Abstract
Gold nanoparticles of different sizes have been synthesized and surface-functionalized with selected RNA probes in order to develop a rapid, low-cost and sensitive method for detection of microRNA146a (miR146a). The strategy is based on the change of colour that can be observed visually after aggregation of the RNA modified-gold nanoparticles (AuNPs) in presence of miR146a. Experimental conditions have been carefully selected in order to obtain a good sensitivity that allows to perform visual detection of microRNA at the nM level, achieving a detection limit of 5 nM. Good repeatability and selectivity versus other sequences that only differ from miR146a in 3 bases was achieved. miR146a has been described as one of the main microRNA involved in the immune response of bovine mastitis, being expressed in tissue, blood and milk samples. The method was successfully applied to the detection of miR146a in raw cow milk samples. The present scheme constitutes a rapid and low-cost alternative to perform highly sensitive detection of microRNA without the need of instrumentation and amplification steps for the early detection of bovine mastitis in the agrofood industry.

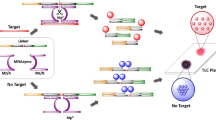
Schematic representation of the assay based on aggregation of RNA-modified gold nanoparticles (blue) in presence of microRNA146a generating a dark blue spot onto a solid support, versus a pink spot observed in absence of miR146a due to dispersed gold nanoparticles (red).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine mastitis is a persistent inflammatory reaction of the udder tissue caused by a wide range of microorganisms infections [1]. It is a highly prevalent and costly disease to the dairy cattle, resulting in large economic losses for the dairy industry [2]. Somatic cell counting and bacteriological culturing of milk samples are the most frequently used diagnostic methods for bovine mastitis [3]. Somatic cell counting suffers from limitations such as the variability on the interpretation of the results between different testers, as well as the high rates of false positive results due to some environmental conditions such as parity, season or stress. [4]. On the other hand, bacteriologic culture is a common diagnostic method to identify the pathogens causing mastitis [5]. It requires between 48 and 72 h to obtain confirmatory results, need to be performed using standardized repeatable methods, and poses a low sensitivity when compared to other novel amplification-based methods [6].
A novel approach in order to perform the detection of mastitis may be the detection of micro RNAs (microRNAs), a noncoding, highly conserved, single-stranded and endogenous RNAs made of 18–25 nucleotides. Discovered in the early 1990s throughout the genome, many publications have demonstrated their involvement in different biological processes, like protein expression and regulation [7]. They are considered as an emerging class of biomarkers for which a growing number of scientific publications report a correlation between the presence of a certain microRNA and a pathological conditions, such as inflammation diseases [8]. In this sense, there are many different microRNA families that are involved in mastitis disease, being present in both, milk samples (such as microRNA223, microRNA142-5p, microRNA21, microRNA146a (miR146a), microRNA155, microRNA222, microRNA383) [9,10,11], and in bovine tissues (such as microRNA144, microRNA451, microRNA7863, microRNA182, microRNA122, microRNA205, microRNA200a, microRNA146a, microRNA146b (miR146b)) [12,13,14]. Particularly, miR146a has been reported as one of the main microRNA involved in the immune response of bovine mastitis, being expressed in tissue, blood and milk samples [9, 11, 12, 15, 16], as well as in other diseases such as tuberculosis [17].
Different detection methods, including northern blotting [18], real-time quantitative polymerase chain reaction (qRT-PCR) [19], microarray [20], sequencing [21], cloning [22] and in situ hybridization [23] have been employed for the detection of microRNAs. However, some of those methods suffer certain limitations including that they are typically time-consuming techniques requiring high volume of sample and multiple experimental steps [8, 24]. Furthermore, many often sensitivity is not good enough for microRNA detection at relevant concentration levels since microRNAs to be detected for rapid diagnosis are at very low concentration levels [25]. Microarray techniques can offer a sensitivity good enough for detection of microRNA [26]. However, they suffer a high degree of false-positive results [27]. Hybridization-based methods such as sequencing, cloning, in situ hybridization and amplification-based methods like qRT-PCR provide high specificity and sensitivity for the detection of genetic targets [28]. Nevertheless, they require delicate and laborious primer design due to the short length of microRNAs, as well as a complex sample preparation which involves multiple experimental steps including modification of microRNAs, requiring highly qualified personnel to perform both, experimental work and data analysis [27], and resulting in higher cost and complexity [29].
In order to ensure a high efficiency in agrofood safety control, dairy products industry and producers would benefit from having reliable and rapid methods of analysis capable to detect mastitis biomarkers in situ and in a fast way to achieve rapid diagnosis of the infectious disease.
In this sense, colorimetric methodologies based on gold nanoparticles (AuNPs) can be a convenient option for rapid and cost-effective microRNA detection. Few colorimetric assays for microRNA detection have been described based on the use of AuNPs. However, most methodologies described so far require long analysis times and include amplification steps based on the use of enzymes [30, 31]. Aggregation of AuNPs followed by Resonance Light Scattering measurements was reported by Ren, M. et al. [32] for microRNA detection with high sensitivity. However, the method is limited for routine analysis by the need of complex and expensive instrumentation.
Building upon these techniques, here it is presented the design, optimization and evaluation of an assay for microRNA visual optical detection that takes advantage of the changes of the surface-plasmon resonance absorption produced after aggregation of AuNPs induced by the target molecule [33]. To ensure selectivity AuNPs are modified with short RNA sequences that are complementary to the sequence of the microRNA of interest. Visualization of the colour difference is enhanced by spotting the samples onto a C18 silica thin-layer chromatography (TLC) plate, retaining the colour of the solution mixture and preventing dissociation of the formed aggregates, allowing a permanent record of the test. This strategy provides a rapid, cost-effective and highly sensitive method for the detection of microRNA, avoiding the design of sophisticated templates, and where the readout can be performed visually, without the need of complex and expensive instrumentation [33].
Materials and methods
Materials and reagents
All reagents were of Analytical grade and used without any further purification unless otherwise mentioned.
The thiolated RNA strands employed to functionalize the AuNPs surface (Probe-1) 5’-SH-A AAA AAA AAA AUU CAG UUC UCA-3′, (Probe-2) 5’-ACA ACC UAU GGA AAA AAA AAA A-HS-3′, and the micro RNAs (miR146a) 5’-UGA GAA CUG AAU UCC AUA GGU UGU-3′, (miR146b) 5’-UGA GAA CUG AAU UCC AUA GGC UGU-3′, (miR146c) 5’-UGA GAA CUG AAU UCC AUA CCU UCU-3′, were obtained from Integrated DNA Technologies (Iowa, United States; www.idtdna.com). Hydrogen Tetrachloroaurate Trihydrate (HAuCl4 ∙ 3H2O), Sodium citrate tribasic dihydrate (Na3C6H5O7 ∙ 2H2O), Magnesium Chloride Hexahydrate (MgCl2 ∙ 6H2O), Trizma® Hydrochloride (NH2C(CH2OH)3 ∙ HCl) and Potassium Chloride (KCl) were purchased from Sigma-Aldrich (St. Louis, USA; www.sigmaaldrich.com). Thiolated Polyethylene Glycol (mPEG-SH1000) from Laysan Bio, Inc. (Huntsville, USA; www.laysanbio.com). TLC Aluminium Sheets Silica gel 60 F254 and Analtech TLC Uniplates™, where the C18-silica gel matrix is immobilized onto a glass support, were purchased from Merck KGaA (Darmstadt, Germany; www.merckmillipore.com). Milk samples were obtained from the Department of Animal Nutrition, Grassland and Forages, Regional Institute for Research and Agro-Food Development (SERIDA, Asturias, Spain). RNA extraction was performed using QIAzol Lysis Reagent from QIAGEN (USA; www.qiagen.com) and mirVana microRNA Isolation Kit and Phosphate-Buffered Saline (PBS) from Thermo Fisher Scientific (USA; www.thermofisher.com).
Synthesis of gold nanoparticles
Gold nanoparticles (AuNPs) were prepared following a procedure described elsewhere [34], and details are given in the Electronic Supplementary Material.
Size, morphology and size dispersion of AuNPs were assessed by Dynamic Light Scattering (DLS) and Transmission Electron Microscopy (TEM) measurements, using only particles with a polydispersity index (PDI) below 0.1. The average diameters of the synthesized AuNPs are 15 ± 1 nm (n = 670) with a PDI of 0.01; 20 ± 1 nm (n = 600) with a PDI of 0.07; and 25 ± 3 (n = 117) with a PDI of 0.1.
Functionalization of AuNPs
Two sets of AuNPs (named SET 1 and SET 2 respectively) were functionalized with two different RNA sequences with a 5′-thiol-modification (Probe-1) and a 3′-thiol-modification (Probe-2). Both sequences are partially complementary to the target analyte miR146a and have a poly-A chain of nucleotides (10 adenines) between the thiol modification and the complementary sequence (see Table 1), acting as a spacer in order to facilitate their binding to the miR146a target analyte. SET 1 and SET 2 of AuNPs were prepared by mixing 20 μL of AuNPs (100 nM) with 20 μL of Probe-1 and Probe-2 (15 μM) respectively, in binding buffer (ultrapure water with 0.01% v/v Tween-20 and 90 mM Trisodium Citrate HCl buffer, pH = 3) for 30 min at room temperature.
Afterwards, in order to improve the colloidal stability of the NPs, thiolated polyethylene glycol (mPEG-SH1000) was added to block any RNA-free areas that can be present on the AuNPs surface. For such purpose, each set of surface-modified AuNPs was mixed with 10 μL of mPEG-SH1000 (2 mM), and the mixture was incubated at 60 °C for 30 min. Finally, a purification step to remove the mPEG-SH1000 excess from the supernatant was carried out by three centrifugation cycles (16,000 g) for 30 min. Finally, a sample dilution with ultrapure water with 0.01% v/v Tween-20 was carried out to obtain a final concentration of 5 nM AuNPs, based on Extinction coefficient of 2.33 × 108 M−1 cm−1 at λ = 521 nm for 15 nm AuNPs.
Detection assay
In a typical experiment, 2 μL of different concentrations of standard solutions of target analyte (0–80 nM), were mixed with 5 μL of AuNPs SET 1 and 5 μL of AuNPs SET 2 (to get a final concentration of 2.5 nM) in 8 μL of buffer assay (300 mM MgCl2 ∙ 6H2O, 0.1 M Trizma® HCl and 0.5 M KCl, pH = 8.3) for 20 min at 60 °C to allow hybridization of the complementary strands.
Sample pre-treatment
Raw cow milk samples were obtained at the Regional Institute for Research and Agrofood Development of the Principality of Asturias (SERIDA). The milk samples were pre-treated with QIAzol Lysis Reagent from QIAGEN (USA) and mirVana microRNA Isolation Kit from Thermo Fisher (USA) following the instructions of the manufacturer.
For each milk sample, 50 mL were centrifuged at 4000 g−1 for 20 min at 4 °C to obtain a pellet with the cells. The top layer containing fat and whey were discarded. Then, the pellet, which corresponds with the milk cells, was washed twice with PBS, and then homogenized in 2 mL of QIAzole reagent. Then, miR146a was added at different concentrations. Finally, total RNA was isolated with the mirVana kit following instructions of the manufacturer. A recovery between 70% of the RNA has been assumed for the extraction [35].
Results and discussion
Choice of materials
With the aim of performing a visual detection of microRNA avoiding the use of instrumentation to perform the measurement of the results, the use of nanoparticles is an excellent alternative when compared to traditional dyes. Optoelectronic properties of certain nanoparticles offer the possibility of observing a change in the colour as consequence of changes on the distance between them. In this sense, the most remarkable and well-known NPs whose optical properties change depending on the distance between them are AuNPs and silver NPs (AgNPs).
Functionalization of the NPs with ARN that is complementary to miR146a is required to specifically detect the microRNA of interest. This process can be easily carried out by using thiolated RNA oligomers and metallic NPs. AuNPs were selected for this purpose due to the highly strength of Au-S bonds [36], which ensure that the surface of the AuNPs is functionalized with RNA strands with a high colloidal stability. Additionally, AuNPs present higher photostability when compared to AgNPs.
In order to observe the differences in the colour of the AuNPs once the assay was performed, the resulting functionalized AuNPs in presence/absence of miR146a were deposited onto a white TLC plate. For this purpose, two types of TLC plates were evaluated, Analtech TLC Uniplates™ and TLC Aluminium Sheets Silica gel 60 F254. Similar results in terms of differentiation of the colour were obtained. However, TLC Aluminium Sheets Silica gel 60 F254 present a very adherent and hard surface that does not crack or blister easily. Therefore, the last ones were more practical from the experimental point of view, and were selected for further experiments.
Design of the assay
First, the design of the assay was carried out in order to perform a visual detection of miR146a. The assay is based on the observation of colour changes of a solution that contains 15 nm AuNPs in presence and absence of the target analyte. For this purpose, AuNPs were synthesized and surface modified as described in the synthesis of AuNPs. The target miR146a, is a microRNA that contains 24 RNA bases, and it has been reported as a mastitis biomarker, since it is one of the microRNAs expressed in tissue, blood and milk samples as consequence of bovine mastitis. In order to achieve a selective aggregation of the nanoparticles in presence of the analyte, the AuNPs surface was modified with two different thiolated RNA sequences which are complementary to the target miR146a following the procedure previously described in the functionalization of AuNPs section.
Then, in order to check the feasibility of the reported scheme for the visual detection of miR146a, appropriate volumes (5 μL) of SET 1 (Probe-1) and SET 2 (Probe-2) of AuNPs were mixed with a sample (2 μL) containing the target analyte miR146a. After an incubation time (20 min) at controlled temperature (60 °C), a portion of the target analyte will hybridize with the complementary strand of the Probe-1, and the other portion will do the same with the strand of Probe-2. Hence, due to this hybridization, AuNPs will become close to each other, and a change on the colour from red to purple/black will take place (see Fig. 1) [33]. Afterwards, 3 μL of the aforementioned mixture were spotted onto a thin-layer chromatography (TLC) plate to facilitate observation of the difference on the colour due to the aggregation of the AuNPs, allowing a more sensitive visual detection [34]. In order to record images of the results obtained in the TLC plate, a digital scanner was employed, minimizing bias due to the influence of the ambient light in the visual perception of the colour. Additionally, measurement of the UV/Vis spectroscopic shift of the wavelength of the absorption surface plasmon resonance peak was used for optimization of the assay.
The RNA:AuNP molar ratio used in the assay has been evaluated in order to ensure that the surface of the AuNPs is fully functionalized with the maximum possible surface density of thiolated RNA strands. For this purpose, different molar concentrations of RNA:AuNP were studied. A range from 0:1 to 300:1 were prepared for 15 nm suzed AuNPs, and afterwards, a gel electrophoresis using 1% agarose was run at 100 V for 1 h for both, Probe-1 and Probe-2. When the RNA:AuNP ratio is low, the bioconjugates migrate faster, and as the ratio increases, they become slower. Hence, once the surface of the AuNPs is fully saturated with thiolated oligomers, they migrate all at the same rate, which indicates that no more RNA strands will be bound to the surface of the AuNPs. A similar study was performed for different sized AuNPs, varying from 0:1 to 1000:1 for AuNPs of 20 nm, and from 0:1 to 2000:1 for AuNPs of 25 nm, Results obtained from this study are shown in Fig. S1 in the Electronic Supplementary Material, and have shown that the optimum RNA:AuNP ratio is 200:1 for AuNPs of 15 nm, 667:1 for AuNPs of 20 nm, and 1333:1 for AuNPs of 25 nm.
With the aim of developing a tool to perform visual detection of microRNA, concentration of AuNPs is a key parameter to be optimized, since a low concentration of AuNPs would provide a weak colour of the solution that may difficult the discrimination between aggregated and non-aggregated. On the other hand, if the concentration of AuNPs is too high, an intense pink colour will be easily observed before interaction of the GNP-based probes with the microRNA, but the presence of small concentrations of the target would produce an aggregation of the AuNPs that may be neglected due to the high concentration of non-aggregated AuNPs. Hence, for this purpose, the effect of the concentration of AuNPs (Probe-1 and Probe-2) over the response for the detection of miR146a was evaluated. In this study, AuNPs concentrations ranging from 10 nM to 1.25 nM were employed, and results showed that the best sensitivity for the detection of the target was obtained for a AuNPs final concentration of 2.5 nM, which was selected for further experiments.
Optimization of the method
Several experimental parameters are crucial to allow an appropriate visual detection of the microRNA. In fact, the presence of salts has a direct impact on the RNA hybridization process as well as on the aggregation of AuNPs, Hybridization of the RNA strands strongly depends on the temperature. Additionally, depending on the size of AuNPs, discrimination between the colour of aggregated and non-aggregated AuNPs can be improved. Hence, the following parameters were optimized: (a) MgCl2 concentration; (b) temperature of the assay; (c) AuNPs size. Respective data and figures are given in the Electronic Supporting Material. The following experimental conditions were found to provide best results:
- (a)
Optimal MgCl2 concentration: a concentration of 30 mM MgCl2 has been chosen for further experiments, since no aggregation of AuNPs is observed in absence of the target (the SPR peak wavelength is lower than 525 nm), and the SRP peak wavelength starts to be shifted towards higher wavelengths at 5 nM target concentration, which means that a visual difference in colour can be observed for lower concentrations of miR146a (see Fig. S2).
- (b)
Optimal temperature: the assay was performed at different temperatures varying from 15 °C to 70 °C, being 60 °C the temperature that provided a better sensitivity selected for further experiments.
- (c)
Optimal AuNPs size: three different sizes of AuNPs (15, 20 and 25 nm respectively) were evaluated in order to select those NPs that provide a better visual sensitivity. AuNPs of 20 nm diameter generate a colour change that is more noticeable at lower target concentration, so they were selected for further experiments (see Fig. S3).
Visual detection of miR146a with AuNPs
In order to perform the detection of miR146a, two sets of AuNPs were mixed with the buffer assay and standards of microRNA with different concentrations as described in the Materials and Methods section. After 20-min incubation at 60 °C, 3 uL of the mixture were placed onto a TLC plate in order to observe the difference in the colour between aggregated and non-aggregated AuNPs, and the remaining solution was diluted in order to measure the shift on the SPR peak wavelength due to aggregation of AuNPs.
The presence of miR146a at concentrations above 5 nM already gives rise to a noticeable aggregation of the 20 nm AuNPs, turning the pink colour into purple spots due to a shift on the SPR peak towards higher wavelengths (up to 565 nm) when target concentration is increased and complete aggregation of the AuNPs takes place. Both, spectroscopic measurements and TLC spots were done in triplicate (n = 3), sitting an optimal limit of detection for 20 nm AuNPs of 5 nM of miR146a. Results obtained are shown in Fig. 2.
a Absorption spectra of AuNPs in presence of different concentrations of miR146a. b SPR wavelength and TLC spots AuNPs of 20 nm at the following miR146a concentrations: 0 nM, 1 nM, 2.5 nM, 5 nM, 7.5 nM, 10 nM, 20 nM, 40 nM, 80 nM. Both, spectroscopic measurements and TLC spots were carried out in triplicate (n = 3)
The method here reported for the detection of miR146a is expected to be easy to use, rapid and visual, without the need of analytical instrumentation to measure the results of the assay. To the best of our knowledge, there are no reported methods for detection of miR146a, neither for detection of any other microRNA in milk samples, based on the use of nanomaterials. There are few methods reported for the detection of microRNA based on aggregation of AuNPS. However, most of them are based on complex amplification schemes that achieve a low detection limit, but require temperature cycles, long assay times and spectrophotometric detection (see Table 2). Only two methods with no amplification strategies have been reported, but both require an instrumental readout using dynamic light scattering and ratiometric spectrophotometry respectively in order to measure the results obtained, and present long times to develop the assay. The method here proposed is not based on complex amplification schemes, and the assay is carried out in 20 min. Additionally, the readout of the results is visual, which is an enormous advantage for a future development of a point-of-care device that allows to carry out the detection of miR146a in milk samples without the need of trained personnel.
Selectivity of the miR146a detection
In order to evaluate the selectivity of this method towards the specific detection of miR146a, two other microRNAs, miR146b and miR146c, with sequences that only differ from miR146a in 1 and 3 bases respectively were evaluated. The sequences of the targets are summarized in Table 1.
For this purpose, those microRNAs were analysed following the general procedure. The detection assay was carried out for each microRNA separately, and by mixing the analyte miR146a with both, miR146b and miR146c. Results obtained are summarized in Fig. 3. The absence of aggregation in presence of miR146c demonstrates the capability to discriminate the target from another microRNA with a three-base mismatch. However, the aggregation of AuNPs in presence of miR146b indicates that it is not possible to differentiate the analyte from another microRNA with a single base mismatch. This issue is not considered as a disadvantage, since miR146b is also involved in dairy cows bacterial infections. Hence, this probe can be potentially applied to the detection of both, miR146a and miR146b. A mixture of all the targets still produces an aggregation of the AuNPs-based probes, which means that a higher concentration of RNA in the medium does not prevent aggregation of the AuNPs, giving rise to a positive result.
Detection of miR146a in milk samples
The usefulness of the method was evaluated for the detection of miR146a in raw milk samples obtained at the Regional Institute for Research and Agrofood Development of the Principality of Asturias (SERIDA). For this purpose, 9 samples of healthy cows were obtained after cow milking and were spiked with different amounts of miR146a. According to the literature, most of the microRNA present in milk samples is found in the pellet obtained after sample centrifugation [40]. Therefore, in order to be able to evaluate the recovery of the detection of the spiked microRNA, milk samples were centrifuged, the fat and serum top layers were discarded, and a known amount of the analyte was added to the pellet: 1.25, 2.5, 5.0, 7.5, 10, 20, 40 and 80 pmol. Then, the extraction protocol was performed following instructions of the manufacturer as it is briefly described in the Materials and Methods section. The total RNA extracted was collected in 100 μL and the detection assay was performed under optimal experimental conditions.
Results obtained are shown in Fig. 4, which indicate that a positive result is obtained at a 7.5 nM miR146a concentration with good repeatability. It is worth to mention that the concentration of the target has been calculated assuming an RNA extraction yield of 100%, but it is well-known that the yield of the extraction process is lower, since several steps are involved and a loss in the total RNA extracted takes place. Assuming a 70% yield of the RNA extraction [35], the concentration where the change of the colour is observed is 5 nM, which is in agreement with the LOD obtained for the method. Hence, it can be considered that this method can be applied to the analysis of miR146a in real milk samples.
a Absorption spectra of AuNPs in presence of different concentrations of miR146a. b SPR wavelength and TLC Spots of AuNPs-miR146a complex at different miR146a standard additions of 0 nM, 1 nM, 2.5 nM, 5 nM, 10 nM, 20 nM, 40 nM, 80 nM to milk samples. Both, the spectroscopic measurements and TLC spots were carried out in triplicate (n = 3)
Conclusions
A simple scheme for efficient and highly sensitive detection of microRNA based on the visual display of the colour change of AuNPs solution induced by the controlled aggregation of the NPs after incubation with the target microRNA in the medium. The approach has been designed for detection of miR146a (a model analyte that has been recognized as possible biomarker of mastitis), obtaining good sensitivity in the detection (detection limit of 5 nM miR146a) without the need of any amplification step.
The high selectivity of the assay (it is possible to discriminate the target microRNA from other species with a 3 bases mismatch) can be ensured by a proper functionalization of the AuNPs with complementary oligonucleotides.
It is worth to mention that the assay is performed in just 20 min under mild isothermal conditions. Additionally, it is possible to perform visual detection of the target without the need of instrumentation, and the readout can be easily interpreted by non-qualified personnel, since only a change in colour has to be observed. Both aspects are important advantages for a potential implementation of the method in the field. Images of the TLC plates were obtained using a digital scanner in order to minimize bias derived from differences in the perception of the colour due to ambient light. However, a digital scanner may not be available when applying this method in field. This issue can be overcome by using a smartphone to register the image, and an appropriate software controlled through an application in order to analyse the colour, allowing to obtain quantitative information, since the degree of aggregation can be related to the concentration of microRNA.
To sum up, this method for the detection of microRNAs is rapid, low cost and highly sensitive, and it has been successfully applied to the detection of miR146a differential expression in raw milk samples, which are important features for a future development of fast and in-situ monitoring device for the detection of bovine mastitis in livestock production industry.
This strategy opens new avenues for development of visual nanoprobes for a wide variety of applications, as it can be easily adapted for detection of other microRNA biomarkers different to the here-tested just by modifying the AuNPs with different appropriate oligomers complementary to the desired microRNA.
References
Yang W, Zerbe H, Petzl W, Brunner RM, Günther J, Draing C, von Aulock S, Schuberth HJ, Seyfert HM (2008) Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-κB in mammary epithelial cells and to quickly induce TNFα and interleukin-8 (CXCL8) expression in the udder. Mol Immunol 45:1385–1397. https://doi.org/10.1016/j.molimm.2007.09.004
Seegers H, Fourichon C, Beaudeau F (2003) Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res 34(5):475–491. https://doi.org/10.1051/vetres:2003027
Lam TJGM, Olde Riekerink RGM, Sampimon OC, Smith H (2009) Mastitis diagnostics and performance monitoring: a practical approach. Ir Vet J 62:34–39. https://doi.org/10.1186/2046-0481-62-S4-S34
Yagi Y, Shiono H, Chikayama Y, Ohnuma A, Nakamura I, Yayou K (2004) Transport stress increases somatic cell counts in milk, and enhances the migration capacity of peripheral blood neutrophils of dairy cows. J Vet Med Sci 66:381–387. https://doi.org/10.1292/jvms.66.381
Svennesen L, Mahmmod YS, Skjølstrup NK, Mathiasen LR, Katholm J, Pedersen K, Klaas IC, Nielsen SS (2018) Accuracy of qPCR and bacterial culture for the diagnosis of bovine intramammary infections and teat skin colonisation with Streptococcus agalactiae and Staphylococcus aureus using Bayesian analysis. Prev Vet Med 161:69–74. https://doi.org/10.1016/j.prevetmed.2018.10.013
Adkins PRF (2018) Methods for diagnosing mastitis bovine mastitis Intramammary infection diagnosis. Vet Clin NA Food Anim Pract 34:479–491. https://doi.org/10.1016/j.cvfa.2018.07.003
Wang J, Chen J, Sen S (2016) MicroRNA as biomarkers and diagnostics. J Cell Physiol 231:25–30. https://doi.org/10.1002/jcp.25056
De Planell-Saguer M, Rodicio MC (2011) Analytical aspects of microRNA in diagnostics: a review. Anal Chim Acta 699:134–152. https://doi.org/10.1016/j.aca.2011.05.025
Lai YC, Fujikawa T, Maemura T et al (2017) Inflammation-related microRNA expression level in the bovine milk is affected by mastitis. PLoS One 12:1–13. https://doi.org/10.1371/journal.pone.0177182
Sun J, Aswath K, Schroeder SG et al (2015) MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genomics 16:1–10. https://doi.org/10.1186/s12864-015-2044-9
Cai M, He H, Jia X, Chen S, Wang J, Shi Y, Liu B, Xiao W, Lai S (2018) Genome-wide microRNA profiling of bovine milk-derived exosomes infected with Staphylococcus aureus. Cell Stress Chaperones 23:663–672. https://doi.org/10.1007/s12192-018-0876-3
Wang XP, Luoreng ZM, Zan L Sen, et al (2016) Expression patterns of miR-146a and miR-146b in mastitis infected dairy cattle. Mol Cell Probes 30:342–344. https://doi.org/10.1016/j.mcp.2016.08.004
Luoreng ZM, Wang XP, Mei CG, Zan L Sen (2018) Expression profiling of peripheral blood miRNA using RNAseq technology in dairy cows with Escherichia coli-induced mastitis. Sci Rep 8:1–10. https://doi.org/10.1038/s41598-018-30518-2
Luoreng ZM, Wang XP, Mei CG, Zan L Sen (2018) Comparison of microRNA profiles between bovine mammary glands infected with Staphylococcus aureus and Escherichia coli. Int J Biol Sci 14:87–99. https://doi.org/10.7150/ijbs.22498
Paterson MR, Kriegel AJ (2017) MiR-146a/b: a family with shared seeds and different roots. Physiol Genomics 49:243–252. https://doi.org/10.1152/physiolgenomics.00133.2016
Naeem A, Zhong K, Moisá SJ, Drackley JK, Moyes KM, Loor JJ (2012) Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with streptococcus uberis1. J Dairy Sci 95:6397–6408. https://doi.org/10.3168/jds.2011-5173
Iannaccone M, Cosenza G, Pauciullo A, Garofalo F, Proroga YT, Capuano F, Capparelli R (2018) Milk microRNA-146a as a potential biomarker in bovine tuberculosis. J Dairy Res 85:178–180. https://doi.org/10.1017/S0022029918000122
Várallyay É, Burgyán J, Havelda Z (2008) MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 3:190–196. https://doi.org/10.1038/nprot.2007.528
Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FM (2003) Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol Med 9:189–195. https://doi.org/10.1016/S1471-4914(03)00047-9
Shingara J, Keiger K, Shelton J, Laosinchai-Wolf W, Powers P, Conrad R, Brown D, Labourier E (2005) An optimized isolation and labeling platform for accurate microRNA expression profiling. Rna 11:1461–1470. https://doi.org/10.1261/rna.2610405
Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4:143–159. https://doi.org/10.1002/emmm.201100209
Mattie MD, Benz CC, Bowers J et al (2006) Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5:1–14. https://doi.org/10.1186/1476-4598-5-24
Nelson PT, Wilfred BR (2009) In situ hybridization is a necessary experimental complement to microRNA (miRNA) expression profiling in the human brain. Neurosci Lett 466:69–72. https://doi.org/10.1016/j.neulet.2009.04.044
Hunt EA, Goulding AM, Deo SK (2009) Direct detection and quantification of microRNAs. Anal Biochem 387:1–12. https://doi.org/10.1016/j.ab.2009.01.011
Pall GS, Codony-Servat C, Byrne J, et al (2007) Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic acids Res 35. https://doi.org/10.1093/nar/gkm112
Wegman DW, Krylov SN (2013) Direct miRNA-hybridization assays and their potential in diagnostics. TrAC - Trends Anal Chem 44:121–130. https://doi.org/10.1016/j.trac.2012.10.014
Qavi AJ, Kindt JT, Bailey RC (2010) Sizing up the future of microRNA analysis. Anal Bioanal Chem 398:2535–2549. https://doi.org/10.1007/s00216-010-4018-8
Espy MJ, Uhl JR, Sloan LM, et al (2006) Real-Time PCR in Clinical Microbiology : Applications for 19:8512. https://doi.org/10.1128/CMR.00022-06
Russell C, Welch K, Jarvius J, Cai Y, Brucas R, Nikolajeff F, Svedlindh P, Nilsson M (2014) Gold nanowire based electrical DNA detection using rolling circle amplification. ACS Nano 8:1147–1153. https://doi.org/10.1021/nn4058825
Li RD, Yin BC, Ye BC (2016) Ultrasensitive, colorimetric detection of microRNAs based on isothermal exponential amplification reaction-assisted gold nanoparticle amplification. Biosens Bioelectron 86:1011–1016. https://doi.org/10.1016/j.bios.2016.07.042
Shi HY, Yang L, Zhou XY et al (2017) A gold nanoparticle-based colorimetric strategy coupled to duplex-specific nuclease signal amplification for the determination of microRNA. Microchim Acta 184:525–531. https://doi.org/10.1007/s00604-016-2030-1
Ren M, Wang S, Cai C et al (2016) A simple and sensitive resonance light scattering method based on aggregation of gold nanoparticles for selective detection of microRNA-21. RSC Adv 6:83078–83083. https://doi.org/10.1039/c6ra12366j
Elghanian R, Storhoff JJ, Mucic RC, et al (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science (80- ) 277:1078–1081. https://doi.org/10.1126/science.277.5329.1078
Storhoff JJ, Elghanian R, Mucic RC et al (1998) One-pot colorimetric differentiation of polynucleotides with Single Base imperfections using gold nanoparticle probes. J Am Chem Soc 7863:1959–1964. https://doi.org/10.1021/ja972332i
Zaporozhchenko IA, Morozkin ES, Skvortsova TE, Bryzgunova OE, Bondar AA, Loseva EM, Vlassov VV, Laktionov PP (2015) A phenol-free method for isolation of microRNA from biological fluids. Anal Biochem 479:43–47. https://doi.org/10.1016/j.ab.2015.03.028
Pakiari AH, Jamshidi Z (2010) Nature and strength of M−S bonds (M = au, Ag, and cu) in binary alloy gold clusters. J Phys Chem A 114:9212–9221. https://doi.org/10.1021/jp100423b
Hakimian F, Ghourchian H, Hashemi A sadat, et al (2018) Ultrasensitive optical biosensor for detection of miRNA-155 using positively charged au nanoparticles. Sci Rep 8:2943. https://doi.org/10.1038/s41598-018-20229-z
Tian R, ZhenG X (2016) Sensitive colorimetric detection of microrna based on target catalyzed double-arm hairpin DNA assembling. Anal Sci 32:751–756. https://doi.org/10.2116/analsci.32.751
Persano S, Guevara ML, Wolfram J, Blanco E, Shen H, Ferrari M, Pompa PP (2016) Label-free isothermal amplification assay for specific and highly sensitive colorimetric miRNA detection. ACS Omega 1:448–455. https://doi.org/10.1021/acsomega.6b00109
Li R, Dudemaine PL, Zhao X et al (2016) Comparative analysis of the miRNome of bovine milk fat, whey and cells. PLoS One 11:1–21. https://doi.org/10.1371/journal.pone.0154129
Acknowledgements
Financial support provided from Projects FC-GRUPIN-IDI/2018/000166 and GRUPIN-IDI/2018/000237 (Principado de Asturias), Project CTQ2016-79412-P (Ministerio de Economía y Competitividad de Economia y Competitividad, Spain) and Project RTA2015-00061/AEI/FEDER (Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria Nacional de Investigaciones Agrarias), co-financed with European Union FEDER funds, is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on IX NyNA 2019. International Congress on Analytical Nanoscience and Nanotechnology
This work was presented at the IX NyNA 2019, International Congress on Analytical Nanoscience and Nanotechnology at Zaragoza (Spain) from 2 - 4 July, 2019. Chairman: Dr. Juan R. Castillo.
Electronic supplementary material
ESM 1
(DOCX 468 kb)
Rights and permissions
About this article
Cite this article
Sánchez-Visedo, A., Gallego, B., Royo, L.J. et al. Visual detection of microRNA146a by using RNA-functionalized gold nanoparticles. Microchim Acta 187, 192 (2020). https://doi.org/10.1007/s00604-020-4148-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-4148-4