Abstract
Süßwassertang, a popular aquatic plant that is sold worldwide in aquarium markets, has been long considered a liverwort because of its ribbon-like thallus. However, its antheridia are remarkably fern-like in morphology. To corroborate the hypothesis that Süßwassertang is a fern gametophyte and to determine its closest relative, we have sequenced five chloroplast regions (rbcL, accD, rps4–trnS, trnL intron, and trnL-F intergenic spacer), applying a DNA-based identification approach. The BLAST results on all regions revealed that Süßwassertang is a polypod fern (order: Polypodiales) with strong affinities to the Lomariopsidaceae. Our phylogenetic analyses further showed that Süßwassertang is nested within the hemi-epiphytic fern genus Lomariopsis (Lomariopsidaceae) and aligned very close to L. lineata. Our study brings new insights on the unexpected biology of Lomariopsis gametophytes—the capacity of retaining a prolonged gametophytic stage under water. It is of great interest to discover that a fern usually known to grow on trees also has gametophytes that thrive in water.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
An aquatic plant called Süßwassertang, which means “freshwater seaweed” in German, has been commercially available on the aquarium market worldwide for a number of years (Fig. 1a). Because of its liverwort-like appearance, it has long been considered to be a liverwort, such as Pellia or Monoselenium. Our observation of Süßwassertang gametangia, which are only rarely produced by the submerged thallus, suggested that this plant is not a liverwort but a fern gametophyte. Its archegonia are fern-like in having short necks, and the venter is immersed partly in the thallus. The antheridia resemble those of polypodialean ferns in that they consist of three cells: a cap cell, a ring cell, and a basal cell (Fig. 1b; Nayar and Kaur 1971). Although gametangia are present occasionally, sporophytes have never been observed in aquaria and even after planting onto soil. It is therefore difficult to identify the plant with certainty.
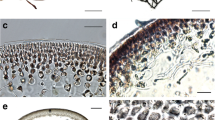
Süßwassertang, its microscopic features, and the habit of Lomariopsis spectabilis. a A portion of the gametophyte thallus showing extensive lateral branching. Bar: 1 cm. b Side view of an antheridium, showing a cap cell (cc), ring cell (rc), and basal cell (bc). Bar: 20 µm. c Scanning electron microscope image of developing lateral branches with rhizoids (arrowhead) and meristems (m) in the rounded apex. Bar: 0.2 mm. d Ribbon-like, branched gametophyte (g) of L. spectabilis bearing a young sporophyte (sp) in a field of Taiwan. It has similar morphology with the mysterious gametophyte. Arrowhead Branch points. Bar: 1 cm
To unravel this mysterious identity, we employed a DNA-based identification approach consisting of sequence comparisons and phylogenetic analyses. We sequenced five chloroplast regions [rbcL, accD, rps4–trnS, trnL intron, and the trnL-F intergenic spacer (IGS)], of which rbcL has already been successfully used to identify an unknown fern gametophyte in a similar study (Schneider and Schuettpelz 2006). The widespread occurrence of rbcL in online databases for all plant lineages makes it well-suited for broad-scale screening. Likewise, the analyses by Quandt et al. (2004) indicated that the trnL-F region (intron and IGS) has the power to relate any sequence via a database comparison on generic level in most land plant lineages. The rapidly evolving gene rps4 (plus rps4–trnS IGS) was included because it represents one of the broadly sequenced regions (together with the trnL-F region and rbcL) in seedless plants (e.g., Quandt and Stech 2003; Schneider et al. 2004). In the approach chosen here, BLAST results pinpointed which clade Süßwassertang belongs to and therefore guided the taxonomic sampling for phylogenetic inferences. To increase the phylogenetic signal, we combined accD with rbcL in the analyses. Once narrowed to a certain lineage, a more precise marker was then used to resolve the position of Süßwassertang among more recently diverged lineages. In this case, the plastid trnL-F IGS was employed to infer inter-species affinities of this supposedly aquatic fern gametophyte.
Materials and methods
Taxonomic sampling for molecular phylogeny
Taxonomic sampling was guided by the MegaBLAST (Zhang et al. 2000) results obtained from the five regions sequenced (rbcL, accD, rps4–trnS, trnL intron, and trnL-F IGS). To obtain more robust confirmation of the relationship of Süßwassertang, we compiled two data sets for phylogenetic analyses. As indicated by the BLAST results, the first data set comprised a representative set of sequences from two coding plastid regions, rbcL and accD, of polypod ferns. In addition to the Süßwassertang sequences, 32 species representing 28 fern genera were included in the analyses. Athyrium niponicum (Mett.) Hance and Matteuccia struthiopteris (L.) Todaro were used as outgroups. The second matrix included all available trnL-F IGS accessions of Lomariopsis plus sequences obtained from two Süßwassertang accessions, L. spectabilis (Kunze) Mett., and Cyclopeltis crenata (Fée) C. Chr. Cyclopeltis crenata and Hypodematium crenatum Kuhn ex v. Deck. were used as outgroups for the second matrix.
DNA extraction, amplification, and sequencing
Total genomic DNA was extracted using either the Plant Genomic DNA Mini kit (Geneaid, Taipei, Taiwan) or the Plant Genomic DNA Purification kit (GeneMark, Taichung, Taiwan). In some cases a modified CTAB (cetyl trimethylammonium bromide) procedure (Wang et al. 2004) was applied. The PCR amplifications, which followed standard PCR protocols, were performed in 50-μl reaction volumes containing 1.5 U Taq DNA polymerase, 1.0 mM dNTPs-Mix, 10× buffer, 1.5 mM MgCl2, 10 pmol of each amplification primer, and 1.0 µl DNA. The PCR primers used for amplification and sequencing were: trnL-F with primers C (or E) and F (Taberlet et al. 1991; modifications according to Quandt and Stech 2004); rps4 (plus rps4–trnS IGS) with rps5′ (Nadot et al. 1994) and trnS (Souza-Chies et al. 1997); rbcL with NM34 (Cox et al. 2000) and M1390 (Lewis et al. 1997); accD with the newly designed primers “FW_accDF” (5′-ACG TCT GTA ACA AAT TGG TTT GAA G-3′) and “FW_accDR” (5′-AAA CTC AAC GTT CCT TCT TGC AT-3′). The PCR products were either directly purified using the GeneMark PCR Clean-Up kit (Taichung, Taiwan) or cleaned via gel extraction employing the Nucleospin PCR Purification kit (Macherey-Nagel, Düren, Germany). Sequencing was done with the amplification primers by Macrogen (Seoul, Korea). In order to corroborate the results, isolation, amplification, and sequencing of all regions were performed independently on two different samples in Taipei and Dresden. Newly obtained sequences and other accessions from GenBank used in the analyses are summarized in the Appendix.
Sequence alignment and phylogenetic analyses
DNA sequences were manually aligned using PhyDE0.995 (Müller et al. 2005). During manual alignment, gap placement was guided by the identification of putative microstructural changes following recently published concepts (Kelchner 2000; Quandt et al. 2003). Identified inversions were positionally separated in the alignments, but they were included as a reverse complement in the phylogenetic analyses, as discussed in Quandt et al. (2003). Phylogenetic reconstructions using parsimony were performed using winPAUP* 4.0b10 (Swofford 2002) in combination with PRAP (Müller 2004). The latter program generates command files for PAUP* that allow parsimony ratchet searches as designed by Nixon (1999). In our study, ten random addition cycles of 200 ratchet iterations each were used, with 25% of the positions being randomly double-weighted. The shortest trees collected from the different tree islands were finally used to compute a strict consensus tree. Heuristic bootstrap searches (BS; Felsenstein 1985) were performed with 1000 replicates, ten random addition cycles per bootstrap replicate, and otherwise the same options in effect as in the ratchet.
For a further measurement of support, posterior probabilities were calculated using MrBayes V3.1 (Ronquist and Huelsenbeck 2003), applying the GTR + Γ + I model. The a priori probabilities supplied were those specified in the default settings of the program. Posterior probability (PP) distributions of trees were created using the Metropolis-coupled Markov chain Monte Carlo (MCMCMC) method and followed the search strategies suggested by Huelsenbeck et al. (2001, 2002). Ten runs with four chains (106 generations each) were run simultaneously. Chains were sampled every ten generations, and the respective trees were written to a tree file. Calculation of the consensus tree and of the PP of clades was performed based upon the trees sampled after the chains converged (within the first 250,000 generations). Consensus topologies and support values from the different methodological approaches were compiled and drawn using TreeGraph (Müller and Müller 2004).
Results
BLAST results of the sequenced markers
The sequences (rbcL, accD, rps4-trnS, trnL intron, and trnL-F) obtained from two independent collections of Süßwassertang were identical. BLAST results indicated that Süßwassertang shares high sequence similarities to leptosporangiate ferns and is closest to the Lomariopsidaceae (except for trnL intron), especially in terms of the reported maximum identity (Table 1). Lomariopsis lineata (C. Presl) Holttum was found to be the best match for the trnL-F IGS, whereas for the three coding regions (rbcL, accD, and rps4), L. spectabilis Mett. or L. marginata (Schrad.) Kuhn. received the highest maximum identity scores. Although members of the Dryopteridaceae were among the best matches in a BLAST search using trnL, these results are biased since this region is currently represented by only few ferns in GenBank.
Phylogenetic analyses
Phylogenetic inferences of a representative set of polypod ferns for each of the single-gene data sets (rbcL and accD) were congruent. The phylogenetic relationships presented are thus based on the analyses using the combined data set. Maximum parsimony and Bayesian inference both clearly positioned the aquatic fern gametophyte within the fern genus Lomariopsis (Lomariopsidaceae), a placement that receives high branch support in the phylogenetic analyses (BSMP = 100, PPMB = 1.0; Fig. 2).
One of two most parsimonious trees [length 1739 steps, consistency index (CI) 0.449, retention index (RI) 0.602, rescaled consistency index (RC) 0.270] retained by the parsimony ratchet analysis performed based on the combined rbcL and accD sequence data. This tree was chosen as it perfectly reflects the Bayesian inferences. The values above the branches refer to posterior probabilities from Bayesian analysis, whereas those below the branches indicate bootstrap support values
Phylogenetic analyses of Lomariopsis based on the trnL–F IGS (340 nt) indicated that Lomariopsis lineata (C. Presl) Holttum is the species closest to Süßwassertang (BSMP = 96, PPMB = 1.0; Fig. 3). The trnL-F sequences from L. lineata and the aquatic fern gametophyte show a 97.6% similarity and share an 8-nt indel (Fig. 3) that is absent in other Lomariopsis species. Despite the strong sequence similarity in the non-coding region of the chloroplast genome, the possibility for the fern gametophyte to be a different species, rather than L. lineata, could not be eliminated. Monophyly of Lomariopsis is robustly supported, in contrast to previous study by Rouhan et al. (2007).
One of 53 most parsimonious trees (length 243 steps, CI 0.774, RI 0.835, RC 0.646) retained by the parsimony ratchet analysis performed on the trnL-F IGS sequence data. The values above the branches refer to posterior probabilities from Bayesian analysis, whereas those below the branches indicate bootstrap support values. The occurrence of both observed inversions as well as two characteristic indels (5 and 8 nt) are indicated on the tree
Interestingly, two hairpin-associated inversions were observed in the spacer approximately 185 nt upstream of trnF, which is different from the previously reported one for bryophytes (Quandt and Stech 2004; Quandt et al. 2004). Inversion 1 (inv 1) is homoplastic and occurs twice in: (1) L. recurvata, L. vestita, L. maxonii, and L. salicifolia as well as (2) L. amydrophlebia and L. wrigthii, whereas inversion 2 unites L. hederacea, L. muriculata, and L. manii. The distribution of both inversions in phylogenetic context is plotted on the tree in Fig. 3.
Morphology of the aquatic fern gametophyte
The thallus of the alleged aquatic gametophyte of Lomariopsis is ribbon-shaped, profusely branched, and one-cell thick throughout, without a midrib or multicellular cushion. Rhizoids are colorless, mostly borne as marginal clusters. It grows indeterminately with active meristematic cells at the rounded apex (Fig. 1c). There are no gemmae, although small lateral branches sometimes detach from the thallus and develop as new individuals. Archegonia and three-celled antheridia are sparsely formed. These characters were also observed in the gametophytes of Lomariopsis spectabilis found in Taiwan (Fig. 1d), although they do not exactly match the strap-shaped Lomariopsis gametophytes described and illustrated by Atkinson (1973).
No associated sporophyte of the Süßwassertang under study has ever been observed. Following transplantation of the gametophyte from water to soil, its growth rate was reduced, and the old portions of the thallus began to die. Unlike the gametophyte in water, rhizoids in soil-grown gametophyte were brown, and numerous antheridia formed along the thallus margins. However, sporophytes did not develop under such conditions either.
Discussion
The utility of different markers in identifying the mysterious gametophyte
The DNA-based identification approach used here shares a similar concept with DNA barcoding, yet the latter tries to utilize more or less universal DNA barcodes. Deciding which barcode to be used for plants is still in progress (e.g., Kress et al. 2005; Chase et al. 2005, 2007; Ford et al. 2009; Hollingsworth et al. 2009). Several of the proposed DNA barcodes, such as the trnL intron (Taberlet et al. 2006), accD (Ford et al. 2009), and rbcL (Schneider and Schuettpelz 2006; Kress and Erickson 2007), were employed in this study for identifying the mysterious thallus; hence, we believe our results could provide a guideline for the future selection of plant barcodes, especially considering the situation of seedless plants.
The maximum identity and E-values from the BLAST results nicely illustrate that the trnL-F IGS is more suitable for species identification in ferns than rbcL, as sequence similarity for rbcL of Süßwassertang and Lomariopsis spectabilis or L. marginata already reaches 96–98%, while sequence identity of the trnL-F IGS of both species with Süßwassertang is only 89%. In addition, the trnL-F IGS amplicon is only about 600 nt compared to 2.1 kb of rbcL and therefore easier to handle in a barcoding approach. However, as the spacer is missing in some green algae (Quandt et al. 2004) and merely reaches 60 nt in derived mosses (Quandt and Stech 2004), its use is limited.
Although more than 21,000 trnL intron sequences (bryophytes >3000, flowering plants >18,000) are recorded in GenBank, ferns are vastly underrepresented, with 342 records (on 2 February 2008), which rendered the database comparison problematic. No trnL intron sequences of Lomariopsis species or Lomariopsidaceae are recorded in GenBank, which explains why the closest matches were found among members of the Dryopteridaceae (Table 1). However, similar to the coding regions, trnL also placed Süßwassertang within polypod ferns. The reported values from BLAST searches representing sequence divergence indicate that the trnL intron resolves more relatively recent divergences compared to rbcL. Likewise, BLAST searches based on the obtained rps4 (plus rps4–trnS IGS) sequence showed only 86% maximal sequence identity of Süßwassertang with L. marginata compared to 96% found for rbcL, indicating the higher potential of rps4-trnS in barcoding approaches compared to rbcL (Table 1). AccD displayed a slightly higher performance than rbcL, with 96% identity to L. spectabilis (Table 1).
Therefore, if rbcL was to be chosen as the DNA barcode, a two-step approach would be favorable. With the similar concept, Kress and Erickson (2007) proposed a two-locus barcode combining trnH–psbA with rbcL for plants. This combination worked well in filmy ferns (Nitta 2008). However, trnH–psbA is absent in black pine (Wakasugi et al. 1994) and since two copies of trnH–psbA can be found in the Adiantum chloroplast genome (Wolf et al. 2003), a careful investigation should be done on whether multiple copies may or may not mislead species identification in ferns. Regardless of the rather conserved rbcL, Lahaye et al. (2008) proposed matK as the prime plant barcode. However, due to the rapidly evolving nature of ferns’ matK and a lack of universal priming sites, especially at the 5′ end (Kuo et al., unpublished data; Wicke and Quandt, unpublished data), it would be problematic to use matK in ferns. Clearly, a comprehensive survey in seedless plants on the utility of different potential barcodes is urgently needed.
An aquatic gametophyte from an epiphytic sporophyte
Fern gametophytes are well known for their extreme tolerance to environmental stresses, such as winter cold (Sato 1982), light deficiency (Johnson et al. 2000), and desiccation (Watkins et al. 2007). As a result, gametophytes in some cases were able to establish populations in sites that were probably far too extreme for sporophytes by exclusively maintaining the gametophyte generation (Farrar 1967, 1990; Dassler and Farrar 1997; Rumsey et al. 1999). Our discovery of Süßwassertang contributes another extraordinary example. Süßwassertang, originally known as a species of bryophytes, has been used to decorate fish tanks. Based on the results of our study involving DNA markers, we have identified Süßwassertang as gametophytes of Lomariopsis, an exclusively hemi-epiphytic fern clade, and found that these gametophytes have an exceptional capability to thrive in water for years without forming their sporophyte counterpart.
References
Atkinson LR (1973) The gametophyte and family relationships. Bot J Linn Soc 67[Suppl 1]:73–90
Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, Savolainen V (2005) Land plants and DNA barcodes:short-term and long-term goals. Philos Trans R Soc Lond B Biol Sci 360:1889–1895
Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madriñán S, Petersen G, Seberg O, Jørgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M (2007) A proposal for a standardised protocol to barcode all land plants. Taxon 56:295–299
Cox CJ, Goffinet B, Newton AE, Shaw AJ, Hedderson TAJ (2000) Phylogenetic relationships among the diplolepideous-alternate mosses (Bryidae) inferred from nuclear and chloroplast DNA sequences. Bryologist 103:224–241
Dassler CL, Farrar DR (1997) Significance of form in fern gametophytes: clonal, gemmiferous gametophytes of Callistopteris baueriana (Hymenophyllaceae). Int J Plant Sci 158:622–639
Farrar DR (1967) Gametophytes of four tropical fern genera reproducing independently of their sporophytes in the southern Appalachians. Science 155:1266–1267
Farrar DR (1990) Species and evolution in asexually reproducing independent fern gametophytes. Syst Bot 15:98–111
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Ford CS, Ayres KL, Toomey N, Haider N, Stahl JV, Kelly LJ, Wikström N, Hollingsworth PM, Duff RJ, Hoot SB, Cowan RS, Chase MW, Wilkinson MJ (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc 159:1–11
Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, Cowan R, Chase MW, Gaudeul M, Hollingsworth PM (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour 9:439–457
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314
Huelsenbeck JP, Larget B, Miller RE, Ronquist F (2002) Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol 51:673–688
Johnson GN, Rumsey FJ, Headley AD, Sheffield E (2000) Adaptions to extreme low light in the fern Trichomanes speciosum. New Phytol 148:423–431
Kelchner SA (2000) The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann Mo Bot Gard 87:482–498
Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:e508
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374
Lahaye R, Bank M, Borgarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105:2923–2928
Lewis LA, Mishler BD, Vilgalys R (1997) Phylogenetic relationships of the liverworts (Hepaticae), a basal embryophyte lineage, inferred from nucleotide sequence data of the chloroplast gene rbcL. Mol Phylogenet Evol 7:377–393
Li CX, Lu SG (2006) Phylogenetic analysis of Dryopteridaceae based on chloroplast rbcL sequences. Acta Phytotax Sin 44:503–515
Müller K (2004) PRAP—calculation of Bremer support for large data sets. Mol Phylogenet Evol 31:780–782
Müller J, Müller K (2004) TreeGraph: automated drawing of complex tree figures using an extensible tree description format. Mol Ecol Notes 4:6–788
Müller K, Quandt D, Müller J, Neinhuis C (2005) PhyDE® 0995. Phylogenetic Data Editor. Available at: http://www.phyde.de
Nadot SN, Bajon R, Lejeune B (1994) The chloroplast gene rps4 as a tool for the study of Poaceae phylogeny. Plant Syst Evol 191:27–38
Nayar BK, Kaur S (1971) Gametophytes of homosporous ferns. Bot Rev 37:295–396
Nitta J (2008) Exploring the utility of three plastid loci for biocoding the filmy ferns (Hymenophyllaceae) of Morea. Taxon 57:725–736
Nixon KC (1999) The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 15:407–411
Quandt D, Stech M (2003) Molecular systematics of bryophytes in context of land plant evolution. In: Sharma AK, Sharma A (eds) Plant genome: biodiversity and evolution, vol 1. Science Publ, Enfield, pp 267–295
Quandt D, Stech M (2004) Molecular evolution and phylogenetic utility of the chloroplast trnT-trnF region in bryophytes. Plant Biol 6:545–554
Quandt D, Müller K, Huttunen S (2003) Characterisation of the chloroplast DNA psbT-H region and the influence of dyad symmetrical elements on phylogenetic reconstructions. Plant Biol 5:400–410
Quandt D, Müller K, Stech M, Hilu KW, Frey W, Frahm JP, Borsch T (2004) Molecular evolution of the chloroplast trnL-F region in land plants. Monogr Syst Bot Mo Bot Gard 98:13–37
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rouhan G, Hanks JG, McClelland D, Moran RC (2007) Preliminary phylogenetic analysis of the fern genus Lomariopsis (Lomariopsidaceae). Brittonia 59:115–128
Rumsey FJ, Vogel JC, Russell SJ, Barrett JA, Gibby M (1999) Population structure and conservation biology of the endangered fern Trichomanes speciosum Willd (Hymenophyllaceae) at its northern distributional limit. Biol J Linn Soc 66:333–344
Sato T (1982) Phenology and wintering capacity of sporophytes and gametophytes of ferns native to northern Japan. Oecologia 55:53–61
Schneider H, Schuettpelz E (2006) Identifying fern gametophytes using DNA sequences. Mol Ecol Notes 6:989–991
Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallón Lupia R (2004) Ferns diversified in the shadow of angiosperms. Nature 428:553–557
Skog JE, Mickel JT, Moran RC, Volovsek M, Zimmer EA (2004) Molecular studies of representative species in the fern genus Elaphoglossum (Dryopteridaceae) based on cpDNA sequences rbcL, trnL-F, and rps4-trnS. Int J Plant Sci 165:1063–1075
Smith AR, Cranfill RB (2002) Infrafamilial relationships of the thelypteroid ferns (Thelypteridaceae). Am Fern J 92:131–149
Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B (1997) Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Syst Evol 204:109–123
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods) Version 40b10. Sinauer, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2006) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14
Tsutsumi C, Kato M (2005) Molecular phylogenetic study on Davalliaceae. Fern Gaz 17:147–162
Tsutsumi C, Kato M (2006) Evolution of epiphytes in Davalliaceae and related ferns. Bot J Linn Soc 151:495–510
Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, Sugiura M (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci USA 91:9794–9798
Wang C-N, Moller M, Cronk QCB (2004) Phylogenetic position of Titanotrichum oldhamii (Gesneriaceae) inferred from four different gene regions. Syst Bot 29:407–418
Watkins JE Jr, Mack MC, Sinclair TR, Mulkey SS (2007) Ecological and evolutionary consequences of desiccation tolerance in tropical fern gametophytes. New Phytol 176:708–717
Wolf PG, Rowe CA, Sinclair RB, Hasebe M (2003) Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA Res 10:59–65
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgment
The authors thank Li-Yaung Kuo for laboratory help, Tien-Chuan Hsu for collecting Taiwanese Lomariopsis spectabilis, and Dr. Wen-Liang Chiou (Taiwan Forestry Research Institute) for valuable comments.
Author information
Authors and Affiliations
Corresponding authors
Appendix
Appendix
Voucher information and GenBank accession numbersa for rbcL, accD, trnL-F and rps4 (plus rps4-trnS IGS) sequences used in this study.
Part 1: rbcL and accD
Taxon—GenBank accessions: rbcL, accD; voucher (collection locality; herbarium) or reference.
Arachniodes aristata (G.Forst.) Tindale—AB232490, AB232418; Tsutsumi and Kato 2006. Araiostegia faberiana (C.Chr.) Ching—AB212688*; Tsutsumi and Kato 2005. Arthropteris backleri (Hook.) Mett.—AB212686*; Tsutsumi and Kato 2005. Athyrium niponicum (Mett.) Hance—AB232413, AB232441; Tsutsumi and Kato 2006. Bolbitis repanda (Blume) Schott—AB232399, AB232427; Tsutsumi and Kato 2006. Colysis wrightii (Hook.) Ching—AB232406, AB232434; Tsutsumi and Kato 2006. Crypsinus enervis (Cav.) Copel.—AB232407, AB232435; Tsutsumi and Kato 2006. Ctenitis eatonii (Baker) Ching—AB232391, AB232419; Tsutsumi and Kato 2006. Cyclopeltis crenata (Fée) C. Chr.—DQ054517+; Li and Lu 2006. Cyclopeltis crenata (Fée) C. Chr+, EU216746; F. W. Li 568 (private garden, originally from Thailand; TAIF). Davallia formosana Hayata—AB212704*; Tsutsumi and Kato 2005. Dryopteris erythrosora (D.C.Eaton) Kuntze—AB232392, AB232420; Tsutsumi and Kato 2006. Elaphoglossum callifolium (Blume) T.Moore—AB232400, AB232428; Tsutsumi and Kato 2006. Goniophlebium persicifolium (Desv.) Bedd.—AB232408, AB232436; Tsutsumi and Kato 2006. Grammitis reinwardtii Blume—AB232398, AB232426; Tsutsumi and Kato 2006. Gymnogrammitis dareiformis (Hook.) Ching ex Tardieu and C.Chr.—AB232409, AB232437; Tsutsumi and Kato 2006. Hypodematium crenatum (Forssk.) Kuhn ssp. fauriei (Kodama) K.Iwats.—AB232414, AB232442; Tsutsumi and Kato 2006. Leucostegia immersa (Wall. ex Hook.) C.Presl—AB232388, AB232416; Tsutsumi and Kato 2006. Leucostegia pallida (Mett.) Copel.—AB232389, AB232417; Tsutsumi and Kato 2006. Lomariopsis marginata (Schrad.) Kuhn.—AY818677, NA; Skog et al. 2004. Lomariopsis sp. (Süßwassertang)—EU216743, EU216744; F. W. Li 569 (unknown origin; TAIF). Lomariopsis sp. (Süßwassertang) —AM946394+; F042 (unknown origin; SING). Lomariopsis spectabilis (Kunze) Mett.—AB232401, AB232429; Tsutsumi and Kato 2006. Loxogramme avenia (Blume) C.Presl—AB232410, AB232438; Tsutsumi and Kato 2006. Matteuccia struthiopteris (L.) Tod.—AB232415, AB232443; Tsutsumi and Kato 2006. Microsorum zippelii (Blume) Ching—AB232411, AB232439; Tsutsumi and Kato 2006. Nephrolepis acuminata (Houtt.) Kuhn—AB232403, AB232431; Tsutsumi and Kato 2006. Nephrolepis cordifolia (L.) C.Presl—AB232404, AB232432; Tsutsumi and Kato 2006. Oleandra pistillaris (Sw.) C.Chr.—AB232405, AB232433; Tsutsumi and Kato 2006. Oleandra wallichii (Hook.) C.Presl—AB212687*; Tsutsumi and Kato 2005. Polybotrya caudata Kunze—AB232393, AB232421; Tsutsumi and Kato 2006. Polystichum fibrilloso-paleaceum (Kodama) Tagawa—AB232394, AB232422; Tsutsumi and Kato 2006. Pyrrosia rasamalae (Racib.) K·H.Shing—AB232412, AB232440; Tsutsumi and Kato 2006. Quercifilix zeylanica (Houtt.) Copel.—AB232395, AB232423; Tsutsumi and Kato 2006. Rumohra adiantiformis (G.Forst.) Ching—AB232396, AB232424; Tsutsumi and Kato 2006. Tectaria phaeocaulis (Rosenst.) C.Chr.—AB232397, AB232425; Tsutsumi and Kato 2006. Teratophyllum wilkesianum (Brack.) Holttum—AB232402, AB232430; Tsutsumi and Kato 2006.
Part 2: trnL-F
Taxon—GenBank accessions: trnL-F; voucher (collection locality; herbarium) or reference.
Lomariopsis: L. amydrophlebia (Sloss. ex Maxon) Holttum—DQ396555; Rouhan et al. 2007. L. boivinii Holttum—DQ396557; Rouhan et al. 2007. L. cordata (Bonap.) Alston—DQ396558; Rouhan et al. 2007. L. crassifolia Holttum—DQ396559; Rouhan et al. 2007. L. guineensis (Underw.) Alston—DQ396560; Rouhan et al. 2007. L. hederacea Alston—DQ396561; Rouhan et al. 2007. L. jamaicensis (Underw.) Holttum—DQ396562; Rouhan et al. 2007. L. japurensis (Mart.) J. Sm.—DQ396567; Rouhan et al. 2007. L. kunzeana (Underw.) Holttum—DQ396570; Rouhan et al. 2007. L. latipinna Stolze—DQ396571; Rouhan et al. 2007. L. lineata (C. Presl) Holttum—DQ396572; Rouhan et al., 2007. L. longicaudata (Bonap.) Holttum—DQ396573; Rouhan et al. 2007. L. madagascarica (Bonap.) Alston—DQ396576; Rouhan et al. 2007. L. mannii (Underw.) Alston—DQ396577; Rouhan et al. 2007. L. marginata (Schrad.) Kuhn—DQ396579; Rouhan et al. 2007. L. maxonii (Underw.) Holttum—DQ396580; Rouhan et al. 2007. L. muriculata Holttum—DQ396582; Rouhan et al. 2007. L. nigropaleata Holttum—DQ396584; Rouhan et al. 2007. L. palustris (Hook.) Mett. ex Kuhn—DQ396585; Rouhan et al. 2007. L. pervillei (Mett.) Kuhn—DQ396587; Rouhan et al. 2007. L. pollicina Willem. ex Kuhn—DQ396588; Rouhan et al. 2007. L. prieuriana Fée—DQ396590; Rouhan et al. 2007. L. recurvata Fée—DQ396592; Rouhan et al., 2007. L. rossii Holttum—DQ396594; Rouhan et al. 2007. L. salicifolia (Kunze) Lellinger—DQ396595; Rouhan et al. 2007. L. sp.—DQ396602; Rouhan et al. 2007. L. sp.—DQ396601; Rouhan et al. 2007. L. sp.—DQ396603; Rouhan et al. 2007. L. sp. (Süßwassertang)—EU216745; F. W. Li 569 (unknown origin; TAIF). Lomariopsis sp. (Süßwassertang) —AM946393; F042 (unknown origin; SING). L. spectabilis—EU216748; F.W. Li 567 (Wulai, Taiwan; TAIF). L. vestita E. Fourn.—DQ396598; Rouhan et al. 2007. L. wrightii Mett. ex D. C. Eaton—DQ396600; Rouhan et al. 2007.
Cyclopeltis crenata (Fée) C. Chr.—EU216746; F. W. Li 568 (private garden, originally from Thailand; TAIF). Hypodematium crenatum (Forssk.) Kuhn—AF425122; Smith and Cranfill 2002.
Part 3: rps4 (plus rps4-trnS IGS)
Lomariopsis sp. (Süßwassertang) — AM947063; F042 (unknown origin; SING).
aAsterisk, The same accession as previously noted; cross, this sequence is available in a different voucher but the same taxon; NA, data are not available for this taxon.
Rights and permissions
About this article
Cite this article
Li, FW., Tan, B.C., Buchbender, V. et al. Identifying a mysterious aquatic fern gametophyte. Plant Syst Evol 281, 77–86 (2009). https://doi.org/10.1007/s00606-009-0188-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-009-0188-2