Abstract
A fluorometric method is described for nucleic acid signal amplification through target-induced catalytic hairpin assembly with DNA-templated copper nanoparticles (Cu NPs). The toehold-mediated self-assembly of three metastable hairpins is triggered in presence of target DNA. This leads to the formation of a three-way junction structure with protruding mononucleotides at the 3′ terminus. The target DNA is released from the formed branched structure and triggers another assembly cycle. As a result, plenty of branched DNA becomes available for the synthesis of Cu NPs which have fluorescence excitation/emission maxima at 340/590 nm. At the same time, the branched structure protects the Cu NPs from digestion by exonuclease III. The unreacted hairpins are digested by exonuclease III, and this warrants a lower background signal. The method can detect ssDNA (24 nt) at low concentration (44 pM) and is selective over single-nucleotide polymorphism. On addition of an aptamer, the strategy can also be applied to the quantitation of thrombin at levels as low as 0.9 nM.

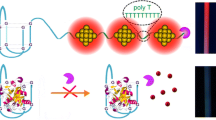
Schematic representation of target-induced catalytic hairpin assembly to form branched DNA template for the in situ synthesis of fluorescent Cu nanoparticles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nucleic acid are promising biomaterials in well-defined nanostructure construction [1]. Utilizing the “bottom-up” self-assembly strategy, several different kinds of dimension DNA nanostructure are obtained that with distinct advantage in vivo and vitro application [2]. Such as the long nicked double-stranded DNA with repeating units, a typical one dimension DNA structure, is obtained through the cascade hybrid chain reaction (HCR) between two metastable hairpins [3], which benefit the signal amplification and payload delivery [4]. The other impressing framework structures, including two-dimension DNA origami and three-dimension DNA tetrahedron, are also favored by researchers for their extraordinary performance in interfacial engineering and molecular recognition [5]. However, to implement all these multifunctional applications, an appropriate labeling is required, which resulting high-cost and time-consuming purification process. Thus, a more efficient method for the fabrication of functional DNA nanostructure is highly desirable.
DNA metallization is an effective approach for functional DNA materials fabrication [6]. Through the in situ reduction, various kinds of metal nanomaterials are formed on the DNA template. It is worth noting that the metal nanoclusters possess unique optical, electrical and biological properties that enabling the widely-range application in the field of sensing, catalysis and imaging [7]. More importantly, recent studies reveal that there is a significant correlation between these properties and the structure of DNA template [8]. Inspired by the assembly of dendrimer-like DNA structure, as a basic building blocks, the branched DNA structure can construct a multifunctional materials in a “plug-and-play” approach as well as stabilize the metal nanocluster [9,10,11]. Based on the classic branched DNA, the silver nanoclusters (Ag NCs) were successfully synthesized on the X-shape and Y-shape DNA [12]. With adding the crossing-linker, the temperature responsive fluorescent hydrogel was formed through the assembly of Y-shape Ag NCs [13]. The Ag-NC hydrogel was also exhibited tunable fluorescent properties by changing the nucleated sequences in Y-shape template. Owing to the advantage of branch structure materials in spatial distribution, the dendrimetric DNA structure was chose as scaffolds for the precisely arrangement of Ag NCs [14]. These super-Ag NCs were not only possess tunable fluorescent properties but also shown excellent biocompatibility with tissue cells. To date, all these metallized branched DNA structures are based on Ag NCs in which other metal nanoparticles are rarely reported.
In addition, compared with the statically regulation, the DNA nanostructure constructed through the dynamic self-assembly is more attractive in sensing application [15]. Especially, considering the sensitivity of the method, many hairpins assemble into long ds DNA polymer via HCR process is an effective method to obtain signal enhancement [16]. Embedding the template sequence in the hairpins, the Ag NC functional nanowires were formed as signal tag for the ultrasensitive detection of protein and micro RNA [17, 18]. Nevertheless, more than two hours incubation time for the synthesis of Ag NC is the big challenge for quick detection [19]. In that case, due to its quick preparation, DNA-templated copper nanoparticles (Cu NPs) is an alternative signal indicator for the fluorescent bioassay [20,21,22]. It is reported that the product of HCR is suitable as the efficient template for the synthesis of Cu NPs [23]. To reduce the background signal, an extra magnetic separation step is needed to remove the hairpins that do not take part in the HCR process [24].
Herein, we have constructed a target-induced catalytic hairpin assembly (CHA) to form branched DNA structure for the synthesis of Cu NPs. The rational design of the three metastable hairpins guarantee the in situ formation of Cu NPs through the toehold mediate strand displacement reaction (SDR) in presence of exonuclease III. The signal amplification mechanism of CHA is also benefit from the selective digestion of exonuclease III that enabling lower background signal. Furthermore, the synthesis strategy of the Cu NPs can be applied for protein detection through regulating the active of toehold domain. To the best of our knowledge, this is the first time to synthesis Cu NPs based on branched DNA structure, which formed through dynamic DNA nanotechnology.
Materials and methods
Reagents and apparatus
All oligonucleotides used in this study were synthesized and ULTRAPAGE purified by Sangon Biotech Co., Ltd. (http://www.sangon.com Shanghai, China). Sequences of the synthesized oligonucleotides are given in Supporting Information Table S1. Thrombin, trypsin, casein, bovine serum albumin, lysozyme, and 3-(N-morpholino) propanesulfonic acid (MOPS) were purchased from Sigma-Aldrich (https://www.sigmaaldrich.com/ St. Louis, MO). Exonuclease III (Exo III) ordered from Thermo Fisher Scientific Inc. (https://www.thermofisher.com/ Shanghai, China). All chemical reagents were of analytical grade and used without further purification. All solutions were prepared with ultra-pure water (18.25 MΩ·cm) from a Millipore system. Transmission electron microscope (TEM) was measurement performed on JEM-2100 instrument (https://www.jeol.co.jp/en/ JEOL Ltd., Japan) at an acceleration voltage of 200 kV. The UV spectrum was recorded on a TU-1901 spectrometer (http://www.pgeneral.com/Persee, Beijing, China). The fluorescence spectrum was measured on a RF-6000 spectrometer (http://www.pgeneral.com/Shimadzu, Japan) with 10 nm band-pass spectrometer slits and scanning rate at 600 nm per min in the range from 510 to 640 nm. X-ray photoelectron spectra (XPS) was recorded on an ESCALAB 250Xi system (Thermo Scientific). Gels were imaged by using a ChemiDoc X-ray diffraction (XRD) system (Bio-Rad). All optical measurements performed at room temperature under ambient conditions, and the fluorescence emission intensity measured at 590 nm with excitation wavelength at 340 nm.
Preparation of fluorescent cu NPs on the branched DNA template
Three hairpin strands (H1, H2, and H3) were annealed in reaction buffer (20 mM Tris-HCl,100 mM NaCl, 25 mM KCl, 10 mM MgCl2, pH = 8.0) at 95 °C for 5 min and cooled down to room temperature for further use over 4 h. 10 μL of target DNA at different concentrations was incubated with 30 μL of three annealed hairpin strands (H1, H2, and H3, each was 700 nM) in 95 μL reaction buffer at 25 °C for 60 min. Subsequently, 5 μL Exo III (20 U μL−1) was added to the mixture at 25 °C for another 40 min incubation. After enzyme digestion, the products were diluted to 380 μL with MOPS buffer (10 mM MOPS, 150 mM NaCl, pH 7.6). Finally, the 2 mM ascorbate and 200 μM Cu2+ were added to give a final volume of 400 μL and was allowed to react 10 min before fluorescence measurement.
Procedure for the thrombin detection
Pretreated H1, H2, and H3 hairpin strands were prepared as above. For the thrombin detection, 100 nM probe was mixed with 400 nM Block DNA in 25 μL of reaction buffer at room temperature for 30 min. Then, 40 μL of thrombin at different concentrations was added to the mixture and incubated at 37 °C for 60 min. Then, three annealed hairpin strands (10 μL of each strands) were added to the above solution and was incubated for another 60 min. Subsequently, 5 μL Exo III (20 U μL−1) was added to the mixture at 25 °C and was incubation for 40 min. After enzyme digestion, the products were diluted to 380 μL with MOPS buffer (10 mM MOPS, 150 mM NaCl, pH 7.6). Finally, the 2 mM ascorbate and 200 μM Cu2+ were added to give a final volume of 400 μL and was allowed to react 10 min before fluorescence measurement.
Agarose gel electrophoresis
Gel electrophoresis analysis was performed on 3% (w/w) agarose gels containing 0.5 μg mL−1 SYBR Gree I and 0.5 μg mL−1 bromophenol blue running in 1× TAE buffer at room temperature. The electrophoresis was performed at a constant potential of 100 V for 50 min after loading 10 μL of each sample into the lanes.
DNA and thrombin detection in human serum
For the real biological sample analysis, the human serum was diluted 100 folds with reaction buffer. Before the detection procedure, different concentrations of DNA and thrombin were spiked in the diluted serum. Three duplicate measurements were performed for all samples. All analytical procedures were identical to those mentioned above.
Results and discussions
Synthesis and characterization of cu NPs
The principle of the designed strategy is illustrated in Scheme 1A. Three metastable hairpins, with protruding 5′ ends and recessed 3′ ends, are selected as the building blocks to form the branched DNA structure through toehold mediated catalytic hairpin assembly. A DNA segment with toehold domain is used as a proof-of-concept nucleic acid target. The domain a* of target hybridizes to the toehold of H1 that initiate branch migration through the SDR, resulting the Target-H1 complex where the domain b* of H1 is exposed. Then, the exposed domain b* hybridizes to the toehold of H2, which opens the H2 and reveals the domain c* of H2. Through the exposed domain c*, the H3 is proceed hybridized and displaced the target to form a three way-junction structure (3WJ). Finally, the released target triggers a new cycle of catalytic assembly and much more 3WJ structures are formed. The formed 3WJ structures with protruding 3′ ends that inhibit the digestion of ds DNA in presence of Exo Ш [25]. By sequentially adding ascorbate and copper ions, the fluorescent Cu NPs can be synthesized in situ on these 3WJ structures. In contrast, in the absence of target, three metastable hairpins are hydrolyzed by Exo Ш and no Cu NPs are formed.
To verify the successfully synthesis of Cu NPs on the branched DNA structure, a series of characterization was performed on the nanoparticles. As shown in Fig. 1a, the TEM image represents the Cu NPs with uniform morphology and well monodisperse. The high-resolution TEM image exhibits well crystal lattice structure of Cu NPs at a magnified observation. According to calculate the size distribution, the Cu NPs show narrow size distribution in the range from 1.68 nm to 4.25 nm with an average size of 2.28 ± 0.56 nm (Fig. 1b). The branched DNA template, the three way-junction structure, bears three dsDNA domains that available for the synthesis of typical dsDNA-templated fluorescent Cu NPs [20]. As a consequence, the synthesized Cu NPs present an obvious red emission at 590 nm when excited at the 340 nm absorbance band, which are consistent with previous reports (Fig. 1c) [26]. The Cu oxidation state was analysis by the X-ray photoelectron spectroscopy (XPS). As depict in Fig. 1d, two peaks at 932 and 952 eV are assign to the feature 2p3/2 and 2p1/2 binding energy of Cu atom, respectively, which indicate the successfully synthesis of Cu NPs [27].
The well-fabricated 3WJ structure is the premise of the synthesis of Cu NPs. Agarose gel electrophoresis analysis was carried out to investigate the mechanism of catalytic hairpin assembly for the construction of branched DNA template (Fig. S1). The 3WJ structure is formed through the toehold mediated strand displacement cascade reaction with three hairpins as the fuel strand. The gel results confirmed that, in presence of target, the 3WJ structure were formed when three hairpins were present, which resulting a slowly motion rate of the smear band (lane 4). There is no different of the motion rate between the other two intermediate complexes of the CHA reaction (lane 2 and lane 3). On the contrary, in the absence of target, the motion rate of three hairpins (lane 5) is the same as that of H1 only (lane 1), which illustrate the branched DNA structure were rarely formed. Due to the vulnerable system of multiple strand displacement, a minimal leakage phenomenon is present in lane 5, which resulting from the undesirable cross-catalysis [28].
Branched DNA-templated cu NPs formation triggered by nucleic acid
Owing to the high efficiency signal amplify of CHA, the sensitive detection of target DNA is benefit from the synthesis strategy of branched DNA-templated Cu NPs. To study the feasibility of nucleic acid sensing, the fluorescence responsive of this system at different conditions were investigated. As shown in Fig. 2a, in present of three hairpins, this system exhibits strong fluorescence emission at 590 nm (curve a). Upon the addition of Exo Ш, the fluorescence intensity was significant decreased, which indicated the hairpin templates were digested (curve b). In present of target, the toehold mediated CHA was triggered and formed intermediate complex of T-H1-H2, resulting a slightly fluorescence recovery (curve c). A dramatic fluorescence enhancement was obtained when three hairpins and target were co-existence in the sensing system (curve d), which demonstrate the branched DNA structure protected Cu NPs from the destruction of Exo Ш. On the other hand, the background signal of the sensing system was decreased by Exo Ш and a higher sensitivity was gained. All these results suggested that the synthesis of Cu NPs on the branched DNA template was available for the detection of trigger DNA via signal amplification.
a The fluorescence responsive of the Cu NPs synthesis under different conditions. a H1 + H2 + H3; b H1 + H2 + H3 + Exo Ш; c T + H1 + H2 + Exo Ш; d T + H1 + H2 + H3 + Exo Ш. b Fluorescence emission spectra at different target concentrations (0, 0.1, 0.5, 1, 1.5, 2, 5, 10, 15, and 20 nM) c Relationship between the ΔF and the concentration of Target. The inset shows the linear relationship over the concentration range from 0.1 to 2 nM. d Selectivity of target DNA analysis. Concentration of H1, H2, and H3: 700 nM; Exo Ш concentration: 100 U; the concentration of target DNA and other mismatch strands were all 2 nM
In order to achieve the best performance of the sensing system, several experimental conditions including the reaction temperature and time of the CHA and the concentration of hairpin strands and Exo Ш were optimized. This toehold mediated strand displacement reaction is highly dependent on the incubation temperature, which accelerate the reaction efficiency as well as reduce the undesirable cross-catalysis between hairpins. As shown in Fig. S2, five typical temperatures commonly used in nucleic acid bioassay were chose to investigate the fluorescence responsive. The maximized fluorescence responsive was achieved at 25 °C, and further increased temperature would affect the stability of unreactive hairpins leading higher background signal. According to the fluorescence responsive of sensing system, the time of CHA and the concentration of hairpins were set as 60 min and 700 nM, respectively. In our design, due to the highly selective of that toward a blunt or recessed 3′ terminus, the Exo Ш do not can only discriminate the hairpin-templated and branch structure-templated Cu NPs but also effective reduce the background signal. Thus, 100 U Exo Ш was selected for the further experiment. Under the optimal experimental condition, the sensitivity of the method was investigated. As shown in Fig. 2b, the fluorescence intensity gradually increased with increasing the concentration of target, indicating that abundant branched DNA-templated Cu NPs were formed. The calibration plot obtained for the determination of target DNA with different concentrations is shown in Fig. 2c, and a liner relationship between the difference of fluorescence intensity (ΔF) and the concentration of target DNA from 0.1 to 2.0 nM (R2 = 0.9848) was obtained. The detection limit was calculated to be 44 pM according to the signal-to-noise ratio of three. The selectivity of the method was evaluated by detecting single nucleotide polymorphism (Fig. 2d), only the target DNA resulted significant fluorescence responsive than that of other mismatch strands. It can be concluded that method has capability to distinguish even single base mismatch. Considering the sample matrix effect, the recovery of the assay was detected in the spike human serum samples. Three concentrations of the target (0.5, 1 and 2 nM) were spiked into 1% human serum. The recovery values were in the range of 99.01–114.57% (Table S2), indicating that the method is capable of analyzing real biological samples without interference.
Toehold regulate the formation of cu NPs for the detection of thrombin
Hitherto, many reports have been demonstrated that the toehold domain of trigger DNA is the key factor that control the kinetics of strand displacement reaction [29]. It is supposed that control the activation of the toehold would realize the non-nucleic acid target sensing through allosterically regulated the probe. As shown in Scheme 2, a sequence of thrombin aptamer was embedded in the 3′ end of the trigger DNA. To inactive the toehold domain, in the absence of thrombin, a blocker DNA was designed to make them double strand. Formation of aptamer-thrombin complex can active the toehold domain of thrombin probe and trigger the subsequent CHA process to form fluorescent Cu NPs.
To validate our design, the typical fluorescence profile of sensing system was recorded with different trigger inputs. As shown in Fig. 3a, an obvious florescence responsive was obtained with the probe sequence as an input, which indicating the probe enable to trigger the CHA and form Cu NPs (curve a). When the blocker was added to make double strand, the hybridization of complementary domain served to inactive the toehold that resulting in a decrease in fluorescence intensity (curve b). Upon the addition of thrombin, the duplex DNA was disturbed by the thrombin-aptamer complex and the reactive toehold was further induced dramatic fluorescence enhancement (curve c). These results demonstrate this toehold mediated CHA induced Cu NPs synthesis can be expanded to the thrombin detection.
a The fluorescence profile of the sensing system. a Probe; b Probe+Bloker-2; c Probe+Bloker-2 + 15 nM thrombin. b Fluorescence emission spectra at different thrombin concentrations (0, 1, 2, 5, 8, 10, 15, 20, 35, and 50 nM) c Relationship between the ΔF and the concentration of thrombin. The inset shows the linear relationship over the concentration range from 1 to 20 nM. d Selectivity of thrombin analysis. Concentration of Probe and Blocker-2 were 100 and 400 nM, respectively; Concentration of H1, H2, and H3: 700 nM; Exo Ш concentration: 100 U; the concentration of thrombin and other proteins were 15 nM and 150 nM, respectively
To achieve better signal to background ratios, the base length of the blocker and the molar ratio between the probe sequences and blocker were optimized (Fig. S3). Under the optimal condition, the fluorescence responsive of sensing system was increased with the increasing concentration of thrombin in the range from 1 to 50 nM (Fig. 3b). The linear regression equation can be expressed as follows: F (a. u.) = 22.76 Cthrombin-10.22 with a correlation coefficient of 0.996 (Fig. 3c). Additionally, the detection limit of the present strategy for thrombin was estimated to be as low as 0.9 nM according to the 3σ criteria. Compare with other reported methods, our strategy shows comparable analysis performance (Table 1). Especially for the materials used in the reference method, most of them require time-consuming conjugation process and complex synthesis environment (high temperature, organic solvent, etc.) that restricting the application of such methods. As contract, branched DNA templated Cu NPs shows advantage in high-efficient preparation, good fluorescent properties, and environmental friendliness. The last but the most, this branched DNA templated Cu NPs is convenient to integrate with other recognition unit, such as aptamer or DNAzyme based recognition capability and entropy drove signal amplification. Furthermore, four kinds of protein with 10-fold concentration than that of thrombin were selected for exploring the selectivity of the method. As shown in Fig. 3d, the more fluorescence enhancement was achieved on thrombin than that of other proteins, which demonstrate the selective recognition for the thrombin of this method. The recovery assay was also studied in the spike human serum samples. Three concentrations of the thrombin (10, 15 and 20 nM) were spiked into 1% human serum. The satisfactory recovery values were obtained in the range of 103.34–112.63% (Table S3) that revealed the tolerability of this method toward the interference from real biological samples. Despite good analytical performance of our method, the catalytic hairpin assembly process is still time-consuming due to its highly dependent on the local concentration of hairpin. Thus, to short the analysis time, the signal amplify method with high efficiency is yet needed to investigate in further work.
ECL: electrochemiluminesence; GO: graphene oxide; 3D-NGO: Three-dimensional nitrogen-doped graphene oxide.
Conclusions
A dynamic hairpin assembly to form a branched DNA template for the synthesis of fluorescent Cu NPs has been constructed. The branched DNA structure can be readily synthesized by a DNA segment triggered catalytic hairpin assembly of three metastable hairpins. Taking advantage of rationally design, with the aid of Exo Ш, the branched DNA templated Cu NPs can also be used as an indicator for the detection of nucleic acid and protein via signal amplification. This strategy also exhibits excellent performance in real biological sample detection. Given that the branched DNA template employed in this strategy can be flexible design, its combing with functional nucleic acid should enable the preparation of fluorescent DNA hydrogel. Moreover, in situ controlled synthesis of metal NPs on DNA nanostructures would ultimately pave the new way to explore the energy transfer and DNA circuits.
References
Li J, Green AA, Yan H, Fan C (2017) Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat Chem 9(11):1056–1067. https://doi.org/10.1038/nchem.2852
Chen YJ, Groves B, Muscat RA, Seelig G (2015) DNA nanotechnology from the test tube to the cell. Nat Nanotechnol 10(9):748–760. https://doi.org/10.1038/nnano.2015.195
Yang D, Tang Y, Miao P (2017) Hybridization chain reaction directed DNA superstructures assembly for biosensing applications. TrAC Trends Anal Chem 94:1–13. https://doi.org/10.1016/j.trac.2017.06.011
Bi S, Yue S, Zhang S (2017) Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem Soc Rev 46(14):4281–4298. https://doi.org/10.1039/c7cs00055c
Yang F, Zuo X, Fan C, Zhang X-E (2018) Biomacromolecular nanostructures-based interfacial engineering: from precise assembly to precision biosensing. Natl Sci Rev 5(5):740–755. https://doi.org/10.1093/nsr/nwx134
Chen Z, Liu C, Cao F, Ren J, Qu X (2018) DNA metallization: principles, methods, structures, and applications. Chem Soc Rev 47(11):4017–4072. https://doi.org/10.1039/c8cs00011e
Chen Y, Phipps ML, Werner JH, Chakraborty S, Martinez JS (2018) DNA Templated metal Nanoclusters: from emergent properties to unique applications. Acc Chem Res 51(11):2756–2763. https://doi.org/10.1021/acs.accounts.8b00366
New SY, Lee ST, Su XD (2016) DNA-templated silver nanoclusters: structural correlation and fluorescence modulation. Nanoscale 8(41):17729–17746. https://doi.org/10.1039/c6nr05872h
Lee JB, Roh YH, Um SH, Funabashi H, Cheng W, Cha JJ, Kiatwuthinon P, Muller DA, Luo D (2009) Multifunctional nanoarchitectures from DNA-based ABC monomers. Nat Nanotechnol 4(7):430–436. https://doi.org/10.1038/nnano.2009.93
Meng HM, Zhang X, Lv Y, Zhao Z, Wang NN, Fu T, Fan H, Liang H, Qiu L, Zhu G, Tan W (2014) DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano 8(6):6171–6181. https://doi.org/10.1021/nn5015962
Schultz D, Gardner K, Oemrawsingh SS, Markesevic N, Olsson K, Debord M, Bouwmeester D, Gwinn E (2013) Evidence for rod-shaped DNA-stabilized silver nanocluster emitters. Adv Mater 25(20):2797–2803. https://doi.org/10.1002/adma.201204624
Park J, Song J, Park J, Park N, Kim S (2014) Branched DNA-based synthesis of fluorescent silver Nanocluster. Bull Kor Chem Soc 35(4):1105–1109. https://doi.org/10.5012/bkcs.2014.35.4.1105
Guo W, Orbach R, Mironi-Harpaz I, Seliktar D, Willner I (2013) Fluorescent DNA hydrogels composed of nucleic acid-stabilized silver nanoclusters. Small 9(22):3748–3752. https://doi.org/10.1002/smll.201300055
Yang L, Yao C, Li F, Dong Y, Zhang Z, Yang D (2018) Synthesis of branched DNA Scaffolded Super-Nanoclusters with enhanced antibacterial performance. Small 14(16):e1800185. https://doi.org/10.1002/smll.201800185
Qing Z, He X, Huang J, Wang K, Zou Z, Qing T, Mao Z, Shi H, He D (2014) Target-catalyzed dynamic assembly-based pyrene excimer switching for enzyme-free nucleic acid amplified detection. Anal Chem 86(10):4934–4939. https://doi.org/10.1021/ac500834g
Wang H, Li C, Liu X, Zhou X, Wang F (2018) Construction of an enzyme-free concatenated DNA circuit for signal amplification and intracellular imaging. Chem Sci 9(26):5842–5849. https://doi.org/10.1039/c8sc01981a
Chen L, Sha L, Qiu Y, Wang G, Jiang H, Zhang X (2015) An amplified electrochemical aptasensor based on hybridization chain reactions and catalysis of silver nanoclusters. Nanoscale 7(7):3300–3308. https://doi.org/10.1039/c4nr06664b
Yang C, Shi K, Dou B, Xiang Y, Chai Y, Yuan R (2015) In situ DNA-templated synthesis of silver nanoclusters for ultrasensitive and label-free electrochemical detection of microRNA. ACS Appl Mater Interfaces 7(2):1188–1193. https://doi.org/10.1021/am506933r
Orbach R, Guo W, Wang F, Lioubashevski O, Willner I (2013) Self-assembly of luminescent Ag nanocluster-functionalized nanowires. Langmuir 29(42):13066–13071. https://doi.org/10.1021/la402888b
Liu R, Wang C, Hu J, Su Y, Lv Y (2018) DNA-templated copper nanoparticles: versatile platform for label-free bioassays. TrAC Trends Anal Chem 105:436–452. https://doi.org/10.1016/j.trac.2018.06.003
Han Y, Zhang F, Gong H, Cai C (2018) Double G-quadruplexes in a copper nanoparticle based fluorescent probe for determination of HIV genes. Microchim Acta 186(1):30. https://doi.org/10.1007/s00604-018-3119-5
Kim S, Kim JH, Kwon WY, Hwang SH, Cha BS, Kim JM, Oh SS, Park KS (2019) Synthesis of DNA-templated copper nanoparticles with enhanced fluorescence stability for cellular imaging. Microchim Acta 186(7):479–475. https://doi.org/10.1007/s00604-019-3620-5
Zhang Y, Chen Z, Tao Y, Wang Z, Ren J, Qu X (2015) Hybridization chain reaction engineered dsDNA for cu metallization: an enzyme-free platform for amplified detection of cancer cells and microRNAs. Chem Commun (Camb) 51(57):11496–11499. https://doi.org/10.1039/c5cc03144c
Song C, Yang X, Wang K, Wang Q, Huang J, Liu J, Liu W, Liu P (2014) Label-free and non-enzymatic detection of DNA based on hybridization chain reaction amplification and dsDNA-templated copper nanoparticles. Anal Chim Acta 827 (0):74–79. doi:https://doi.org/10.1016/j.aca.2014.04.006
Chen J, Zhou S (2016) Label-free DNA Y junction for bisphenol a monitoring using exonuclease III-based signal protection strategy. Biosens Bioelectron 77:277–283. https://doi.org/10.1016/j.bios.2015.09.042
Cao Q, Li J, Wang E (2019) Recent advances in the synthesis and application of copper nanomaterials based on various DNA scaffolds. Biosens Bioelectron 132:333–342. https://doi.org/10.1016/j.bios.2019.01.046
Wang Z, Y-e S, Yang X, Xiong Y, Li Y, Chen B, Lai W-F, Rogach AL (2018) Water-soluble biocompatible copolymer Hypromellose grafted chitosan able to load exogenous agents and copper Nanoclusters with aggregation-induced emission. Adv Funct Mater 28(34):1802848. https://doi.org/10.1002/adfm.201802848
Wang B, Thachuk C, Ellington AD, Winfree E, Soloveichik D (2018) Effective design principles for leakless strand displacement systems. Proc Natl Acad Sci U S A 115(52):E12182–E12191. https://doi.org/10.1073/pnas.1806859115
Zhang DY, Seelig G (2011) Dynamic DNA nanotechnology using strand-displacement reactions. Nat Chem 3(2):103–113
Xu N, Wang Q, Lei J, Liu L, Ju H (2015) Label-free triple-helix aptamer as sensing platform for “signal-on” fluorescent detection of thrombin. Talanta 132:387–391. https://doi.org/10.1016/j.talanta.2014.09.031
Li L, Liang Y, Zhao Y, Chen Z (2018) Target binding and DNA hybridization-induced gold nanoparticle aggregation for colorimetric detection of thrombin. Sensors Actuators B Chem 262:733–738. https://doi.org/10.1016/j.snb.2018.02.061
Chung S, Moon J-M, Choi J, Hwang H, Shim Y-B (2018) Magnetic force assisted electrochemical sensor for the detection of thrombin with aptamer-antibody sandwich formation. Biosens Bioelectron 117:480–486. https://doi.org/10.1016/j.bios.2018.06.068
Sui N, Wang L, Xie F, Liu F, Xiao H, Liu M, Yu WW (2016) Ultrasensitive aptamer-based thrombin assay based on metal enhanced fluorescence resonance energy transfer. Microchim Acta 183(5):1563–1570. https://doi.org/10.1007/s00604-016-1774-y
Khonsari YN, Sun S (2018) Electrochemiluminescent aptasensor for thrombin using nitrogen-doped graphene quantum dots. Microchim Acta 185(9):430–410. https://doi.org/10.1007/s00604-018-2942-z
Wang L, Yang W, Li T, Li D, Cui Z, Wang Y, Ji S, Song Q, Shu C, Ding L (2017) Colorimetric determination of thrombin by exploiting a triple enzyme-mimetic activity and dual-aptamer strategy. Microchim Acta 184(9):3145–3151. https://doi.org/10.1007/s00604-017-2327-8
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (31801636), National Key Research and Development Program of China (2017YFC1600603),, and Shanghai Sailing Program (Grant No. 18YF1417300).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1035 kb)
Rights and permissions
About this article
Cite this article
Ye, T., Peng, Y., Yuan, M. et al. Target-induced in-situ formation of fluorescent DNA-templated copper nanoparticles by a catalytic hairpin assembly: application to the determination of DNA and thrombin. Microchim Acta 186, 760 (2019). https://doi.org/10.1007/s00604-019-3927-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3927-2