Abstract
The biology of the late twentieth century was marked by the discovery in 1993 of a new class of small non-coding ribonucleic acids (RNAs) which play major roles in regulating the translation and degradation of messenger RNAs. These small RNAs (18–25 nucleotides), called microRNAs (miRNAs), are implied in several biological processes such as differentiation, metabolic homeostasis, or cellular apoptosis and proliferation. The discovery in 2008 that the presence of miRNAs in body fluids could be correlated with cancer (prostate, breast, colon, lung, etc.) or other diseases (diabetes, heart diseases, etc.) has made them new key players as biomarkers. Therefore, miRNA detection is of considerable significance in both disease diagnosis and in the study of miRNA function. Until these days, more than 1200 miRNAs have been identified. However, traditional methods developed for conventional DNA does not apply satisfactorily for miRNA, in particular due to the low expression level of these miRNA in biofluids, and because they are very short strands. Electrochemical biosensors can provide this sensitivity and also offer the advantages of mass fabrication, low-cost, and potential decentralized analysis, which has wide application for microRNAs sensing, with many promising results already reported. The present review summarizes some newly developed electrochemical miRNA detection methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
MicroRNAs: a new class of biomarkers
It may be pertinent to quickly define what is a microRNA (miRNA), compared to other RNAs. miRNAs are a class of tiny single-stranded RNAs of 18–25 nucleotides long that were discovered 27 years ago by Ambros and colleagues [1,2,3,4]. miRNAs play an important role as regulators of gene expression and therefore have important functions in various biological mechanisms. They do not code protein synthesis, in contrast to messenger RNAs (mRNAs), with which they must not be confused. Indeed, mRNAs are produced during transcription of DNA and expelled from the nucleus to the cytoplasm as a single strand, where it is read by tRNAs (transcription RNAs), which translate it into proteins with the help of the ribosomal RNA (rRNA). In the last years, miRNA have taken a new dimension with the discovery that they can be present in body fluids in correlation with diseases, including cancer [5,6,7,8]. This has opened up the possibility of using them as new non-invasive biomarkers [9, 10]. The ability to dispose of technology able to profile circulating miRNAs give a real hope for cancer diagnosis, classification, prognosis, prediction of therapeutic efficacy, surveillance following surgery, and forecast of cancer recurrence [5, 11, 12]. Even if ways to detect RNA are essentially similar to those used for DNA, significant differences between the two nucleic acids may guide the detection strategies: DNA is double-stranded while miRNA is single-stranded; RNA is more chemically unstable than DNA; DNA is present in cells while it is more diverse for miRNA, which first localize in the cytoplasm (for mature miRNA) but are also present in the exosomes, i.e., are expelled from the cell for cell-to-cell communication. For this reason, miRNA can be found freely circulating in body fluids; miRNAs are much shorter strands compared to DNA. As the miRNA field continues to evolve (Fig. 1) [6, 9, 13,14,15], it is an essential step to develop efficient and reliable detection strategies to better understand the functions of miRNAs in diverse regulatory pathways, which eventually influence the development of miRNA-based therapies and new targets in drug discovery. First, the small size of miRNAs make the conventional polymerase chain reaction (PCR) technique inadequate because the primers used in most conventional PCR are similar in length to miRNAs, which means that very short primers would be required for assay design, which affects the PCR efficiency due to a very low melting temperature [15,16,17]. As for hybridization-based detection, it is difficult to label short probes for selective detection of miRNAs. In addition to that, the melting temperature of short probe-target duplex is too low to allow a sufficient stringency of the hybridization conditions and significantly increases the risk of false positive signal, so that new methods are necessary for improving the specificity of miRNA detection. Second, miRNA concentrations are extremely low (in the range of the fM and lower) [18] and the high sequence similarity among family members makes the specific detection even more difficult. Third, because miRNA concentrations can be extremely variable from one sample to another, the dynamic range of quantification must be large, over more than four decades [14]; thus, a wide dynamic range of detection is required for miRNA assay. Meanwhile, a single gene can be simultaneously regulated by multiple miRNAs, which requires methods to detect multiple miRNA in a single sample. Another challenge is intracellular or in situ detection. A non-invasive monitoring approach is needed to provide detailed spatial expression patterns for specific miRNAs in vivo and facilitate the translation of miRNA detection methods into clinical practice. In Table 1 below are given all the acronyms and abbreviation used in this review.
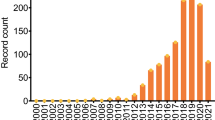
a Annual trend in the number of publications these last 10 years. Research made under keywords “microRNA detection” and “microRNA bioanalysis” on Scopus. b Timeline for microRNA discovery and detection. Reproduced from [13] with permission. Copyright 2008 from Wiley VCH
Current methods for microRNA detection
The usual methods for miRNA analysis are based on conventional DNA detection techniques [17] such as cloning [3, 19, 20], enzymatic ligation assays, Northern blot analysis [2, 21], oligonucleotide microarrays [22, 23], quantitative real-time polymerase chain reaction (qRT-PCR) [24,25,26], in situ hybridization [27, 28], and deep sequencing [29]. Most of these different techniques rely on an optical detection and are relatively laborious [30]. A fragment of DNA carrying a covalently bound digoxigenin, an antigen having a high affinity for its anti-digoxigenin antibody, is used as specific probe and binds the DNA strand to detect. Digoxigenin is itself tagged so to be detected using colorimetry, chemiluminescence, or fluorescence techniques. Therefore, there is an important bioanalytical challenge to develop alternative technologies. These emerging detection devices should have more advantages than traditional methods such as simplicity, reproducibility, sensitivity, mass fabrication, and multiplexing capabilities [17].
Northern blot
Northern blot is still widely used [2, 21]. It involves the use of electrophoresis to separate RNA samples by size, followed by transfer on a membrane onto which they are hybridized with a complementary labeled probe. To date, the northern blot remains the gold standard technique. However, Northern blot is not well-adapted to detect a large number of different miRNAs, requires too large quantities of material, and is not sufficiently sensitive, a drawback which was solutioned by the use of locked nucleic acid (LNA) probes, developed by Valoczi et al. [31]. The strategy proposed by Ramkissoona et al. [32], for which short RNA sequences are labeled with 3′-digoxigenin, the hybridization being revealed using anti-digoxigenim antibodies, can also be applied to identify target miRNAs and also participates to improve sensitivity. However, it is unlikely to be used as a routine method for diagnostic purposes [27].
Real-time reverse transcription polymer chain reaction
Due to its high level of sensitivity, accuracy, and practical ease, real-time reverse transcription polymer chain reaction (qRT-PCR) is a powerful technique, which has been applied not only for DNA detection but to miRNA detection as well. The first miRNA real-time PCR approach was reported by Schmittgen et al. [25]. More recently, Balcells et al. [33] have described a PCR method for quantification of microRNAs based on a single reverse transcription reaction for all microRNAs combined with real-time PCR with two microRNA-specific DNA primers (Fig. 2A). Today, qRT-PCR is capable of detecting miRNAs present in only picograms of total RNA. The method is also easily adapted to 384-well plates, or even microarrays so that it is possible to carry out high-throughput screening. The transduction used in qRT-PCR is also optical, generally using a fluorescent label. The typical example is the TaqMan probe. It uses the TaqDNA polymerase which cleaves a probe sequence carrying both a fluorescent label and a fluorescence quencher. Upon cleavage, the label/quencher couple is split, which reveals fluorescence.
A Scheme of the qRT-PCR method to detect miRNAs in purified RNA samples via a single reverse transcription reaction combined with real-time PCR. Reproduced with permission from [33], Copyright 2011 Springer Nature. B Scheme describing a typical microarray method for miRNA profiling via the following steps: (a) capture probe immobilization, (b) isolation of miRNAs from samples, (c) labeling of miRNAs with fluorescent dye; (d) hybridization and (f) reading from a scanned microarray. Reproduced with permission from [22], Copyright 2009 Springer Nature and from [34], the RNA Society, available under Creative Commons License 4.0. C Diagram illustrating an enzyme-linked assay to detect of miRNAs using magnetic beads platform for RNA capture probe immobilization, p19 protein (His-tagged p19) to recognize RNA duplexes and monoclonal anti-His-tag antibody conjugated with HRP for signal generation. Reprinted from [35], Copyright 2011, with permission from Elsevier. D SPRi measurement for miRNA detection based on combining poly(A) and AuNPs-amplified SPRi for miRNA detection: (i) hybridization of LNA probe with miRNA; (ii) growth of the poly(A) tails at the 3′-end of miRNAs target using poly(A) polymerase; and (iii) hybridization of poly(A) probes with T30 targets-coated Au nanoparticles for detection by SPRi. Reprinted with permission from [36]. Copyright 2006 American Chemical Society
Microarrays
The microarray technology was first developed in 1995, on the basis of the ability to perform multiple hybridizations in parallel with the oligo probes pre-spotted on a glass or a quartz plate. Microarrays most often use a fluorescent-labeled probe targeting the sequences bound on the array, and a reader (a scanner) able to read optically each microspot independently. This strategy was later adapted and modified for miRNA profiling (Fig. 2B). Most miRNA arrays to date use conventional capture probes, i.e., oligonucleotides. However, as cited above, other probes already demonstrated their efficiency, as peptide nucleic acids (PNA) and locked nucleic acids (LNAs), which allowed to improve sensitivity and, most of all, specificity. Above all, the main drawback of microarrays is that they are designed for identification but not for quantification, while it is the quantity of circulating miRNAs which is of medical interest.
Mass spectroscopy
Conventional analytical methods keep their advantages in terms of accuracy and robustness and can facilitate detection and quantification of miRNAs in high-throughput clinical studies. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is an example. It has been used mainly for proteomics research but also, in the past two decades, also in the analysis of DNA and RNA. To quantify microRNAs by this technique, they should be modified by organic dyes, such as small organic compound, often known as “matrix,” which readily undergoes desorption on contact with UV laser and assists the ionization of the analyte [37,38,39,40,41]. This technique is generally used as an assistance technique for others methods, such as microarrays, RT-PCR or Northern blot [31].
Surface plasmon resonance
Another powerful conventional analytical tool is surface plasmon resonance (SPR). Detection of miRNA using SPR has been reported by Sipova et al. [42]. This approach can be used for fast and direct miRNA detection; however, it is less sensitive than other techniques (limit of detection of only 100 pM). Nasheri et al. [35] have presented a SPR-based miRNA sensing method where RNA probes are immobilized on gold surfaces demonstrating the use of p19 protein in recognition of miRNA-bound probes (Fig. 2C). In this work, the 3′-biotinylated RNA probe against miR-122 is attached to the surface of streptavidin-coated magnetic beads then small RNAs extracted from hepatoma cells harboring miR-122 are incubated with the beads. His-tagged p19 is added to allow capping onto the duplexed probe on the beads then a monoclonal anti-His-tag HRP-conjugated antibody is added to bind to the His-tagged p19. Using a suitable HRP substrate, miRNAs are detected by bioluminescence on multiwell plates. This allows detection of miRNAs in the nanomolar range. To increase the sensitivity down to 0.1 nM, a bead-based enzyme immunoassay was performed, which allows a linear dynamic range from 1 pmol to 1 fmol in a volume of 10 μL. In addition, Corn’s laboratory [36, 43] has developed a surface plasmon resonance imaging (SPRi) method for microRNA detection. They described a miRNA profiling technique that employs a combination of surface poly(A) enzyme chemistry and nanoparticle-amplified SPRi measurements on a microarray platform. miRNAs are first hybridized onto LNA microarrays, then poly(A) sequences are polymerized at the end of the miRNAs so to extend their length and to be more easily detected by SPR (Fig. 2D). This approach allows detection of multiple miRNAs with a limit of detection of 10 fM.
Surface-enhanced Raman spectroscopy
Surface-enhanced Raman scattering (SERS) has been used for rapid, accurate identification of miRNAs [44, 45] but is not sufficiently sensitive for medical applications [45] and, above all, the required experimental setups limit its use to research laboratories.
Electrochemical approaches for microRNA detection
Electrochemical detection methods are known to be simpler and to require mostly inexpensive simple electronics, which are among their key advantages. This allows rapid measurements, in miniaturized easy-to-use portable systems. Electrochemical methods even allow time-resolved measurements, in contrast to most optical techniques. Such real-time quantification is not only advantageous for applicative purposes, but it also allows to access kinetics information. Compared to their optical counterparts, they are not sensitive to pollution light, and can be used on turbid samples. However, the problematic of direct electrochemical methods for nucleic acid detection is to find a way to generate an electrical signal whereas there is no charge transfer reaction during hybridization [46].
Indirect detection approach
Most of electrochemical miRNAs sensors require miRNA targets or DNA probes labeling with redox active and/or electrocatalytic molecules [47], electrocatalytic nanoparticle tags like osmium oxide [48], ruthenium oxide [49], or redox enzymes [50]. For example, DNA capture probes have been labeled by electroactive redox molecules such as methylene blue (MB) [51]. The miRNA sensor was developed based on the self-assembly of a triple-stem DNA structure conjugated with MB and immobilized on the surface of a gold microelectrode (50 μm diameter) functionalized with terminal thiol (-SH) groups (Fig. 3A). This approach allows to sensitively detect miRNA-122 with a LoD of 0.1 fM. To further enhance the sensitivity of electrochemical-based miRNAs sensors, nanostructures were introduced to increase the active surface area and the amount of immobilized electroactive substances [52]. For example, catalytic Fe3O4/CeO2@Au magnetic nanoparticles and hairpin assemblies have been used to develop an electrochemical microRNA-21 biosensor. The Fe3O4/CeO2@Au NPs not only allowed to adsorb a large amount of electroactive substances–methylene blue (MB)–but also acted as nanocatalyst to directly catalyze the reduction of MB for amplifying the electrochemical signal (Fig. 3B). Gao and Yu [47] have modified miRNA by a redox active and electrocatalytic complex, Ru(PD)2Cl2 (PD = 1,10-phenanthroline-5,6-dione), acting as a catalyst towards the oxidation of hydrazine. This catalytic amplification strategy allows a LoD as low as 0.20 pM and a dynamic range between 0.50 and 400 pM. The major drawback of this technique is the fact that the labeling process is only effective on G and A bases so that the labeling intensity depends on their occurrence in the sequence, which makes quantitative analysis of miRNAs difficult. Other indirect electrochemical methods need adding in solution a redox reporter such as Fe(CN)64−/Fe(CN)63− [45] or Fe-Ru redox pair [58, 59]. A highly sensitive and selective label-free impedimetric miRNA biosensor was developed by Ren et al. [53] (Fig. 3C). Briefly, thiolated DNA capture probes (CPs) are immobilized onto a gold electrode through self-assembly then hybridized with the target miRNA and a conventional isothermal amplification cycle is performed. Changes in electrochemical impedance spectra between a control and the cleaved biosensor allows label-free (but using a reagent in solution) detection of miRNA down to the fM range. Another approach for the development of indirect electrochemical miRNA biosensors is based on interaction of redox indicators with DNA/miRNA, RNA/miRNA, or PNA/miRNAs hybrids upon hybridization. For example, Tian et al. [54] reported an electrochemical biosensor for microRNA-21 detection based on an accumulation of toluidine blue (TB) onto RNA/RNA hybrids (Fig. 3D). Firstly, polypyrrole-coated gold nanoparticles (AuNPs) superlattice was used as a support material for increasing the active surface area and the surface concentration of single-stranded RNA(ss-RNA) probes. miRNA targets were hybridized, and toluidine blue added. Cyclic voltammetry and differential pulse voltammetry were used to quantify TB and deduce the miRNA concentration within the range 100 aM to 1 nM, with a LoD of 78 aM. Jolly et al. used positively charged AuNPs as reporters for detection of PNA/miRNA hybrids (Fig. 3E) [55]. In this approach, non-Faradaic electrochemical impedance spectroscopy (EIS) was used to monitor the changes in capacitance upon hybridization, without the need for any redox reporter. The accuracy of the method was compared to a Faradaic approach where thiolated ferrocene was introduced to bind AuNPs, and SWV used to quantify the miRNA concentration. Both approaches allowed a LoD below 1 fM and a dynamic range from 1 fM to 100 nM. The use of conducting polymer nanowires was proposed by Gao’s group in a label-free approach for RT-PCR-free miRNA detection [60], based on electrostatic interactions. For that, PNA (hence neutral) capture probes were used in-between interdigitated microelectrodes. After hybridization with charged miRNA, polyaniline nanowires were grown along the charged duplex, which resulted in a decrease of resistance correlated with the amount of hybridized miRNA (Fig. 3F). Using EIS with Fe(CN)64−/Fe(CN)63− as redox probe, the authors demonstrated a LoD of 5 fM and a dynamic range between 10 fM and 20 pM [56]. Indirect detection mode can be developed using an insulating polymer instead of conducting nanowires [49, 57], using the same strategy which consisted in using neutral probes, here morpholino capture probes. Upon hybridization, the neutral surface of the biosensor becomes anionic due to accumulation of miRNA strands. The deposition of an insulating polymer, poly(3,3′-dimethoxybenzidine) (PDB) was then carried out. The insulating polymer generates a decrease in conductivity proportional to the quantity of miRNA, measured using EIS with Ru(NH3)62+/3+ as redox probe. miRNA were quantified with a LoD of 2.0 fM (Fig. 3G) and a dynamic range between 5 fM and 2 pM [57]. The developed electrochemical miRNAs biosensors have been summarized in Table 2.
A Labeled approach for development of an electrochemical miRNA biosensor on gold microelectrode using a triple-stem DNA capture probe-conjugated with methylene blue (MB) as redox probe. Reprinted with permission from [51]. Copyright 2015 American Chemical Society. B Electrochemical miR-21 biosensor based on Fe3O4/CeO2@AuNPs nanocomposite as a nanocatalyst for electrochemical signal application. Reproduced from [52] with permission. Copyright 2018 from Elsevier. C EIS let-7b miRNA biosensor based on using a duplex-specific nuclease for cleaving off any miRNA/DNA duplex and Fe(CN)64−/Fe(CN)63− as a redox probe for EIS measurements. Adapted with permission from [53]. Copyright 2013 American Chemical Society. D Combination of AuNPs superlattice and toluidine blue for electrochemical miRNA biosensor signal amplification. Reproduced from [54] with permission. Copyright 2018 from Elsevier. e An EIS miR-145 biosensor using PNA as capture probes; AuNPs and thiolated ferrocene as the electrochemical tags. Reproduced from [55], Creative Commons CC BY license. F Conductance electrochemical let-7b miRNA biosensor using PNA as capture probe, where the formed miRNA/PNA duplexes (with anionic charge on the miRNA target) interacted with cationic aniline, which leads to deposition of polyaniline (PAn) nanowires onto the miRNA/PNA duplexes. Reprinted with permission from [56]. Copyright 2007 American Chemical Society. G EIS let-7c miRNA biosensor based on an amine-terminated morpholino capture probes (MCPs) and 3,3′-dimethoxybenzidine (DB) monomer to make an isolating poly(3,3′-dimethoxybenzidine) layer by HRP-catalyzed polymerization of DB in the presence of H2O2. EIS was measured using Ru(NH3)6 2+/3+ as redox probe. Reprinted with permission from [57]. Copyright 2013 American Chemical Society
Direct detection approach
Lusi and coworkers [70] were the first in 2009 to realize a direct and label-free approach to miRNA detection. For that, they adapted a label-free detection method already described for DNA detection and based on guanine oxidation. After hybridization of the miRNA target with an inosine-substituted capture probe, the electrooxidation of guanine on the electrode surface generated a current measured by differential pulse voltammetry (Fig. 4a). The detection limit was 5 nM. To improve the sensitivity of this approach, the electrode has been modified by ionic liquids or graphene [74, 75]. Conducting polymers carrying quinone derivatives are recognized in the literature for their remarkable properties [76,77,78], namely good biocompatibility, simple synthesis, easy bio-functionalization, and easy deposition over various electrode substrates using electrochemical methods. These properties have been advantageously used by our group to probe biomolecular interactions in several studies [71, 79,80,81,82,83]. For example, Tran et al. [71] have reported a simple electrochemical method for reagentless and label-free detection of miRNA, using a bifunctional conjugated copolymer poly(5-hydroxy-1,4-naphthoquinone-co-5-hydroxy-2-carboxyethyl-1,4-naphthoquinone) (poly(JUG-co-JUGA)). This conducting polymer is able to probe neighboring biomolecular interactions due to its sensitivity to any changes in its cation-exchange process. Typically, upon hybridization, it has been demonstrated that some room is liberated at the solution/polymer interface, which accelerates the diffusion processes, itself transduced into a current increase, measure by EIS, conventional CV, or square wave voltammetry (SWV) in the cathodic domain of potential where quinones are electroactive, avoiding most redox interferences. The main working concept of this developed miRNA sensor is based on the change in conformation of the DNA probes upon hybridization with the target miRNA. For that, the surface density of ODN probes has been finely tuned (ca. 10±5 pM cm−2) to guaranty that probes are closely packed together. Because these single-stranded probes behave as random coils, they generate a strong steric hindrance on the electrode surface which decreases the apparent diffusion coefficient of counter-ions going through the polymer/electrolyte interface, therefore decreases the faradic current density measured by SWV. Upon hybridization, the double strand becomes straight, which lowers its steric hindrance, allows better ion diffusion, and leads to an improved quinone electroactivity (Fig. 4b). Majd et al. [72] have reported a label-free electrochemical biosensor for miRNA-155 by using a field-effect transistor (FET) via drop-casting of a molybdenum disulfide (MoS2) flacks suspension onto the FET surface. The DNA probes were immobilized on the surface of the MoS2 FET device for miRNs-155 target hybridization (Fig. 4c). The LoD was very low, 0.03 fM, with a dynamic range between 0.1 fM and 10 nM. Zhang et al. [73] have reported a completely different approach, using a field-effect transistor based on PNA-functionalized silicon nanowires for direct and ultrasensitive detection of miRNA. In this system, the resistivity of the nanowires was monitored before and after hybridization between PNA probes and complementary miRNA targets. The detection limit was 1 fM but suffered for a lack of reproducibility (Fig. 4d). Table 3 summarizes several typical label-free and reagentless developed electrochemical miRNAs biosensors, for which advantages and limitations are indicated.
a A developed direct and label-free electrochemical miRNA biosensor by using inosine-modified DNA capture probe (guanine-free DNA probe). The electrochemical signal was generated by oxidation of guanine contained in the RNA/DNA hybrid, by DPV. Reprinted with permission from [70]. Copyright 2009 American Chemical Society. b A label-free and reagentless electrochemical miR-141 biosensor based on interaction between DNA capture probe and quinone groups on poly(JUG-co-JUGA) platform. Reprinted by permission from [71], Copyright 2014 Springer Nature. c Working scheme of molybdenum disulfide (MoS2)-based FET miR-155 biosensor. Reproduced from [72] with permission. Copyright 2018 from Elsevier. d A FET miRNA biosensor based on PNA capture probe-functionalized silicon nanowires for miRNA detection. Reproduced from [73] with permission. Copyright 2009 from Elsevier
New trends for the development of electrochemical microRNA biosensors
Nanomaterials application
To improve sensitivity and selectivity of electrochemical miRNAs biosensors, nanomaterials have been used, following four main approaches: (i) nanostructured electrodes for amplifying electrochemical signals and therefore improving sensitivity [15, 37, 72, 73, 83]; (ii) nanoparticles for preparing signal tags or signal reporters, such as AuNPs [32, 61, 80] or quantum dots [32]); (iii) nanomaterials as catalytic tags conjugated with reporters such as AgNPs [80]; and (iv) highly conductive nanostructured platforms such as carbon nanotubes [32, 44], graphene [61, 69, 80], graphene oxide [66, 79], or metal oxide nanoparticles (e.g. Fe3O4 [59]). Herein, we summarize these approaches.
Noble metal-nanostructured microelectrodes
Kelley’s group [59] has succeeded in developing ultrasensitive microRNA sensors with sub-aM sensitivity. In their work, a palladium nanostructure was electrochemically deposited on gold microelectrode then thiolated PNA probes were immobilized for miRNA hybridization. Electrochemical measurements were carried out by cyclic voltammetry (CV) and differential pulse voltammetry (DPV) techniques in an aqueous solution containing [Ru(NH3)6]3+, [Fe(CN)6]3−, sodium phosphate and NaCl (Fig. 5a) [59]. This approach presented a very high sensitivity. Currents were extremely low, however (in the nA range), which made measurements difficult. Similarly, this group has also developed gold nanorods as nanoelectrodes for miRNA detection with [Ru(NH3)6]2+ and [Fe(CN)6]3− as electrochemical redox markers (Fig. 5b) [58]. This strategy has been also extended to prepare a hierarchical flower-like Au nanostructure on ITO electrode (Fig. 5c) [61]. Metallic nanoparticles were also used to decorate the electrodes surface [45, 62, 66, 68, 69, 84, 90, 91]. For example, Wan et al. [86] reported an electrochemical miRNA sensor based on AuNPs deposited on the surface of a MoSe2-modified electrode for thiol-modified hairpin probe immobilization. When adding target miRNA, probe DNA hybridizes on the complementary sequence and unfolds the stem-and-loop structure. Hybridization with an assistance DNA at the terminus of capture DNA results in the formation of a supersandwich structure (Fig. 5d) [86]. Other examples of AuNPs platforms were published, for example on nitrogen-doped graphene sheets (AuNPs@N-G)-modified electrodes (Fig. 5e) [87] or gold nanoparticle-decorated MoS2 nanosheet (AuNPs@MoS2) [89]. Wu et al. [88] have developed a label-free amperometric biosensor for detection of microRNA-155 based on a conducting self-assembled multilayer of Nafion, thionine, and PdNPs. Nafion was firstly dropped on a bare glassy carbon electrode (Fig. 5f), then thionine was absorbed on Nafion. PdNPs, used to immobilize the target biomolecules, were immobilized with thionine as linker. The resulting biosensor presented high sensitivity, good stability, a LoD of 2 pM, and a broad dynamic range from 6 pM to 0.6 μM. Su et al. [89] reported a dual-mode electrochemical miRNA-21 biosensor based on a “sandwich” structure with AuNPs-decorated MoS2 nanosheets (AuNPs@MoS2), where DNA-modified AuNPs@MoS2 nanoprobes were immobilized on a first capture DNA. [Fe(CN)6]3−/4- and [Ru(NH3)6]3+ were used as redox indicators (Fig. 5g) [89]. However, even if sandwich-type architectures have been often used for their sensitivity, this approach is extremely tedious, which limits its application.
a, b Nanostructured microelectrodes for development of miRNA electrochemical biosensors: a Au nanowire electrode [58], and b Pd nanostructured on Au nanoelectrodes [59], respectively. In these works, the thiolated PNA was used as capture probes and [Ru(NH3)6]3+/[Fe(CN)6]3− as redox probes. Reprinted with permission from [58]. Copyright 2009 American Chemical Society, and adapted from [59] with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Copyright 2009. c A hierarchical flower-like AuNPs nanostructured electrode for electrochemical miR-21 detection. The cationic Ru(NH3)63+ ions as a redox probe were stoichiometrically bound with the anionic phosphates of DNA/miRNA strands, thereby resulting in enhanced SWV signals. Reproduced from [61] with permission from WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Copyright 2016. d A sandwich-type electrochemical miRNA biosensor based on AuNPs decorated on MoSe2-modified electrode for thiol-modified hairpin DNA capture probe immobilization and the hemin/G-quadruplexes functionalized the nitrogen-doped graphene/gold nanoparticles (NG-AuNPs as electrochemical tags. Reproduced from [86] with permission from the Royal Society of Chemistry, © 2018. e A working principle of an electrochemical miR-122 biosensor based on a multifunctional iron-based metal-organic frameworks assembled palladium nanoparticles (PdNPs@Fe-MOFs) as an electrochemical tags and AuNP-functionalized nitrogen-doped graphene sheets (AuNPs@N-G) as a detection platform. Reproduced from [87] with permission. Copyright 2018 from Elsevier. f A scheme of an electrochemical miR-155 biosensor using palladium nanoparticles (PdNPs)-modified electrodes for immobilization of RNA capture probes. Reproduced from [88] with permission from the Royal Society of Chemistry, © 2013. g A dual-mode electrochemical miRNA-21 biosensor based on a “sandwich” structure using AuNP-decorated MoS2 nanosheet (AuNPs@MoS2) for immobilization of the first DNA1 capture probe and DNA2-modified AuNPs@MoS2 nanoprobe as the second probe. [Fe(CN)6]3−/4- and [Ru(NH3)6]3+ were used as electrochemical indicators. Reproduced from [89] with permission. Copyright 2017 from Elsevier
Carbon-based nanomaterials
Carbon-based nanomaterials such as carbon nanotubes (CNTs), graphene or graphene oxide (Gr, GO), carbon nanofibers, and carbon quantum dots (QDs) offer attractive opportunities for developing novel sensors and refining the analytical performance of already existing platforms. The electronic properties of carbon-based nanomaterials (semi-conducting, metallic or superconducting) make them particularly well-indicated to improve electron transport, while their nanometric dimensions favor peak effects and improve electron transfer. Besides, their large surface-to-volume ratio allows to increase surface densities of various biomolecules on the electrodes surface. These properties make carbon-based nanomaterials excellent candidates in electrochemical miRNA biosensing [8, 10, 66, 82, 83, 92,93,94,95]. Based on the same strategy as developed in ref. [71], but using multiwalled carbon nanotubes (MWCNTs) included in a quinone-based electroactive polymer, Tran et al. demonstrated a porous nanostructured film with a well-defined electroactivity in neutral aqueous medium (Fig. 6a) [82], electroactivity which was enhanced upon hybridization. An application was made with miR-141 target (a prostate cancer biomarker), giving a very low LoD of 8 fM [71]. One may also cite the work from Liu et al., who combined the action of graphene sheets and dendrimers of a biopolymer (PAMAM, polyaminoamine) containing clusters of gold and silver, to achieve a better electron collection, therefore a better sensitivity. They reached a LoD of 0.8 fM [94].
a An illustration of a label-free and reagentless electrochemical miRNA-141 sensor based on a nanostructured poly(JUG-co-JUGA)/CNTs composite-modified electrode. Inset: SEM images of (i) poly(JUG-co-JUGA)/GCE; (ii) o-MWCNTs/GCE, and (iii) poly(JUG-co-JUGA) o-MWCNTs/GCE, respectively. Reproduced from [82] with permission. Copyright 2013 from Elsevier. b An illustration of miRNA assay using electrocatalytic activity of the OsO2 nanoparticles. Reprinted with permission from [48]. Copyright 2006 American Chemical Society. c Following step for development of an electrochemical miRNA biosensor using oligonucleotide encapsulated Ag-NCs and the specific DPV response curves of the developed biosensor. Reprinted with permission from [96]. Copyright 2012 American Chemical Society. d Scheme illustrating a three amplification steps electrochemical miRNA biosensor based on dendritic gold nanostructure decorated on graphene modified electrodes. Reproduced from [97] with permission. Copyright 2012 from Elsevier
Nanoparticles as electrochemical tags
This approach uses NP-conjugated oligonucleotide probe as reporter [48, 62, 67, 86, 89, 96,97,98,99,100,101,102]. For example, Gao et al. [48] immobilized isoniazid-capped osmium oxide (OsO2) NPs on hybridized miRNA through a condensation reaction. These NPs were able to catalyze the oxidation of hydrazine at −0.10 V, a potential at which few interfering reactions can occur (Fig. 6b). The LoD was 80 fM, with a dynamic range up to 200 pM. Dong [96] designed and developed an electrochemical sensor for microRNA detection using a functional oligonucleotide probe onto which silver nanoclusters (Ag-NCs) were immobilized, acting as catalyst for H2O2 (Fig. 6c). Their sensor employs molecular beacon (MB) probes: after hybridization between the target and the functional probe, the template oligonucleotides bearing Ag-NCs are brought to the electrode surface and catalyze H2O2 electroreduction. The LoD was 67 fM, with a linear range over five decades. To amplify the electrochemical signal, nanostructured electrochemical tags have been developed, as illustrated by ref. [97] (Fig. 6d). In this example, LNA and biotin-functionalized DNA were immobilized onto AuNPs then on dendritic gold nanostructures and graphene nanosheets. A catalytic cycle with HRP and the quinone/hydroquinone couple was used to monitor miRNA-21 hybridization, providing a LoD of 0.06 pM. It was applied to practical medical cases such as detection of hepatocarcinoma cells [97].
Recognition based on antibodies to RNA/DNA duplexes
In the last decade, a new approach for specific detection of miRNA has been developed based on the use of specific antibodies able to recognize RNA/DNA and RNA/RNA duplexes. Monoclonal and polyclonal antibodies recognizing RNA/DNA and RNA/RNA duplexes have been developed and used in hybridization-based assays [103,104,105,106,107]. Anti-RNA/DNA antibodies used to recognize miRNAs were previously reported by S.H. Leppla’s group [108] based on fluorescence assays or H. Sipova et al. [42] and A.J. Qavi et al. [107] based on SPR detection. This immunological approach is very original and efficient. Indeed, it combines the selectivity of antibodies with the sensitivity of hybridization. Sipova et al. used SPR combined to such DNA/RNA antibodies. Thiolated capture oligonucleotides were used and hybridization was followed by incubation with the dedicated antibody. The large size of this antibody provided amplification of the SPR signal down to a LoD of 2 pM (the LoD was 100 pM without using the antibodies). Qavi et al. used a similar approach but coupled to a photonic microring resonator. The increase in mass due to the antibodies improved the LoD down to 10 pM. Tran et al. [83] have developed a three-detection mode electrochemical miRNAs sensor using antibodies as checkers. In this work, they have used non sequence-specific RNA/DNA antibodies (S9.6 antibody) to improve their microRNA detection. S9.6 antibodies are very original antibodies that recognize specifically DNA/RNA hybrids. The use of these antibodies present a considerable advantage: after the specific signal-on obtained after hybridization, binding of RNA/DNA antibodies on the electrode surface will, conversely, generate strong steric hindrance that is specific and gives a signal decrease (signal-off). At the end, when free diffusing RNA/DNA hybrids are added into solution, they compete with the bound hybrid and make the antibodies going off the surface (Fig. 7a). With this approach, the authors implemented a sequential three detection mode in a single experiment: signal-on, signal-off then signal-on again. This on-off-on procedure is very useful as a triple check to make the hybridization results definitely sure [83]. Following a different approach, but still using antibodies, Zouari et al. [109] described an original disposable device based on AuNPs-modified electrodes onto which DNA–miRNA-21 heteroduplex were immobilized and bound by antibodies, themselves further bound by Protein A (ProtA) coupled to HRP. This catalytic amplification provided a LoD of 29 fM at a dynamic range of detection between 0.1 and 25 pM (Fig. 7b) [109]. Combining the S9.6 antibody with nanomaterials such as carbon nanotubes (CNTs) and reduced graphene oxide (rGO), Tran and his colleagues have reported an indirect electrochemical miRNA biosensor [92]. This type of electrochemical miRNA worked similarly to an ELISA-test for microRNA detection. For that, a composite of rGO and MWCNTs was drop-casted on the electrode’s surface to play the role of immobilization platform for DNA probes. If a sample containing the complementary miRNA is added, hybridization between DNA and miRNA targets was probed by the S9.6 antibody. The last step was an electrochemical ELISA-like amplification using a HRP-conjugated secondary antibody (Fig. 7c). This architecture, compared to classical optical detection, lowers the detection limit down to 10 fM [92]. To improve the sensitivity, “sandwich-type” assays were also proposed. Wang et al. [110] reported a photoelectrochemical (PEC) biosensor for the detection of microRNA-396a based on a MoS2/g-C3N4/black TiO2 heterojunction as the photoactive material and gold nanoparticles carrying Histostar antibodies (Histostar@AuNPs) for signal amplification (Fig. 7d). Briefly, electrodes were functionalized with MoS2 and black TiO2 materials, then gold nanoparticles and probe DNA. Hybridization with the target miRNA was identified by S9.6 antibodies. Amplification was provided by using secondary IgG antibodies conjugated to HRP. This approach provided a LoD of 0.13 fM and a dynamic range from 0.5 to 5 pM. A similar approach by Wang et al. consisted in using alkaline phosphatase-labeled IgG (ALP-IgG) (Fig. 7e), for similar results [111].
a A three-detection mode electrochemical miRNAs sensor using S9.6 antibody as a checker. Reprinted with permission from [83]. Copyright 2013 American Chemical Society. b An amperometric immunosensor for miR-21 detection using AuNPs-modified electrode and S9.6 anti-DNA.RNA hybrid antibodies conjugated with horseradish peroxidase (HRP). Reproduced from [109] with permission. Copyright 2018 American Chemical Society. c An ELISA-like assay for electrochemical immunosensor detection of miR-141 based on rGO/CNTs modified screen-printed gold electrodes (SPGEs). Reproduced from [92] with permission. Copyright 2014 from Elsevier. d A labeled approach for fabrication of electrochemical immunosensor to detect miR-396a using S9.6 antibodies for linking between miRNA/DNA duplexes with MoS2/g-C3N4/black TiO2 heterojunction with Histostar@AuNPs as a signal amplification. Reproduced from [110] with permission. Copyright 2019 from Elsevier. e An electrochemical immunosensor approach for miRNA-319a detection based on the use of S9.6 anti-DNA/RNA hybrid antibody conjugated with alkaline phosphatase (ALP)-labeled goat anti-mouse IgG as an enzymatic signal amplifier. Reproduced from [111] with permission. Copyright 2015 from Elsevier
Recognition based on proteins to RNA/RNA duplexes
Beside the S9.6 antibody which has been used for recognition of RNA/DNA hybrids as mentioned above, a novel approach consists in using protein 19 (p19) [91, 112, 113]. Protein p19 is a 19-kDa fusion protein which is a RNA silencing suppressor and binds with high affinity only to double stranded/duplex RNA (dsRNA) (i.e., it does not bind ssRNA, tRNA, rRNA, ssDNA, or dsDNA) in a size-specific and sequence-independent manner [112, 113]. The p19 viral suppressor of the RNA silencing protein has useful applications in biotechnology due to its high affinity for binding to small RNAs such as small interfering RNAs (siRNAs) and miRNAs. Also, its applications for the study and modulation of microRNAs are actively expanding [114, 115]. The p19 binding affinity is determined by the miRNA duplex region length. For example, it has the highest affinity for 21–26 nucleotides (nt) dsRNA, which progressively becomes lower for 19-nt and smaller. Hybridization of a miRNA-specific probe to a single-stranded target miRNA creates dsRNA that tightly binds the p19 fusion protein [112]. Labib et al. [96] have developed an indirect detection mode based on this principle (Fig. 8a), offering a LoD of 5 aM and a dynamic range between 10 aM and 1 μM, without PCR amplification. Killic et al. [113] also used p19 (Fig. 8b), with a LoD of 160 nM. Ramnani et al. [112] proposed a more original approach, combining p19 with the use of a carbon nanotube-based field-effect transistor (Fig. 8c), which gave a wide dynamic range up to 10−14 M and a LoD of 1 aM. To improve sensitivity, nanomaterials have been used. Tu et al. [116] reported a photoelectrochemical (PEC) biosensor using a AuNP-decorated ZnSe-COOH nanoflakes platform as the first signal amplification and using p19 protein as a second signal amplification (Fig. 8d). As reported, this approach can detect miRNA-122a with a dynamic range from 350 fM to 5 nM and a LoD of 150 fM. Authors also reported that this miRNA sensor was successfully applied to analyze the level of miRNA-122a in HeLa cell, which is promising for early diagnosis of tumor. A new structure for miRNA detection based on using p19 has been reported by Li et al. [117]. In this work, p19 protein-functionalized magnetic beads were used. Then, acridone derivative 5, 7-dinitro-2-sulfo-acridone (DSA) has been used as the electrochemical reporter (Fig. 8e). This strategy allows detection of miRNAs at concentrations as low of 6 aM [117]. Table 4 summarizes several updated applications of S9.6 antibodies or p19 for development of miRNA electrochemical biosensors, with advantages and limitations.
a Three modes electrochemical miRNA biosensor based on thiol-modified RNA as capture probe immobilized on the AuNP-modified SPEs and p19 protein as a reporter to bind with miR-21/pRNA-21 duplexes on the electrode surface and with free miR-200/pRNA-200 duplexes in solution. Reprinted from [91] with permission. Copyright 2013 American Chemical Society. b Electrochemical biosensor for miRNA detection based on using of p19 protein: (A) non-binding between p19 protein with RNA (miR-21 target); (B) non-binding between p19 protein with DNA/DNA duplexes which were produced from DNA miR-21 and its DNA capture probe (anti-miR-21); and (C) p19 protein binds to hybrids of RNA/RNA duplexes, which were made from a hybridization of miR-21 (RNA) and its RNA capture probe (anti-miR-21). Reproduced from [113] with permission. Copyright 2013 from Elsevier. c p19-functionalized CNTs-FET for miRNA detection and I–V curves corresponding to the fabrications steps. Reproduced from [112] with permission. Copyright 2013 American Chemical Society. d A photoelectrochemical miR-122a biosensor based on dual signal amplification using AuNP-enhanced ZnSe nanoflakes and p19 protein. Reproduced from [116] with permission. Copyright 2016 American Chemical Society. e An electrochemical miRNA biosensor based on the p19 protein immobilized on magnetite beads (MB) for capturing of miRNA/RNA duplex, using of 5,7-dinitro-2-sulfo-acridone (DSA)—an acridone derivative—as indicator and a DNA concatemer for signal amplification. Reproduced from [117] with permission. Copyright 2015 from Elsevier
Conclusions
As shown in this review, miRNAs are crucial in many biological processes. In particular, they play a significant role in controlling the cell cycle and the deregulation of their expression is often a cause of cancer. Therefore, measuring miRNAs level in biological fluids can improve cancer diagnosis at early stages. Among the various techniques available to make miRNA biosensors, electrochemical strategies could be a good choice. Indeed, their detection and transduction principle offer the advantage of requiring inexpensive and simple electronics having low power requirements and of being easily miniaturized, which makes electrochemical transduction methods well-adapted for implantable and portable hand-held devices. In addition, electrochemical processes are independent from matrix turbidity. The problem with electrochemical DNA detection is that using the direct electroactivity of the nucleobases is not an option: slow, irreversible and poorly sensitive, it is also dependent on the miRNA sequence and makes quantification uneasy. The challenge is therefore to find a way to achieve an electrical signal whereas no charge transfer reaction is involved during hybridization. The simplest way to transduce miRNA hybridization on a DNA or ODN probe is, as for most DNA sensors, to use an indirect method in which DNA strands are labeled with electroactive tags. The latter could be simple redox-active molecules (ferrocene, methylene blue, or any other molecule or complex presenting a reversible redox process and of low electrooxidation potential), covalently coupled to the oligonucleotide probe strand in a hairpin configuration, or able to form non-covalent interactions with the double strand formed between the ODN probe and the miRNA target. This strategy is among the simplest ones but does not bring any amplification, which is absolutely necessary to deal with the extremely low expression level of miRNAs, however. This amplification can be achieved by using electrocatalytic tags such as inorganic complexes or redox enzymes, as reported for many DNA sensors. It is here that nanomaterials are promising options for enhancing sensitivity. First, they bring a much higher surface area to the sensing electrodes, which increase the probe density and the number of hybridization events. Second, when dispersed in a less conducting matrix (organic semiconductors, graphite, etc.), they significantly increase the electron transfer rate, so the measured current. Their intrinsic catalytic activity makes them a good candidate to replace enzymes in catalytic amplification strategies, which increases the robustness of the device and reduces the risk of false negatives or false positives due to the presence of chemical species which interfere with enzymes. The recent immunosensing approach developed for RNA/DNA hybrids detection is extremely promising. Indeed, it extends strategies routinely developed for conventional immunosensors, which afford the selectivity and sensitivity needed. Other attractive approaches, not cited in this review, are those relying on biomolecular techniques. For example, Chen et al. [119] gave an example of amplification through T7-exonuclease activity which allowed an extremely low LoD in the fM range. Rolling circle amplification (RCA) is another biomolecular technique, developed for DNA detection, which can be successfully translated to RNA detection [120]. More original is the approach proposed by Bai et al., who used a RNA:DNA-specific nuclease for signal amplification [121]. DNA origami are also interesting as nanostructured substrate onto which DNA probes can be addressed onto precise locations, which avoids random distribution and improves the DNA:RNA hybrids density [68]. However, one of the most recent biotechnology applicable to miRNA detection is probably the CRISPR/Cas approach, exemplified by Wang et al. in 2020 [122], which promised great developments. Concerning electrochemistry versus other detection techniques, many different approaches have already been reported, derived from DNA sensors or immunosensors. The actual trend is to move to new detection platforms not necessarily amperometric such as transistors, in particular new forms of transistors such as electrolyte-gated [123, 124], floating-gate, lateral-gate [125], nanowire ,or graphene transistors [126]. Significant advances have been also made on logic computation based on DNA (or even RNA) [127]. Such microtechnologies improvements can help bring incredibly low LoD. Ion channels are another track to follow. For example, Zhao et al. [128] have immobilized DNA probes onto ion channel membranes and detected DNA:RNA hybrids in the attomolar range through current-voltage curves. More generally, combination of the most recent microtechnologies with the most recent molecular biotechnologies is certainly the core of future developments. One of the best examples is the CRISPR/Cas system (clustered regular interspaced short palindromic repeats with its CRISPR-associated protein), which was recently introduced as a powerful biotechnology tool of RNA recognition [129]. To conclude, the reader can notice that all the above reviewed works are related to academic researches which did not find concrete applications. There are no patents specifically dedicated to electrochemical detection of miRNA, except the one from Pingarron et al., 2017 [130], which proposes a methodology for detecting and quantifying miRNA by capture of DNA/RNA duplex on suspended antibody-modified magnetic particles and electrochemical detection assisted by a HRP previously coupled to the capture DNA strand. The LoD is 2.4 pM. To our knowledge, there is no other dedicated patent, and no commercial applications yet on the market
References
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854. https://doi.org/10.1016/0092-8674(93)90529-Y
Quintana ML, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–885. https://doi.org/10.1126/science.1064921
Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294(5543):862–864. https://doi.org/10.1126/science.1065329
Ye J, Xu M, Tian X, Cai S, Zeng S (2019) Research advances in the detection of miRNA. J Pharm Anal 9(4):217–226. https://doi.org/10.1016/j.jpha.2019.05.004
de Planell-Saguer M, Rodicio MC (2011) Analytical aspects of microRNA in diagnostics: a review. Anal Chim Acta 699(2):134–152. https://doi.org/10.1016/j.aca.2011.05.025
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:154–158. https://doi.org/10.1093/nar/gkm952
Lautner G, Gyurcsányi RE (2014) Electrochemical detection of miRNAs. Electroanalysis 26(6 Special Issue: Electroanalysis-Based Clinical Diagnostics):1224–1235. https://doi.org/10.1002/elan.201400055
Masud MK, Umer M, Hossain SA, Yamauchi Y, Nguyen N-T, Shiddiky MJA (2019) Nanoarchitecture frameworks for electrochemical miRNA detection. Trends Biochem Sci 44(5):433–452. https://doi.org/10.1016/j.tibs.2018.11.012
Brase JC, Wuttig D, Kuner R, Sültmann H (2010) Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer 9:306. https://doi.org/10.1186/1476-4598-9-306
Jamali AA, Pourhassan-Moghaddam M, Dolatabadi JEN, Omidi Y (2014) Nanomaterials on the road to microRNA detection with optical and electrochemical nanobiosensors. TrAC Trends Anal Chem 55:24–42. https://doi.org/10.1016/j.trac.2013.10.008
Wittmann J, Jäck HM (2010) Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta 1806:200–207. https://doi.org/10.1016/j.bbcan.2010.07.002
Mujica ML, Gallay PA, Perrachione F, Montemerlo AE, Tamborelli LA, Vaschetti VM, Reartes DF, Bollo S, Rodríguez MC, Dalmasso PR, Rubianes MD, Rivas GA (2020) New trends in the development of electrochemical biosensors for the quantification of microRNAs. J Pharm Biomed Anal 189:113478. https://doi.org/10.1016/j.jpba.2020.113478
Wark AW, Lee HJ, Corn RM (2008) Multiplexed detection methods for profiling microRNA expression in biological samples. Angew Chem Int Ed 47:644–652. https://doi.org/10.1002/anie.200702450
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X (2013) MicroRNA: function, detection, and bioanalysis. Chem Rev 113(8):6207–6233. https://doi.org/10.1021/cr300362f
Cissell KA, Shrestha S, Deo SK (2007) MicroRNA detection: challenges for the analytical chemist. Anal Chem 79(13):4754–4761. https://doi.org/10.1021/ac051726m
Qavi AJ, Kindt JT, Bailey RC (2010) Sizing up the future of microRNA analysis. Anal Bioanal Chem 398(6):2535–2549. https://doi.org/10.1007/s00216-010-4018-8
Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP (2003) The microRNAs of Caenorhabditis elegans. Genes Dev 17:991–1008. https://doi.org/10.1101/gad.1074403
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129(7):1401–1414. https://doi.org/10.1016/j.cell.2007.04.040
Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, van Zonneveld AJ, Mano H, Plasterk R, Cuppen E (2006) Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res 16(10):1289–1298. https://doi.org/10.1101/gr.5159906
Sempere LF, Freemantle S, Rowe IP, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5:R13. https://doi.org/10.1186/gb-2004-5-3-r13
Li W, Ruan K (2009) MicroRNA detection by microarray. Anal Bioanal Chem 394(4):1117–1124. https://doi.org/10.1007/s00216-008-2570-2
Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11:241–247. https://doi.org/10.1261/rna.7240905
Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM (2005) Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11(11):1737–1744. https://doi.org/10.1261/rna.2148705
Schmittgen TD, Jiang J, Liu Q, Yang L (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32(4):43e–443e. https://doi.org/10.1093/nar/gnh040
Jiang J, Lee EJ, Gusev Y, Schmittgen TD (2005) Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res 33(17):5394–5403. https://doi.org/10.1093/nar/gki863
Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH (2005) MicroRNA expression in zebrafish embryonic development. Science 309(5732):310–311. https://doi.org/10.1126/science.1114519
Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RHA (2006) In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods 3:27–29. https://doi.org/10.1038/nmeth843
Creighton CJ, Reid JG, Gunaratne PH (2009) Expression profiling of microRNAs by deep sequencing. Brief Bioinform 10(5):490–497. https://doi.org/10.1093/bib/bbp019
Hunt EA, Goulding AM, Deo SK (2009) Direct detection and quantification of microRNAs. Anal Biochem 387(1):1–12. https://doi.org/10.1016/j.ab.2009.01.011
Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32(22):e175. https://doi.org/10.1093/nar/gnh171
Ramkissoon SH, Mainwaring LA, Sloand EM, Young NS, Kajigaya S (2006) Nonisotopic detection of microRNA using digoxigenin labeled RNA probes. Mol Cell Probes 20(1):1–4. https://doi.org/10.1016/j.mcp.2005.07.004
Balcells I, Cirera S, Busk PK (2011) Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol 11:70. https://doi.org/10.1186/1472-6750-11-70
Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E (2005) An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA 11(9):1461–1470. https://doi.org/10.1261/rna.2610405
Nasheri N, Cheng J, Singaravelu R, Wu P, McDermott MT, Pezacki JP (2011) An enzyme-linked assay for the rapid quantification of microRNAs based on the viral suppressor of RNA silencing protein p19. Anal Biochem 412(2):165–172. https://doi.org/10.1016/j.ab.2011.01.030
Fang S, Lee HJ, Wark AW, Corn RM (2006) Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J Am Chem Soc 128(43):14044–14046. https://doi.org/10.1021/ja065223p
Jurinke C, Oeth P, Boom DVD (2004) MALDI-TOF mass spectrometry. Mol Biotechnol 26(2):147–163. https://doi.org/10.1385/MB:26:2:147
Joyner JC, Keuper KD, Cowan JA (2013) Analysis of RNA cleavage by MALDI-TOF mass spectrometry. Nucleic Acids Res 41(1):e3. https://doi.org/10.1093/nar/gks811
Goldman JM, Zhang LA, Manna A, Armitage BA, Ly DH, Schneider JW (2013) High affinity γPNA sandwich hybridization assay for rapid detection of short nucleic acid targets with single mismatch discrimination. Biomacromolecules 14(7):2253–2261. https://doi.org/10.1021/bm400388a
Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T (2009) Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23:433–438. https://doi.org/10.1101/gad.1761509
Chen QR, Yu LR, Tsang P, Wei JS, Song YK, Cheuk A, Chung JY, Hewitt SM, Veenstra TD, Khan J (2011) Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res 10(2):479–487. https://doi.org/10.1021/pr1006697
Sípová H, Zhang S, Dudley AM, Galas D, Wang K, Homola J (2010) Surface plasmon resonance biosensor for rapid label-free detection of microribonucleic acid at subfemtomole level. Anal Chem 82(24):10110–10115. https://doi.org/10.1021/ac102131s
Zhou WJ, Chen Y, Corn RM (2011) Ultrasensitive microarray detection of short RNA sequences with enzymatically modified nanoparticles and surface plasmon resonance imaging measurements. Anal Chem 83:3897–3902. https://doi.org/10.1021/ac200422u
Driskell JD, Seto AG, Jones LP, Jokela S, Dluhy RA, Zhao YP, Tripp RA (2008) Rapid microRNA (miRNA) detection and classification via surface-enhanced Raman spectroscopy (SERS). Biosens Bioelectron 24(4):917–922. https://doi.org/10.1016/j.bios.2008.07.060
Driskell JD, Tripp RA (2010) Label-free SERS detection of microRNA based on affinity for an unmodified silver nanorod array substrate. J Artic Chem Commun 46:3298–3300. https://doi.org/10.1039/C002059A
Drummond TG, Hill MG, Barton JK (2003) Electrochemical DNA sensors. Nat Biotechnol 21:1192–1199. https://doi.org/10.1038/nbt873
Gao Z, Yu YH (2007) Direct labeling microRNA with an electrocatalytic moiety and its application in ultrasensitive microRNA assays. Biosens Bioelectron 22:933–940. https://doi.org/10.1016/j.bios.2006.04.020
Gao Z, Yang Z (2006) Detection of microRNAs using electrocatalytic nanoparticle tags. Anal Chem 78:1470–1477. https://doi.org/10.1021/ac051726m
Peng YF, Gao ZQ (2011) Amplified detection of microRNA based on ruthenium oxide nanoparticle-initiated deposition of an insulating film. Anal Chem 83(3):820–827. https://doi.org/10.1021/ac102370s
Pöhlmann C, Sprinzl M (2010) Electrochemical detection of microRNAs via gap hybridization assay. Anal Chem 82(11):4434–4440. https://doi.org/10.1021/ac100186p
Wang T, Viennois E, Merlin D, Wang G (2015) Microelectrode miRNA sensors enabled by enzymeless electrochemical signal amplification. Anal Chem 87:8173–8180. https://doi.org/10.1021/acs.analchem.5b00780
Liu S, Yang Z, Chang Y, Chai Y, Yuan R (2018) An enzyme-free electrochemical biosensor combining target recycling with Fe3O4/CeO2@Au nanocatalysts for microRNA-21 detection. Biosens Bioelectron 119:170–175. https://doi.org/10.1016/j.bios.2018.08.006
Ren Y, Deng H, Shen W, Gao Z (2013) A highly sensitive and selective electrochemical biosensor for direct detection of microRNAs in serum. Anal Chem 85(9):4784–4789. https://doi.org/10.1021/ac400583e
Tian L, Qian K, Qi J, Liu Q, Yao C, Song W, Wang Y (2018) Gold nanoparticles superlattices assembly for electrochemical biosensor detection of microRNA-21. Biosens Bioelectron 99:564–570. https://doi.org/10.1016/j.bios.2017.08.035
Jolly P, Batistuti MR, Miodek A, Zhurauski P, Mulato M, Lindsay MA, Estrela P (2016) Highly sensitive dual mode electrochemical platform for microRNA detection. Sci Rep 6:36719. https://doi.org/10.1038/srep36719
Fan Y, Chen X, Trigg AD, Tung C, Kong J, Gao Z (2007) Detection of microRNAs using target-guided formation of conducting polymer nanowires in nanogaps. J Am Chem Soc 129(17):5437–5443. https://doi.org/10.1021/ja067477g
Gao Z, Deng H, Shen W, Ren Y (2013) A label-free biosensor for electrochemical detection of femtomolar microRNAs. Anal Chem 85(3):1624–1630. https://doi.org/10.1021/ac302883c
Fang Z, Kelley SO (2009) Direct Electrocatalytic mRNA detection using PNA-nanowire sensors. Anal Chem 81(2):612–617. https://doi.org/10.1021/ac801890f
Yang H, Hui A, Pampalakis G, Soleymani L, Liu FF, Sargent EH, Kelley SO (2009) Direct, electronic microRNA detection for the rapid determination of differential expression profiles. Angew Chem Int Ed 48:8461–8464. https://doi.org/10.1002/anie.200902577
Cosnier S (2003) Biosensors based on electropolymerized films: new trends. Anal Bioanal Chem 377(3):507–520. https://doi.org/10.1007/s00216-003-2131-7
Su S, Wu Y, Zhu D, Chao J, Liu X, Wan Y, Su Y, Zuo X, Fan C, Wang L (2016) On-electrode synthesis of shape-controlled hierarchical flower-like gold nanostructures for efficient interfacial DNA assembly and sensitive electrochemical sensing of microRNA. Small 12(28):3794–3801. https://doi.org/10.1002/smll.201601066
Zhou C, Cui K, Liu Y, Li L, Zhang L, Xu M, Ge S, Wang Y, Yu J (2020) Ultrasensitive lab-on-paper device via Cu/Co double-doped CeO2 nanospheres as signal amplifiers for electrochemical/visual sensing of miRNA-155. Sensors Actuators B Chem 321:128499. https://doi.org/10.1016/j.snb.2020.128499
Wang D, Hu L, Zhou H, Abdel-Halim ES, Zhu J-J (2013) Molecular beacon structure mediated rolling circle amplification for ultrasensitive electrochemical detection of microRNA based on quantum dots tagging. Electrochem Commun 33:80–83. https://doi.org/10.1016/j.elecom.2013.04.030
Moccia M, Caratelli V, Cinti S, Pede B, Avitabile C, Saviano M, LisaImbriani A, Moscone D, Arduini F (2020) Paper-based electrochemical peptide nucleic acid (PNA) biosensor for detection of miRNA-492: a pancreatic ductal adenocarcinoma biomarker. Biosens Bioelectron 165:112371. https://doi.org/10.1016/j.bios.2020.112371
Xu S, Chang Y, Wu Z, Li Y, Yuan R, Chai Y (2020) One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens Bioelectron 149:111848. https://doi.org/10.1016/j.bios.2019.111848
Kasturi S, Eom Y, Torati SR, Kim CG (2020) Highly sensitive electrochemical biosensor based on naturally reduced rGO/Au nanocomposite for the detection of miRNA-122 biomarker. J Ind Eng Chem 93:186–195. https://doi.org/10.1016/j.jiec.2020.09.022
Mandli J, Mohammadi H, Amine A (2017) Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles. Bioelectrochemistry 116:17–23. https://doi.org/10.1016/j.bioelechem.2017.03.002
Han S, Liu W, Yang S, Wang R (2019) Facile and label-free electrochemical biosensors for microRNA detection based on DNA origami nanostructures. ACS Omega 4(6):11025–11031. https://doi.org/10.1021/acsomega.9b01166
Bharti A, Mittal S, Rana S, Dahiya D, Agnihotri N, Prabhakar N (2020) Electrochemical biosensor for miRNA-21 based on gold-platinum bimetallic nanoparticles coated 3-aminopropyltriethoxy silane. Anal Biochem 609:113908. https://doi.org/10.1016/j.ab.2020.113908
Lusi EA, Passamano M, Guarascio P, Scarpa A, Schiavo L (2009) Innovative electrochemical approach for an early detection of microRNAs. Anal Chem 81(7):2819–2822. https://doi.org/10.1021/ac8026788
Tran HV, Piro B, Reisberg S, Duc HT, Pham MC (2014) An innovative strategy for direct electrochemical detection of microRNAs biomarker. Anal Bioanal Chem 406:1241–1244. https://doi.org/10.1007/s00216-013-7292-4
Majd SM, Salimi A, Ghasemi F (2018) An ultrasensitive detection of miRNA-155 in breast cancer via direct hybridization assay using two-dimensional molybdenum disulfide field-effect transistor biosensor. Biosens Bioelectron 105:6–13. https://doi.org/10.1016/j.bios.2018.01.009
Zhang GJ, Chua JH, Chee RE, Agarwal A, Wong SM (2009) Label-free direct detection of MiRNAs with silicon nanowire biosensors. Biosens Bioelectron 24(8):2504–2508. https://doi.org/10.1016/j.bios.2008.12.035
Yaralı E, Kanat E, Erac Y, Erdem A (2019) Ionic liquid modified single-use electrode developed for voltammetric detection of miRNA-34a and its application to real samples. Electroanalysis 31:1–11. https://doi.org/10.1002/elan.201900353
Kilic T, Kaplan M, Demiroglu S, Erdem A, Ozsoze M (2016) Label-free electrochemical detection of microRNA-122 in real samples by graphene modified disposable electrodes. J Electrochem Soc 163(6):B227–B233. https://doi.org/10.1149/2.0481606jes
Pham MC, Hachemi A, Dubois JE (1984) An apparently totally electroactive polymer film obtained by electropolymerizing 5-hydroxy-1,4-naphthoquinone onto graphite. J Electroanal Chem Interfacial Electrochem 161(1):199–204. https://doi.org/10.1016/S0022-0728(84)80262-4
Audebert P, Bidan G (1987) Electrochemical study of poly(phas) in acetonitrile and water + acetonitrile electrolytes. J Electroanal Chem Interfacial Electrochem 238(1–2):183–195. https://doi.org/10.1016/0022-0728(87)85173-2
Piro B, Bazzaoui EA, Pham MC, Novak P, Haas O (1999) Multiple internal reflection FTIR spectroscopic (MIRFTIRS) study of the redox process of poly(5-amino-1,4-naphthoquinone) film in aqueous and organic media. Electrochim Acta 44(12):1953–1964. https://doi.org/10.1016/S0013-4686(98)00304-1
Reisberg S, Piro B, Noël V, Pham MC (2005) DNA electrochemical sensor based on conducting polymer: dependence of the “signal-on” detection on the probe sequence localization. Anal Chem 77(10):3351–3356. https://doi.org/10.1021/ac050080v
Tran HV, Yougnia R, Reisberg S, Piro B, Serradji N, Nguyen TD, Tran LD, Dong CZ, Pham MC (2012) A label-free electrochemical immunosensor for direct, signal-on and sensitive pesticide detection. Biosens Bioelectron 31(1):62–68. https://doi.org/10.1016/j.bios.2011.09.035
Zhang QD, March G, Noel V, Piro B, Reisberg S, Tran LD, Hai LV, Abadia E, Nielsen PE, Sola C, Pham MC (2012) Label-free and reagentless electrochemical detection of PCR fragments using self-assembled quinone derivative monolayer: application to Mycobacterium tuberculosis. Biosens Bioelectron 32(1):163–168. https://doi.org/10.1016/j.bios.2011.11.048
Tran HV, Piro B, Reisberg S, Tran LD, Duc HT, Pham MC (2013) Label-free and reagentless electrochemical detection of micro RNAs using a conducting polymer nanostructured by carbon nanotubes: application to prostate cancer biomarker miR-141. Biosens Bioelectron 49:164–169. https://doi.org/10.1016/j.bios.2013.05.007
Tran HV, Piro B, Reisberg S, Duc HT, Pham MC (2013) Antibodies directed to RNA/DNA hybrids: a novel electrochemical immunosensor for miRNAs detection using graphene-composite electrodes. Anal Chem 85:8469–8474. https://doi.org/10.1021/ac402154z
Kangkamano T, Numnuam A, Limbut W, Kanatharana P, Vilaivan T, Thavarungkul P (2018) Pyrrolidinyl PNA polypyrrole/silver nanofoam electrode as a novel label-free electrochemical miRNA-21 biosensor. Biosens Bioelectron 102:217–225. https://doi.org/10.1016/j.bios.2017.11.024
Wang Y, Zheng D, Tan Q, Wang MX, Gu L-Q (2011) Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol 6:668–674. https://doi.org/10.1038/nnano.2011.147
Wang Y-H, Huang K-J, Wu X, Ma Y-Y, Song D-L, Du C-Y, Chang S-H (2018) Ultrasensitive supersandwich-type biosensor for enzyme-free amplified microRNA detection based on N-doped graphene/Au nanoparticles and hemin/G-quadruplexes. J Mater Chem B 6:2134–2142. https://doi.org/10.1039/C8TB00061A
Li Y, Yu C, Yang B, Liu Z, Xia P, Wang Q (2018) Target-catalyzed hairpin assembly and metal-organic frameworks mediated nonenzymatic co-reaction for multiple signal amplification detection of miR-122 in human serum. Biosens Bioelectron 105:307–315. https://doi.org/10.1016/j.bios.2017.11.047
Wu X, Chai Y, Yuan R, Su H, Han J (2013) A novel label-free electrochemical microRNA biosensor using Pd nanoparticles as enhancer and linker. Analyst 138:1060–1066. https://doi.org/10.1039/C2AN36506E
Su S, Cao W, Liu W, Lu Z, Zhu D, Chao J, Weng L, Wang L, Fan C, Wang L (2017) Dual-mode electrochemical analysis of microRNA-21 using gold nanoparticle-decorated MoS2 nanosheet. Biosens Bioelectron 94:552–559. https://doi.org/10.1016/j.bios.2017.03.040
Goda T, Masuno K, Nishida J, Kosaka N, Ochiya T, Matsumoto A, Miyahara Y (2012) A label-free electrical detection of exosomal microRNAs using microelectrode array. Chem Commun 48:11942–11944. https://doi.org/10.1039/C2CC36111F
Labib M, Khan N, Ghobadloo SM, Cheng J, Pezacki JP, Berezovski MV (2013) Three-mode electrochemical sensing of ultralow microRNA levels. J Am Chem Soc 135(8):3027–3038. https://doi.org/10.1021/ja308216z
Tran HV, Piro B, Reisberg S, Huy Nguyen L, Dung Nguyen T, Duc HT, Pham MC (2014) An electrochemical ELISA-like immunosensor for miRNAs detection based on screen-printed gold electrodes modified with reduced graphene oxide and carbon nanotubes. Biosens Bioelectron 62:25–30. https://doi.org/10.1016/j.bios.2014.06.014
Salimi A, Kavosi B, Navaee A (2019) Amine-functionalized graphene as an effective electrochemical platform toward easily miRNA hybridization detection. Measurement 143:191–198. https://doi.org/10.1016/j.measurement.2019.05.008
Wan Z, Umer M, Lobino M, Thiel D, Nguyen N-T, Trinchi A, Shiddiky MJA, Gao Y, Li Q (2020) Laser induced self-N-doped porous graphene as an electrochemical biosensor for femtomolar miRNA detection. Carbon 163:385–394. https://doi.org/10.1016/j.carbon.2020.03.043
Wang F, Chu Y, Ai Y, Chen L, Gao F (2019) Graphene oxide with in-situ grown Prussian Blue as an electrochemical probe for microRNA-122. Microchim Acta 186:116. https://doi.org/10.1007/s00604-018-3204-9
Dong H, Jin S, Ju H, Hao K, Xu LP, Lu H, Zhang X (2012) Trace and label-free MicroRNA detection using oligonucleotide encapsulated silver nanoclusters as probes. Anal Chem 84(20):8670–8674. https://doi.org/10.1021/ac301860v
Yin H, Zhou Y, Zhang H, Meng X, Ai S (2012) Electrochemical determination of microRNA-21 based on graphene, LNA integrated molecular beacon, AuNPs and biotin multifunctional bio bar codes and enzymatic assay system. Biosens Bioelectron 33(1):247–253. https://doi.org/10.1016/j.bios.2012.01.014
Li Y, Tian R, Zheng X, Huang R (2016) Amplified electrochemical detection of nucleic acid hybridization via selective preconcentration of unmodified gold nanoparticles. Anal Chim Acta 934:59–65. https://doi.org/10.1016/j.aca.2016.06.035
Fang CS, Kim K, Yu B, Jon S, Kim M-S, Yang H (2017) Ultrasensitive electrochemical detection of miRNA-21 using a zinc finger protein specific to DNA–RNA hybrids. Anal Chem 89(3):2024–2031. https://doi.org/10.1021/acs.analchem.6b04609
Yu LD, Wen YX, Zhang XY, Li NB, Luo HQ (2020) Signal-off photoelectrochemical determination of miRNA-21 using aptamer-modified In2O3@Cu2MoS4 nanocomposite. Microchim Acta 187:561. https://doi.org/10.1007/s00604-020-04540-z
Su S, Hao Q, Yan Z, Dong R, Yang R, Zhu D, Chao J, Zhou Y, Wang L (2019) A molybdenum disulfide@Methylene Blue nanohybrid for electrochemical determination of microRNA-21, dopamine and uric acid. Microchim Acta 186:607. https://doi.org/10.1007/s00604-019-3678-0
Sun Y, Jin H, Jiang X, Gui R (2020) Black phosphorus nanosheets adhering to thionine-doped 2D MOF as a smart aptasensor enabling accurate capture and ratiometric electrochemical detection of target microRNA. Sensors Actuators B Chem 309:127777. https://doi.org/10.1016/j.snb.2020.127777
Schwartz EF, Stollar BD (1969) Antibodies to polyadenylate-polyuridylate copolymers as reagents for double strand RNA and DNA-RNA hybrid complexes. Biochem Biophys Res Commun 35(1):115–120. https://doi.org/10.1016/0006-291x(69)90490-2
Stollar BD (1970) Double-helical polynucleotides: immunochemical recognition of differing conformations. Science 169:609–611. https://doi.org/10.1126/science.169.3945.609
Rudkin GT, Stollar BD (1977) High resolution detection of DNA–RNA hybrids in situ by indirect immunofluorescence. Nature 265:472–473. https://doi.org/10.1038/265472a0
Yoshichika K, Stollar BD (1982) Comparison of poly(A)·poly(dT) and poly(I)·poly(dC) as immunogens for the induction of antibodies to RNA-DNA hybrids. Mol Immunol 19(3):413–420. https://doi.org/10.1016/0161-5890(82)90207-3
Qavi AJ, Kindt JT, Gleeson MA, Bailey RC (2011) Anti-DNA:RNA antibodies and silicon photonic microring resonators: increased sensitivity for multiplexed microRNA detection. Anal Chem 83(15):5949–5956. https://doi.org/10.1021/ac201340s
Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH (2006) An antibody-based microarray assay for small RNA detection. Nucleic Acids Res 37(7):e52. https://doi.org/10.1093/nar/gkl142
Zouari M, Campuzano S, Pingarrón JM, Raouafi N (2018) Amperometric biosensing of miRNA-21 in serum and cancer cells at nanostructured platforms using anti-DNA-RNA hybrid antibodies. ACS Omega 3(8):8923–8931. https://doi.org/10.1021/acsomega.8b00986
Wang M, Yin H, Zhou Y, Sui C, Wang Y, Meng X, Waterhouse GIN, Ai S (2019) Photoelectrochemical biosensor for microRNA detection based on a MoS2/g-C3N4/black TiO2 heterojunction with Histostar@AuNPs for signal amplification. Biosens Bioelectron 128:137–143. https://doi.org/10.1016/j.bios.2018.12.048
Wang M, Li B, Zhou Q, Yin H, Zhou Y, Ai S (2015) Label-free, ultrasensitive and electrochemical immunosensing platform for microRNA detection using anti-DNA:RNA hybrid antibody and enzymatic signal amplification. Electrochim Acta 165:130–113. https://doi.org/10.1016/j.electacta.2015.03.011
Ramnani P, Gao Y, Ozsoz M, Mulchandani A (2013) Electronic detection of microRNA at attomolar level with high specificity. Anal Chem 85(17):8061–8064. https://doi.org/10.1021/ac4018346
Kilic T, Topkaya SN, Ozsoz M (2013) A new insight into electrochemical microRNA detection: a molecular caliper, p19 protein. Biosens Bioelectron 48:165–171. https://doi.org/10.1016/j.bios.2013.04.011
Ahmed N, Foss DV, Powdrill MH, Pezacki JP (2019) Site-specific cross-linking of a p19 viral suppressor of RNA silencing protein and its RNA targets using an expanded genetic code. Biochemistry 58(33):3520–3526. https://doi.org/10.1021/acs.biochem.9b00428
Campuzano S, Pedrero M, Pingarrón JM (2016) Viral protein-based bioanalytical tools for small RNA biosensing. TrAC Trends Anal Chem 79:335–343. https://doi.org/10.1016/j.trac.2015.09.012
Tu W, Cao H, Zhang L, Bao J, Liu X, Dai Z (2016) Dual signal amplification using gold nanoparticles-enhanced zinc selenide nanoflakes and P19 protein for ultrasensitive photoelectrochemical biosensing of microRNA in cell. Anal Chem 88(21):10459–10465. https://doi.org/10.1021/acs.analchem.6b02381
Li C, Liu Z, Cai S, Wen F, Wu D, Liu Y, Wu F, Lan J, Han Z, Chen J (2015) An electrochemical microRNA biosensor based on protein p19 combining an acridone derivate as indicator and DNA concatamers for signal amplification. Electrochem Commun 60:185–189. https://doi.org/10.1016/j.elecom.2015.09.012
Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M (2010) Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol 5:807–814. https://doi.org/10.1038/nnano.2010.202
Chen Z, Xie Y, Huang W, Qin C, Yu A, Lai G (2019) Exonuclease-assisted target recycling for ultrasensitive electrochemical detection of microRNA at vertically aligned carbon nanotubes. Nanoscale 11:11262–11269. https://doi.org/10.1039/C9NR02543J
Zhang C, Li D, Li D, Wen K, Yang X, Zhu Y (2019) Rolling circle amplification-mediated in situ synthesis of palladium nanoparticles for the ultrasensitive electrochemical detection of microRNA. Analyst 144:3817–3825. https://doi.org/10.1039/C9AN00427K
Bai Y-Y, Wu Z, Xu C-M, Zhang L, Feng J, Pang D-W, Zhang Z-L (2020) One-to-many single entity electrochemistry biosensing for ultrasensitive detection of microRNA. Anal Chem 91(1):853–858. https://doi.org/10.1021/acs.analchem.9b03492
Wang G, Tian W, Liu X, Ren W, Liu C (2020) New CRISPR-derived microRNA sensing mechanism based on Cas12a self-powered and rolling circle transcription-unleashed real-time crRNA recruiting. Anal Chem 92(9):6702–6708. https://doi.org/10.1021/acs.analchem.0c00680
Kergoat L, Piro B, Berggren M, Pham MC, Yassar A, Horowitz G (2012) DNA detection with a water-gated organic field-effect transistor. Org Electron 13:1–6. https://doi.org/10.1016/j.orgel.2011.09.025
Pallu J, Avci-Adali M, Mackeben P, Mohammadnejad L, Mattana G, Noël V, Piro B (2019) A DNA hydrogel gated organic field effect transistor. Org Electron 75:105402. https://doi.org/10.1016/j.orgel.2019.105402
White SP, Dorfman KD, Frisbie CD (2015) Label-free DNA sensing platform with low-voltage electrolyte-gated transistors. Anal Chem 87(3):1861–1866. https://doi.org/10.1021/ac503914x
Campos R, Borme J, Guerreiro JR, Machado G Jr, Cerqueira MF, Petrovykh DY, Alpuim P (2019) Attomolar label-free detection of DNA hybridization with electrolyte-gated graphene field-effect transistors. ACS Sens 4(2):286–293. https://doi.org/10.1021/acssensors.8b00344
Matsuura S, Ono H, Kawasaki S, Kuang Y, Fujita Y, Saito H (2018) Synthetic RNA-based logic computation in mammalian cells. Nat Commun 9:4847. https://doi.org/10.1038/s41467-018-07181-2
Zhao X-P, Liu F-F, Hu W-C, Younis MR, Wang C, Xia X-H (2019) Biomimetic nanochannel-ionchannel hybrid for ultrasensitive and label-free detection of microRNA in cells. Anal Chem 91(5):3582–3589. https://doi.org/10.1021/acs.analchem.8b05536
Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, Dincer C, Urban GA (2019) CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater 31(51):1905311. https://doi.org/10.1002/adma.201905311
Pingarron JM, Campuzano S, Ruiz-Valdepenas-Montiel V, Torrente-Rodriguez RM, Montoya JJ (2017) Magnetic beads-based electrochemical biosensor. Patent n°WO2017137192A1
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tran, H.V., Piro, B. Recent trends in application of nanomaterials for the development of electrochemical microRNA biosensors. Microchim Acta 188, 128 (2021). https://doi.org/10.1007/s00604-021-04784-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04784-3