, USA. The fresh leaves were frozen with liquid nitrogen and were stored at −80 °C until DNA extraction.
Genomic DNA was extracted from frozen leaf material using CTAB protocol of Murray and Thompson (1980) with minor modifications. Quality and quantity of DNA extracted were determined by electrophoresis on 1 % gel agarose with unrestricted lambda DNA and confirmed by spectrophotometric measurement.
PCR amplification and sequencing
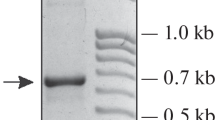
The entire intergenic spacer (IGS) of rDNA was amplified using two universal primers, LR12R and invSR1R, cited by Vilgalys et al. (1994) (Table 1). Primer invSR1R was developed from 5′ end of 18S rDNA gene and 3′ end of primer LR12R was located 256 bp upstream from the 3′ terminus of 28S rDNA gene. However, we used modified invSR1R primer with changes in ninth nucleotide (A–G residue) (Table 1). The presence of the large enough section of the highly conserved 28S gene in the PCR product allowed us to ensure that the amplified PCR product represented the desired region. PCR mixture contained 1× Pfu buffer, 1.3 mM MgSO4, 0.3 mM dNTPs, 0.4 μM of each primer, 2 Units of Taq DNA polymerase (Fermentas, Vilnius, Lithuania), 0.16 Unit of pure Pfu DNA polymerase (Peqlab Co., Germany) and 45 ng of template DNA in a total volume of 45 μl. PCR amplifications were carried out by incubation at 95 °C for 3 min, 35 cycles of denaturation at 95 °C for 1 min, primer annealing at 56 °C for 1 min and primer extension at 68 °C for 5 min, followed by a final extension step of 10 min at 68 °C on a Eppendorf thermal cycler (Germany). The PCR products were visualized on 1 % agarose gel as a single band with a length of approximately 4,000 bp.
The amplified DNA fragments were purified using QIAquick PCR Purification kit, (QIAGEN), according to the manufacturer’s instructions, and were sequenced by the primers LR12R and invSR1R, using the ABI 3730XL sequencer (Macrogen Co., Korea). The sequence data from these external primers were used to design novel internal primers, with overlap of about 100–200 bp, to complete the IGS sequence (Table 1). Furthermore, we developed two primers, GPF4 and GPR4, to sequence the two ends of 4 kb amplified PCR products (Table 1). The specific primers were designed by Oligo software ver.5. To eliminate all ambiguities, each sequence was repeated twice, especially in places where they do not define any reverse primers.
Sequence analysis
DNA sequences for total IGS were assembled into contig file by aligning the overlapped sequences. The complete sequence IGS of Malas-Danezard Isfahan cultivar appears in EMBL databank under the accession No. JX121275. Partial IGS sequences of other cultivars of P. granatum are available via GenBank: Dare-Loshan (accession nos. JQ782224 and JQ782221), Anar-Zinati Golsefid (JQ782225 and JQ782222), and Toryu-Shibori (JQ782226 and JQ782223). Multisequence alignments were applied by the program MEGA5 using ClustalW method (Higgins et al. 1994) with gap opening penalty of 15 and gap extension penalty of 6. Pairwise sequence comparisons were performed by the global alignment using Needleman–Wunsch algorithm (Needleman and Wunsch 1970). We used the Tandem Repeat Finder 4 (trf400) program (Benson 1999) which identified DNA motifs by calculating the number and distribution of the motifs. The prediction of secondary structures of the IGS sequence was done by means of MFOLD software, which was based on Zuker (2003).
Results
PCR amplification and sequencing of the intergenic spacer region
Amplification of the pomegranate genomic DNA with the universal primers (LR12R and invSR1R) resulted in a clear single PCR product. The amplified PCR products in different pomegranate genotypes showed an identical size at approximately 4,000 bp in length. This could reveal that there is no significant length heterogeneity within the studied genotypes. Sequencing of the PCR products via designated internal primers (Table 1) presented the complete nucleotide sequence of the intergenic region between the 18S and 28S rDNA of pomegranate (Fig. 1). The flanked coding regions were determined by alignment with the previously reported sequences for other plants. Results showed that the total IGS region is 3,712 bp long, presenting a 67.8 % GC content. Furthermore, the end point of the 28S rRNA coding region was 277 bp in length and 63.2 % GC. In addition, the results of the partial sequencing of IGS region in three cultivars of pomegranate (Dare-Loshan, Anar-Zinati Golsefid, Toryu-Shibori) and the two ends of IGS sequence of Malas-Danezard Isfahan cultivar revealed a high similarity.
The entire nucleotide sequence of rDNA IGS region of Punica granatum cultivar Malas-Danezard Isfahan. The repeated sequences are underlined with: double line for repeat type A; single line for repeat block B; dashed line for repeat sequence C. The TIS sequence is in dashed boxed and an arrow indicates the initiating A residue. The motif GAAAAT is marked with oval. The box shows the 5′ end of 18S rDNA gene
Transcription initiation and termination sites of rRNA in IGS sequence
Sequence analysis of the pomegranate IGS represented three distinct regions: a central repeated region and two unique regions flanking the repeats. In the first unique region, just after the 3′ end of the 28S, we found a pyrimidine-rich sequence (CCCCCTCCCCTCC); in addition, they contained two A residues between CTCC motifs. This region is highly similar to the beginning of the IGS in other plants (Borisjuk et al. 1997; Gruendler et al. 1991; Kelly and Siegel 1989; Maggini et al. 2008; Perry and Palukaitis 1990; Polanco and Pérez De La Vega 1994) and based on previous reports, it might function as transcription termination site (TTS) (Vincentz and Flavell 1989; Zentgraf et al. 1990).
In the third unique region, after the central repeated sequences, we could identify the putative transcription initiation site (TIS) with a single sequence TCTTTAGGGGGGTAG (from base 2,561 to 2,575), which fitted the reports of TIS in other plants (Hemleben et al. 1988; Toloczyki and Feix 1986). The initiation site of RNA transcript at +1 position is the adenine residue at position of 2,566 (Fig. 1). This is in agreement with most previous reports in other IGS sequences (Delcasso-Tremousaygue et al. 1988; Doelling et al. 1993; Gerstner et al. 1988; Kato et al. 1990; Maggini et al. 2008).
Structure of the repeat sequences and methylation sites
In the central region of IGS sequence, two types of tandemly arranged repeats were identified, termed as A and B. In addition, two direct repeats are located downstream of the TIS sequence in ETS region, designated as C (Fig. 1). The first type of repeat named repeat A consists of two subrepeats (A1 and A2) with length of 68 and 67 bp, respectively, and an average GC content of 57 %. Repeats A1 and A2 are separated by 12 bp. Alignment of the two repeats showed a value of 73.6 % homology (Fig. 2b).
The second type of repeat, named repeat B, is extended from nucleotide position 816 to 2,485. The base pattern of poly (N) runs in block B specifically shows a high frequency of poly (G/C) runs. In the subrepeats of other eukaryotes, the high frequency of poly (G/C) is found, as in African clawed frog [Xenopus laevis (Daudin 1802)] (Moss et al. 1980), rice (Oryza sativa L.) (Takaiwa et al. 1990) and mosquito [Aedes albopictus (Skuse 1894)] (Baldridge and Fallon 1992). The repeat unit B is the GC-richest region in the IGS sequence of pomegranate and presents a 71.3 % GC. Sequence analysis by trf400 software found a high number of 20 bp short direct repeats with consensus sequence of GGTCACTGGGCACGATCACG in the repeat family B that is repeated tandemly from position 954 to 2,371 from 5′ end of IGS.
The repeat unit C consists of two short repeats (C1 and C2) that were separated by an inserted sequence as long as 180 bp. Alignment between two subrepeats, C1 and C2, revealed the high homology (87.5 %) (Fig. 2c).
It is worth noting that the pomegranate IGS sequence includes a large number of GCGC and CCGG motifs (30 and 74, respectively) that are irregularly distributed along the entire sequence. In some places, the motifs are located tandemly. The presence of a large number of the methylable motifs has also been reported in other studies (Maggini et al. 2008; Polanco and Pérez De La Vega 1994).
Discussion
Characterization of IGS
In the present study, we report the complete nucleotide sequence of the IGS region in Punica granatum for the first time. Indeed, the IGS structure is attractive to study because of its importance for regulation of transcription levels of rRNA genes by the signals of stop and start for transcription of the rDNA units that are located in it (Fernández et al. 2000; Hemleben and Zentgraf 1994). Analysis of pomegranate IGS sequence revealed that its organization is similar to the most of the previous studies on IGS and contains the repetitive elements, initiation and termination sites of transcription.
Structure of promoter and terminator sequences and ETS region
The IGS sequence included the motifs involved in signaling RNA transcription and processing like CCCTCC motif that is located just after the 3′ terminus of the 25S. However, this motif is repeated multiple times in the entire sequence of pomegranate IGS, especially near the supposed transcription initiation site. The presence of the similar motif near the TIS region has been previously described in other eukaryotes such as rice (Oryza sativa L.), common oat (Avena sativa L.), maize (Zea mays L.), wheat (Triticum aestivum L.), African clawed frog (Xenopus laevis) and cucumber (Cucumis sativus L.) (Moss and Stefanovsky 1995; Polanco and Pérez De La Vega 1994; Zentgraf et al. 1990) that probably acts as a proximal terminator (Piller et al. 1990).
The putative TIS sequence in pomegranate IGS is identified in the present study. Based on other studies, sequences surrounding the TIS from −5 to +6 have important functions and are highly invariant (Doelling and Pikaard 1995). In most plants, the TATA and GGGG boxes are common elements at the 5′ end of pre-rRNA (Hemleben et al. 1988; Perry and Palukaitis 1990; Toloczyki and Feix 1986; Zentgraf et al. 1990). However, in pomegranate, the putative TIS sequence missed TATA motif in the upstream of the conserved A residue. But it is interesting to note that the core sequence around the initiating A is highly similar to the TIS region of European oak trees [Quercus petraea (Matt.) Liebl. and Q. robur L.] (Bauer et al. 2009), especially the sequence upstream of the A residue (TCTTT), demonstrating the same changes in the TATA motifs in pomegranate and oak. This is in agreement with the suggestion that these changes are probably as a result of different evolutionary path of tree species (Bauer et al. 2009). However, more studies on tree species are required.
In general, the gene promoters are preceded by AT-rich sequence elements (Gerstner et al. 1988; Hemleben and Zentgraf 1994; Reeder 1989; Zentgraf and Hemleben 1992). In pomegranate IGS, the long AT-rich sequence cannot be detected; however, we could find short AT-rich region just upstream of the TIS sequence. This region contains the GAAAAT motif. However, this motif is also repeated irregularly throughout the entire IGS sequence. It is repeated in each short repeat unit A, occurring two times in the 5′ ETS region after the TIS. It is worth to mention that the presence of the similar sequences in the subrepeats and in the promoter region and even downstream from the TIS in the IGS is discussed in other studies and it has been shown that they may function as enhancers for promoter (Barker et al. 1988; Delcasso-Tremousaygue et al. 1988; Echeverria et al. 1992; Gruendler et al. 1991; Polanco and Pérez De La Vega 1994; Tremousaygue et al. 1992; Zentgraf and Hemleben 1992). In addition, the existence of this motif before TIS was reported previously in other plants such as legume species (Fernández et al. 2000), Eruca sativa L. (Lakshmikumaran and Negi 1994), radish (Raphanus sativus L.) (Echeverria et al. 1992); according to Echeverria et al. (1992), the GAAAAT motif may have the function to bind to the homologous nuclear protein fraction. However, experimental data are needed to demonstrate the function of this motif.
The exact ends of pomegranate promoter are yet to be identified. However, further studies with alignment between the total sequence of pomegranate IGS and the promoter region of the European oak (Quercus petraea) revealed the maximum homology with position 2,504–2,572 of pomegranate IGS, in agreement with the position of putative promoter region in the present study. Indeed, comparing the −80 to +10 region of pomegranate promoter with the available promoter of Quercus petraea (Bauer et al. 2009) shows a homology of 50.5 % and some conserved motifs (Fig. 2a).
Further analysis revealed the possible secondary structure at the putative TIS region of the pomegranate with minimum free energy of −7.74 (Fig. 3a). This structure is similar to previous studies in plant species (Vicia faba L. and Pisum sativum L.), which was also found in rat, mouse and human rDNA (Financsek et al. 1982; Kato et al. 1990). In addition, upstream of the TIS region of pomegranate includes the inverted sequence that can construct the stem-loop structure before the initiation site and CCTCC motif involves formation of this structure (Fig. 3b).
In the ETS region, a dyad symmetry was identified just upstream the 5′ end of 18S rRNA, which forms the stem-loop structure with the length of 77 bp (Fig. 3c). The similar stem–loop structure occurs near the 5′ extreme of the 18S rRNA, in far-related plant species (oat (Avena sativa L.), pea (Pisum sativum L.), maize (Zea mays L.), soybean (Glycine max L. Merr.), lentil (Lens culinaris Medik.), faba bean (Vicia faba L.), Hordeum bulbosum L., rice (Oryza sativa L.) and also in animals. This may show the relevance of this structure to pre-rRNA processing (Barker et al. 1988; Fernández et al. 2000; Piller et al. 1990; Polanco and Pérez De La Vega 1994).
Repeat sequences and methylation sites
Sequence analysis of pomegranate IGS was resulted to identify the three types of repeat family (denoted as A, B and C) as shown in Fig. 1. Repeat family A revealed the motif GGATTT that is repeated twice in each A subrepeats with a point mutation in the second nucleotide (G to A); it is also found in downstream of the TIS in two other short directs of C repeats. This could show the conserve nucleotides among the two types of repeats A and C. The repeat family B contains the large number of the motifs GGGCAC and GGTCAC, that are repeated 58 and 50 times, respectively, without any changes in the block B region. Also, these motifs are present in the 20 bp consensus sequence of GGTCACTGGGCACGATCACG that are repeated tandemly in B family. It seems that the B block region was generated by proliferation and duplication of a short sequence motif (GGGCAC). However, the presence of the variation amongst the short repeats was generated as a result of the later point mutations.
Comparing the three types of repeat family demonstrated the relatively low homology (data not shown). However, we applied the drop out poly (N) runs method based on Ryu et al. (2008), by deletion of all consecutive bases in each poly (N) run expect one base, resulting in the increase in the similarity values. Indeed, the high frequency of the poly (N) runs is one of the major factors in the divergence of primary sequence (Ryu et al. 2008). With drop out poly (N) method, the three types of repeat reveal the common short sequence (GATC). It is striking to note that the sequence GATC has been repeated highly in IGS sequence, being found mostly in block B (93.18 %). This could suggest that this short sequence may be an ancestral sequence to raise the repeat families in IGS of pomegranate. However, more comparative studies, especially with close related species to Punica, are needed.
The IGS of pomegranate revealed the abundant number of methylable sites (74 CCGG and 30 GCGC) that are unevenly distributed along the sequence. Indeed, the methylation levels have demonstrated the relation in the transcription of the rRNA genes in several instances (review in Hemleben and Zentgraf 1994; Komarova et al. 2004; Santoro et al. 2002; Sardana et al. 1993). In comparison with Avena sativa, the spacer of P. granatum showed the same number of GCGC sites (30 vs. 30) (Polanco and Pérez De La Vega 1994), which is more than wheat (Triticum aestivum L.) and common olive (Olea europaea L.) (18 and 17, respectively). Punica IGS demonstrated more CCGG motifs than Olea (46), Avena (22) and Triticum (10). In pomegranate, most of the CCGG sites are located in the block B repeats (42 sites), and the less number is found in the first and third unique regions (14 and 15, respectively), while the GCGC motifs are presented mostly in the downstream of TIS (5′ ETS region) and in the B repeat family (13 and 11, respectively). The methylable sites are irregularly distributed in Avena, Olea and Triticum (Maggini et al. 2008; Polanco and Pérez De La Vega 1994; Sardana et al. 1993). In Punica, 56.7 % of the CCGG motifs are found in B block that is similar to B subrepeats of Avena, but the density of the GCGC motif in block B (36.6 %) is lower than Avena B subrepeats. However, the number of these motifs in block B is relatively high and it may play an important function in the expression of rDNA loci according to the results of Polanco and Pérez De La Vega (1997) and Sardana et al. (1993).
The present study is the first report on complete sequence of IGS in pomegranate and also in Myrtales order. Analysis of Pomegranate IGS sequence showed that its organization is similar to the most of the previous studies on other plant IGS. It contains the repetitive elements, initiation and termination sites of transcription and revealed some conserved motifs. The presence of conserved motifs in the IGS region of variable plant taxa could support the hypothesis that these motifs may act as functional sequences to control transcription or processing of rRNA. However, further studies involving the comparative analysis with the IGS of pomegranate will address the homologous features and identification of similar ancestral sequences by sequencing the entire IGS of other species that are closely related to pomegranate, especially the species in Lythraceae family.
Abbreviations
- rDNA:
-
Ribosomal DNA
- IGS:
-
Intergenic spacer
- ETS:
-
External transcribed spacer
- ITS:
-
Internal transcribed spacer
- NOR:
-
Nucleolus organizer region
- NTS:
-
Non-transcribed spacer
- TIS:
-
Transcription initiation site
- TTS:
-
Transcription termination site
- PCR:
-
Polymerase chain reaction
- CTAB:
-
Cetyltrimethylammonium bromide
References
Appels R, Honeycutt RL (1986) rDNA: evolution over a billion years. In: Dutta SK (ed) DNA systematics, vol 2. CRC Press, Boca Raton, pp 81–135
Baldridge GD, Fallon AM (1992) Primary structure of the ribosomal DNA intergenic spacer from the mosquito, Aedes albopictus. DNA Cell Biol 11:51–59
Baldridge GD, Dalton MW, Fallon AM (1992) Is higher-order structure conserved in eukaryotic ribosomal DNA intergenic spacers? J Mol Evol 35:514–523
Barker RF, Harberd NP, Jarvis MG, Flavell RB (1988) Structure and evolution of the intergenic region in a ribosomal DNA repeat unit of wheat. J Mol Biol 201:1–17
Bauer N, Horvat T, Biruš I, Vičić V, Zoldoš V (2009) Nucleotide sequence, structural organization and length heterogeneity of ribosomal DNA intergenic spacer in Quercus petraea (Matt.) Liebl. and Q. robur L. Mol Genet Genomics 281:207–221
Bennett RI, Smith AG (1991) The complete nucleotide sequence of the intergenic spacer region of an rDNA operon from Brassica oleracea and its comparison with other crucifers. Plant Mol Biol 16:1095–1098
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Bhatia S, Negi MS, Lakshmikumaran M (1996) Structural analysis of the rDNA intergenic spacer of Brassica nigra: evolutionary divergence of the spacers of the three diploid Brassica species. J Mol Evol 43:460–468
Borisjuk L, Hemleben V (1992) Nucleotide sequence of potato rDNA intergenic spacer. Plant Mol Biol 21:381–384
Borisjuk NV, Davidjuk YM, Kostishin SS, Miroshnichenco GP, Velasco R, Hemleben V, Volkov RA (1997) Structural analysis of rDNA in the genus Nicotiana. Plant Mol Biol 35:655–660
Carvalho A, Guedes-Pinto H, Lima-Brito J (2011) Intergenic spacer length variants in Old Portuguese bread wheat cultivars. J Genet 90(2):203–208
Chang KD, Fang SA, Chang FC, Chung MC (2010) Chromosomal conservation and sequence diversity of ribosomal RNA genes of two distant Oryza species. Genomics 96:181–190
Conti E, Litt A, Wilson PG, Graham SA, Briggs BG, Johnson LAS, Sytsma KJ (1997) Interfamilial relationships in Myrtales: molecular phylogeny and patterns of morphological evolution. Syst Bot 22:629–647
Da Rocha PSCF, Bertrand H (1995) Structure and comparative analysis of the rDNA intergenic spacer of Brassica rapa: implications for the function and evolution of the Cruciferae spacer. Eur J Biochem 229:550–557
Dahlgren R, Thorne RF (1984) The order Myrtales: circumscription, variation, and relationships. Ann Missouri Bot Gard 71:633–699
Delcasso-Tremousaygue D, Grellet F, Panabieres F, Ananiev ED, Delseny M (1988) Structure and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur J Biochem 172:767–776
Doelling JH, Pikaard CS (1995) The minimal ribosomal RNA gene promoter of Arabidopsis thaliana includes a critical element at the transcription initiation site. Plant J 8:683–692
Doelling JH, Gaudino RJ, Pikaard CS (1993) Functional analysis of Arabidopsis thaliana rRNA gene and spacer promoters in vivo and by transient expression. Proc Natl Acad Sci USA 90:7528–7532
Echeverria M, Delcasso-Tremousaygue D, Delseny M (1992) A nuclear protein fraction binding to dA/dT-rich sequences upstream from the radish DNA promoter. Plant J 2:211–219
Ercisli S, Agar G, Orhan E, Yildirim N, Hizarci Y (2007) Interspecific variability of RAPD and fatty acid composition of some pomegranate cultivars (Punica granatum L.) growing in southern Anatolia region in Turkey. Biochem Syst Ecol 35:764–769
Ercisli S, Kafkas E, Orhan E, Kafkas S, Dogan Y, Esitken A (2011) Genetic characterization of pomegranate (Punica granatum L.) genotypes by AFLP markers. Biol Res 44:345–350
Fernández M, Polanco C, Ruiz ML, Pérez De La Vega M (2000) A comparative study of the structure of the rDNA intergenic spacer of Lens culinaris Medik. and other legume species. Genome 43:597–603
Financsek I, Mizumoto K, Mishima Y, Muramatsu M (1982) Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci USA 79:3092–3096
Gerstner J, Schiebel K, von Waldburg G, Hemleben V (1988) Complex organization of the length heterogeneous 5′ external spacer of mung bean (Vigna radiata) ribosomal DNA. Genome 30:723–733
Gruendler P, Unfried I, Pascher K, Schweizer D (1991) rDNA intergenic region from Arabidopsis thaliana structural analysis, intraspecific variation and functional implications. J Mol Biol 221:1209–1222
Hemleben V, Zentgraf U (1994) Structural organization and regulation of transcription by RNA polymerase I of plant nuclear ribosomal RNA genes. In: Nover L (ed) Results and problems in cell differentiation 20 Plant promoters and transcription factors. Springer, Berlin, pp 3–24
Hemleben V, Ganal M, Gerstner J, Schiebel K, Torres RA (1988) Organization and length heterogeneity of plant ribosomal RNA genes. In: Kahl G (ed) The architecture of eukaryotic genes. VHC, Weinheim, pp 371–383
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Johnson LAS, Briggs BG (1984) Myrtales and Myrtaceae-a phylogenetic analysis. Ann Missouri Bot Gard 71:700–756
Jorgansen RA, Cluster PD (1988) Modes and tempos in the evolution of nuclear ribosomal DNA: new characters for evolutionary studies and new markers for genetic and population studies. Ann Mo Bot Gard 75:1238–1247
Kahl G (1988) Architecture of eukaryotic genes. VCH Verlagsgesellschaft, Weinheim
Kato A, Nakajima T, Yamashita J, Yakura K, Tanifuji S (1990) The structure of the large spacer region of rDNA in Vicia faba and Pisum sativum. Plant Mol Biol 14:983–993
Kelly RJ, Siegel A (1989) The Cucurbita maxima ribosomal DNA intergenic spacer has a complex structure. Gene 80:239–248
Kim KJ, Mabry TJ (1991) Phylogenetic and evolutionary implications of nuclear ribosomal DNA variation in dwarf dandelions (Krigia, Lactuceae, Asteraceae). Plant Syst Evol 177:53–69
King K, Torres RA, Zentgraf U, Hemleben V (1993) Molecular evolution of the intergenic spacer in the nuclear ribosomal RNA genes of Cucurbitaceae. J Mol Evol 36:144–152
Koehne E (1881) Lythraceae monographice describuntur. Botanische Jahrbücher für Systematik 1:142–157
Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA (2004) Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol Biol 56:439–463
Lakshmikumaran M, Negi MS (1994) Structural analysis of two length variants of the rDNA intergenic spacer from Eruca sativa. Plant Mol Biol 24:915–927
Levin GM (1994) Pomegranate (Punica granatum L.) plant genetic resources in Turkmenistan. Plant Genet Resour Newsl 97:31–36
Maggini F, Tucci G, Demartis A, Gelati MT, Avanzi S (1992) Ribosomal RNA genes of Phaseolus coccineus I. Plant Mol Biol 18:1073–1082
Maggini F, Gelati MT, Spolverini M, Frediani M (2008) The intergenic spacer region of the rDNA in Olea europaea L. Tree Genet Genomes 4:293–298
Melgarejo P, Martínez JJ, Hernández F, Martínez R, Legua P, Oncina R, Martínez-Murcia A (2009) Cultivar identification using 18S–28S rDNA intergenic spacer-RFLP in pomegranate (Punica granatum L.). Sci Hortic 120:500–503
Morris JA (2007) A molecular phylogeny of the Lythraceae and inference of the evolution of heterostyly. Ph.D thesis, Kent State University, USA, p 115
Moss T, Stefanovsky VY (1995) Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Nucleic Acids Res Mol Biol 50:25–66
Moss T, Boseley PG, Birnstiel ML (1980) More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res 8:467–485
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453
Perry KL, Palukaitis P (1990) Transcription of tomato ribosomal DNA and the organization of the intergenic spacer. Mol Gen Genet 221:102–112
Piller KJ, Baerson SR, Polans NO, Kaufman LS (1990) Structural analysis of the short length ribosomal DNA variant from Pisum sativum L. cv. Alaska. Nucleic Acids Res 18:3135–3145
Poczai P, Hyvonen J (2010) Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Mol Biol Rep 37:1897–1912
Polanco C, Pérez De La Vega M (1994) The structure of the rDNA intergenic spacer of Avena sativa L.; a comparative study. Plant Mol Biol 25:751–756
Polanco C, Pérez De La Vega M (1997) Intergenic ribosomal spacer variability in hexaploid oat cultivars and landraces. Heredity 78:115–123
Reeder RH (1989) Regulatory elements of the generic ribosomal gene. Curr Opin Cell Biol 1:466–474
Rogers SO, Bendich AJ (1987) Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
Ryu S, Do Y, Fitch DHA, Kim W, Mishra B (2008) Dropout alignment allows homology recognition and evolutionary analysis of rDNA intergenic spacers. J Mol Evol 66:368–383
Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32:393–396
Sardana R, O’Dell M, Flavell RB (1993) Correlation between the size of the intergenic regulatory region, the status of cytosine methylation of rRNA genes and nucleolar expression in wheat. Mol Gen Genet 236:155–162
Schaal BA, Learn GHJ (1988) Ribosomal DNA variation with and among plant populations. Ann Mo Bot Gard 75:1207–1216
Schiebel K, von Waldburg G, Gerstner J, Hemleben V (1989) Termination of transcription of ribosomal RNA genes of mung bean occurs within a 175 bp repetitive element of the spacer region. Mol Gen Genet 218:302–307
Seeram NP, Schulman RN, Heber D (2006) Pomegranate: ancient roots to modern medicine. CRC Press and Taylor and Francis Group, Boca Raton
Takaiwa F, Kikuchi S, Oono K (1990) The complete nucleotide sequence of the intergenic spacer between 25S and 17S rDNAs in rice. Plant Mol Biol 15:933–935
Toloczyki C, Feix G (1986) Occurrence of nine homologous repeat units in the external spacer of a nuclear maize rRNA gene unit. Nucleic Acids Res 14:4969–4986
Tremousaygue D, Laudie M, Grellet F, Delseny M (1992) The Brassica oleracea rDNA spacer revisited. Plant Mol Biol 18:1013–1018
Vilgalys R, Hopple JS Jr, Hibbett DS (1994) Phylogenetic implications of generic concepts in fungal taxonomy: the impact of molecular systematic studies. Mycol Helv 6:73–91
Vincentz M, Flavell RB (1989) Mapping of ribosomal RNA transcripts in wheat. Plant Cell 1:579–589
Viuda-Martos M, Fernández-López J, Pérez-Álvarez JA (2010) Pomegranate and its many functional components as related to human health: a review. Comp Rev Food Sci Food Saf 9:635–654
Wang WL, Tsay JS, Yeuh CS, Chen RS, Tsay JG (2007) Phylogenetic relationships of pomegranate (Punica granatum L.) accessions by using rDNA ITS, RAPD and ISSR molecular markers. J Taiwan Soc Hortic Sci 53:157–172
Weider LJ, Elser JJ, Crease TJ, Mateos M, Cotner JB, Markow TA (2005) The functional significance of ribosomal (r) DNA variation: impacts on the evolutionary ecology of organisms. Annu Rev Ecol Evol Syst 36:219–242
Yakura K, Kato A, Tanifuji S (1984) Length heterogeneity in the large spacer of Vicia faba rDNA is due to the differing number of a 325 bp repetitive sequence element. Mol Gen Genet 193:400–405
Zentgraf U, Hemleben V (1992) Complex formation of nuclear proteins with the RNA polymerase I promoter and repeated elements in the external transcribed spacer of Cucumis sativus ribosomal DNA. Nucleic Acids Res 20:3685–3691
Zentgraf U, Ganal M, Hemleben V (1990) Length heterogeneity of the rRNA precursor in cucumber (Cucumis sativus). Plant Mol Biol 15:465–474
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
This work was supported by Isfahan University of Technology, Iran. We thank the Agricultural Research Center of Isfahan, Iran, and the USDA-ARS, National Clonal Germplasm Repository, Davis, California, USA, for providing the plant materials.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Parvaresh, M., Talebi, M. & Sayed-Tabatabaei, BE. Molecular characterization of ribosomal DNA intergenic spacer (IGS) region in pomegranate (Punica granatum L.). Plant Syst Evol 300, 899–908 (2014). https://doi.org/10.1007/s00606-013-0928-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-013-0928-1