Abstract
Aminopropyltrimethoxysilane (APTMS)-functionalized zinc oxide (ZnO) nanorods and carboxylated graphene nanoflakes (c-GNF) were used in a composite that was electrophoretically deposited on an indium tin oxide (ITO) coated glass substrate. The modified ITO electrodes were characterized using various microscopic and spectroscopic techniques which confirm the deposition of the APTMS-ZnO/c-GNF composite. The electrodes have been used for the covalent immobilization of an Escherichia coli O157:H7 (E. coli)-specific DNA prob. Impedimetric studies revealed that the gene sensor displays linear response in a wide range of target DNA concentration (10−16 M to 10−6 M) with a detection limit of 0.1 fM. The studies on the cross-reactivity to other water-borne pathogens show that the bioelectrode is highly specific.

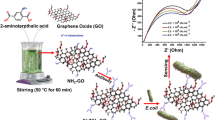
Schematic illustration for fabrication of nucleic acid biosensor for E. coli DNA detection using an ITO electrode modified with siloxane-functionalized zinc oxide (ZnO) nanorods and carboxylated graphene nanoflakes (c-GNFs).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nanostructured materials have gathered interest as functional materials in various aspects of designing biomedical devices [1,2,3,4,5]. Nanostructured metal oxides (NMOs) have been widely used in biosensing applications due to their various characteristics, such as large surface to volume ratio, high electrocatalytic activity, their small band gap that enhances their electron conductivity and the adsorption ability [6] which helps in increasing the loading of biomolecules per unit mass of particles. The employment of NMOs may open the possibility of new signal transduction due to their sub-micrometer dimension and can be useful for in vivo analysis. The NMOs reported to have their potential application in the field of biosensor are of zinc, iron, cerium, tin, zirconium, titanium and magnesium [7]. Among these zinc oxide (ZnO), has been widely used for the development of various sensing platforms. ZnO has gained much interest due to its novel characteristics such as biocompatibility, catalytic efficiency, non-toxicity, as well as chemical and electrical stability [8,9,10,11]. ZnO shows various nanostructures of different morphologies such as nanowires, nanoflakes, nanorods, nanobelts, nanoneedles and nanocombs [12,13,14,15,16].
Though NMOs have shown excellent properties as materials for biosensing fabrication, but there are some limitations such as low conductivity as compared to carbon materials, low surface area and high resistance. So, it will be logical to develop a composite material containing both carbon and transition metal oxide as the electrode material, combining the merits and mitigating the shortcomings of both the individual components [17, 18]. The use of graphene in nanostructured carbon-metal oxides composites not only provides physical support to metal oxides but also serve as channels for charge transport due to its high surface area and the robust conductive structure. In graphene-metal oxide composite, the graphene provides the benefit of high electron conductivity while electro-activities of metal oxides contribute to high specific capacitance. The performance of biosensors based on such materials is governed by the interface between the nanostructured composite material and biomolecule [19]. The nano bio-interface formation and its properties depend upon nature of the nanostructured material and also other parameters related to effective surface area, roughness, porosity, conductance, functional groups etc. The biomolecules are said to bind the metal nanocomposites generally by physical adsorption or chemical binding [20] and the activity of the biomolecule can be retained by establishing a biocompatible microenvironment on the biointerface [21, 22]. Thus the use of nanostructured composite material would decide the sensitivity, selectivity, detection limit, cost and shelf-life of the biosensor.
The enter-hemorrhagic stereotype of bacterium Escherichia coli is E. coli O157:H7 and its infection can lead to kidney failure and hemorrhagic diarrhea [23]. Various methods such as culture and colony counting are available for the detection of the pathogen. The traditional methods require an excess of time and the results are also ambiguous [24]. Some methods involve enrichment of culture by growing it for a specific time and then separated and identified using polymerase chain reaction or immunological methods of identifying genes. These methods are quite complicated and costly in terms of requirement of labeled antibodies and their pre-separation or pre-enrichment. Thus, there is an urgent requirement for a rapid, sensitive and cost-effective technique to overcome these problems. An alternative technique to the methods mentioned above is the use of biosensors for pathogenic bacterial genome detection [25, 26].
The biosensors based on nanostructured composite materials for nucleic acid detection have aroused much more interest due to their efficiency of converting the hybridization event into an analytical signal. Wang et al. reported a biosensor based on ZnO nanowires, multiwalled carbon nanotubes (MWCNT) and gold nanoparticles composite utilizing thiolated single-stranded DNA probe to immobilize it on the gold nanoparticle surface. The detection limit of this DNA sensor was found to be 0.35 pM using [Ru(NH3)6]3+ as a redox indicator [27]. Sol-gel derived nanostructured ZrO2 film for the detection of 16 s rRNA coding region for E.coli has been reported with a sensitive determination in the range of 10−6 to 106 pM of complementary DNA [28]. Yang et al. reported a composite based on nano ZrO2, Chitosan and MWCNT for the development of DNA sensor on glassy carbon electrode. The composite provided good charge transport characteristics with high surface area favoring the increase in loading capacity and enhanced selectivity with a detection limit of 75 pM [29]. For point of care analysis, an ideal detection method should be such that it may detect the disease directly from food and water samples with minimal requirement of sample preparation. However, the drawbacks with nucleic acid based biosensor are that the extraction of DNA from culture samples and processing involve laborious pre–treatment steps. Thus efforts are being made to develop affinity based biosensors (using antibody–antigen interactions) and aptasensors to allow the detection of E. coli O157:H7 cells directly from food and water samples but till date they are not very applicable and have not been fully developed into technology. Hence, it becomes imperative to design such materials which can overcome these drawbacks.
The present work reports studies conducted to exploit the properties of ZnO nanorods by combining it with a new class of graphene material known as carboxylated graphene nanoflakes (c-GNF) [30]. The ZnO nanorods were firstly modified with aminopropyltrimethoxysilane (APTMS) to introduce NH2 groups on the surface of ZnO nanorods. Then the electrostatic interaction of this APTMS functionalized ZnO nanorods (APTMS-ZnO) with the carboxylic acid groups on c-GNF was done for the formation of APTMS functionalized ZnO nanorods-carboxylated graphene nanoflakes composite (APTMS-ZnO/c-GNF) composite. Further, the composite was electrophoretically deposited using magnesium (II) electrolyte which provided overall positive charge to the colloid suspension for the fabrication of impedimetric biosensor for E. coli DNA detection.
Chemicals and materials required
Zinc(II) nitrate hexahydrate (www.alfa.com), aminopropyltrimethoxysilane (www.sigmaaldrich.com), glutaraldehyde (www.spectrochem.co.in) and sodium hydroxide pellets (www.sdfine.com). The specific sequence of E. coli probe (17 mer.) has been identified from the 16 s rRNA coding region of the E. coli genome. The complementary, non-complementary and one-base mismatch target sequences were procured from Sigma Aldrich (www.sigmaaldrich.com). Sequences of oligonucleotide probes used are:
Probe I- probe DNA (pDNA): amino-5′-GGT CCG CTT GCT CTC GC-3′.
Probe II- complementary DNA (cDNA): 5’-GCG AGA GCA AGC GGA CC-3′.
Probe III- non-complementary DNA: 5’-CTA GTC GTA TAG TAG GC-3′.
Probe IV- one-base mismatch DNA: 5’-GCG AGA GAA AGC GGA CC-3′.
The solution of oligonucleotide was made in Tris–EDTA buffer (1 M Tris–HCl, 0.5 M EDTA, pH 8.0) (www.merck.com) and was stored at −20o C prior to use.
Apparatus and measurements
X-ray diffraction (XRD) analysis was performed using D8 Advance/Discover Bruker, Germany, Diffractometer with Cu Kα. The UV-vis spectra was recorded using Hitachi U-3900 spectrophotometer while the transmission electron microscopy (TEM) and energy dispersive X-ray analysis (EDX) were carried out using TECNAI 12 G2FEI microscope, operating at 120 kV instrument. FT-IR spectroscopic study was done using PerkinElmer spectrometer (Spectrum two, UK), the scanning electron microscopy (SEM) images were recorded using a JEOLJSM-6700F field-emitting scanning electron microscope (FESEM, 15 kV) while the contact angle was measured using contact angle meter (Data physics OCA15EC). Electrochemical characterization were carried out on an Autolab potentiostat/galvanostat (www.metrohm-autolab.com) using a three-electrode system with ITO as working electrode, Ag/AgCl as a reference electrode, and platinum foil as a counter electrode in phosphate buffer solution (PBS of 100 mM concentration with pH 7.0 in 0.9% NaCl).
Experimental section
Synthesis of APTMS functionalized ZnO nanorods
The synthesis of ZnO nanorods are discussed in section 1 of supp. Material [31]. For functionalization of the ZnO nanorods, APTMS was used. For amino functionalization of ZnO, 0.2 g ZnO powder was ultrasonicated in 20 mL toluene followed by addition of 400 μL of APTMS under nitrogen atmosphere in a round bottom flask and refluxed at 110 °C for 6 h to get APTMS functionalized ZnO nanorods (APTMS-ZnO) [32, 33].
Synthesis of APTMS functionalized ZnO nanorods-carboxylated graphene nanoflakes composite
The c-GNF were synthesized as reported in the literature [30]. A dispersion of c-GNF was made using DI water (0.5 mgmL−1) via ultrasonication for 2 h. Further 10 mg of APTMS-ZnO nanorods were added to the above dispersion and ultrasonicated for 2 h followed by subsequent stirring (4 h). Then the suspension was centrifuged at 10,000 rpm for 1 h to obtain APTMS-ZnO nanorods-c-GNF composite (APTMS-ZnO/c-GNF). Finally, the obtained material was washed with distilled water and then dried at 70 °C under vacuum (Scheme 1a ).
Electrophoretic deposition of APTMS-ZnO/c-GNF
Prior to deposition, the ITO coated glass electrode were hydrolyzed by immersing in H2O2/NH4OH/H2O solution (1:1:5, v/v) for about 30 min at 80 °C. For the deposition, APTMS-ZnO/c-GNF (5 mg) were dispersed in 10 mL acetonitrile. The electrophoretic deposition was carried using the DC source by applying a voltage of 120 V for 2 min. 10−4 M of Mg(NO3)2.6H2O was added to the colloidal suspension (APTMS-ZnO/c-GNF in acetonitrile) acting as an electrolyte (for creating a charge on the colloids).
Immobilization and hybridization of probe DNA on APTMS-ZnO/c-GNF/ITO electrode
The coupling of amine terminal of probe DNA (pDNA) and APTMS-ZnO/c-GNF/ITO electrode was done by immobilizing 20 μL of pDNA on the modified electrode using glutaraldehyde as cross linker (0.1%, v/v; 4 h). The fabricated pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode was employed for the detection of E. coli complementary DNA (cDNA). The different concentrations of pDNA and cDNA were prepared in Tris-EDTA buffer (1 M Tris−HCl, 0.5 M EDTA) of pH 8.0 from the stock solution of 1 × 10−6 M concentration. The pDNA/APTMS-ZnO/c-GNF/ITO was subjected to different concentrations of cDNA for 30 min and the corresponding Rct value was measured using electrochemical impedance spectroscopy. The different steps involved in the fabrication of APTMS-ZnO/c-GNF/ITO and pDNA/APTMS-ZnO/c-GNF/ITO has been depicted in scheme 1b.
Extraction and pretreatment of DNA from bacterial clinical samples
The extraction of DNA was carried out using the panel of strains comprising E. coli, Salmonella Typhimurium, Neisseria meningitides and Klebsiella pneumonia. The DNA was extracted by suspending the bacterial colonies and vortexing and pouring them into 100 mL of MilliQ water. The suspension was further boiled for 10 min and centrifuged at 10,000 rpm for 5 min and equal volume (100 mL) of 24:1 (v/v) chloroform–iso-amyl alcohol was added to the boiled and separated suspension. This solution was again centrifuged at 12,000 rpm for 10 min, which resulted in the formation of an aqueous layer containing DNA. This aqueous layer was carefully separated out and kept at -20 °C prior to use. For denatured single-stranded DNA, the DNA samples extracted from the above-mentioned method were heated on water bath (95 °C) for 5 min and were immediately chilled in ice. These aliquots of samples were subjected to sonication for 15 min at 120 V to break the long DNA strands into smaller fragments.
Results and discussion
Morphological characterization of the deposited materials
The morphology of the electrodeposited APTMS-ZnO/c-GNF on ITO electrode was studied using SEM (Fig. 1). The SEM image of APTMS-ZnO/c-GNF/ITO shows dendritic structure that might be due to the assembly of APTMS-ZnO nanorods with c-GNF on applying potential during the electrodeposition (Fig. 1a and b at different magnification). Even the rods of the ZnO have not lost its morphology and dendritic structure (Fig. 1c) would lead to a greater surface area of the APTMS-ZnO/c-GNF/ITO which will provide an excellent platform for pDNA attachment. While in the case of electrodeposition of APTMS-ZnO no such assembly is observed and smoothsheet-like morphology can be seen for APTMS-ZnO/ITO (Fig. 1d).
Electrochemical characterization
Electrochemical impedance spectroscopy
The electrochemical properties of the deposited material are evaluated using electrochemical impedance spectroscopic (EIS) studies since it is a powerful tool for observing the charge transfer processes at the electrode-electrolyte interface [25]. The impedimetric spectra are seen in Fig. 2a called as the Nyquist plot which consists of two parts one semicircular part at high frequency and the other linear plot at a lower frequency. The semicircular high-frequency part signifies the electron transfer mediated process. The linear lower frequency part corresponds to the diffusion-limited process. The impedimetric spectrum of the fabricated biosensor was fitted using a modified Randles equivalent circuit. This circuit is made up of different elements such as electrolyte resistance (Rs), Warburg impedance (ZW), double layer capacitance (CPE) and the electron transfer resistance (Rct). The Rs is the resistance which is observed between the working and the reference electrode. ZW is the element associated with the process at the electrode-electrolyte interface due to diffusion of redox probe ions. CPE is related to the double layer capacitance. The numerical value of Rct is helpful in deriving the electrochemical response of the fabricated biosensor. The measurements of the impedance were made to carry out at the open circuit voltage and the test frequency was in the range of 100 kHz to 0.1 Hz with amplitude of ±5 mV [25].
(a) Nyquist diagram (Zi vs. Zr) for the Faradic impedance in the frequency range from 100 kHz to 0.1 Hz for (i) APTMS-ZnO/c-GNF/ITO electrode, (ii) pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode, and (iii) cDNA on pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode., (b) Cyclic voltammogram of APTMS-ZnO/c-GNF/ITO electrode and (c) pDNA/APTMS-ZnO/c-GNF/ITO electrode (in 5 mM [Fe(CN)6]3−/4-, PBS, 100 mM, pH 7.0, 0.9% NaCl)
The EIS technique was adopted to investigate the electrochemical behavior of APTMS-ZnO/c-GNF/ITO electrode, pDNA/APTMS-ZnO/c-GNF/ITO electrode and pDNA/APTMS-ZnO/c-GNF/ITO electrode incubated with E. coli cDNA by comparing their Rct values (Fig. 2a). The semicircular region of the impedance curve reflects the numerical value of Rct, derived by fitting a Randles and Ershler model circuit. Also, the presence of a functional group on the composite material APTMS-ZnO/c-GNF might chemisorb the complementary target of the oligonucleotide. To avoid the false positive signal due to chemisorption, there is a need to optimize the concentration of pDNA. The optimized concentration of pDNA was found to be 10−6 M, which showed the leveling of the Rct value (optimization data not shown).
Cyclic voltammetry
The cyclic voltammetry studies were used to investigate the electrocatalytic behavior of the fabricated electrodes towards the redox behavior of [Fe(CN)6]3−/4- [25].The outcome of the cyclic voltammetry as the function of scan rate (υ) for APTMS-ZnO/c-GNF/ITO and pDNA/APTMS-ZnO/c-GNF/ITO can be seen in Fig. 2b and c. The anodic and cathodic peak corresponds to oxidation and reduction of [Fe(CN)6]3−/4- redox couple respectively. The anodic and cathodic peak currents were found to vary linearly with υ in the range of 10 to 300 mVs−1 indicating the electrochemical reaction of the redox pair at the working electrode to be diffusion-limited (Fig. S7 A and B).
Electrochemical response study
The electrochemical response of the DNA hybridization was studied using the EIS technique [34]. Upon immobilization of pDNA on APTMS-ZnO/c-GNF/ITO an increase in the Rct value (754 Ω) was observed because of the repulsion between the [Fe(CN)6]3−/4- electrolytic redox couple and the phosphate backbone of the pDNA (Fig. 3a curve (i)). There was a further increase in Rct value after the hybridization of complementary DNA (889 Ω) due to an increase in the number of phosphate backbone in the double-stranded DNA (Fig. 3a curve (ii-xii)). It can be inferred that with a change in the negative charge and conformation transition, the interfacial properties changes and this becomes the basis of sensitive determination of cDNA of E. coli. The signal used for the measurement was the difference in the Rct value of pDNA immobilized electrode and the electrode after hybridization with cDNA {∆Rct = Rct(cDNA) – Rct(pDNA)}. There was a linear relationship between the analytical signal (∆Rct) and the log of concentration of cDNA target ranging from 10−16 to 10−6 M (Fig. 3b) and follows the following regression eq. (1):
with linear regression of 0.9952. The detection limit of the fabricated pDNA/APTMS-ZnO/c-GNF/ITO based biosensor is 0.1 fM. As the APTMS-ZnO/c-GNF contain ample functional groups which provide a conducive platform for the immobilization of pDNA, leading to higher loading of pDNA on to the APTMS-ZnO/c-GNF/ITO electrode, resulting in increased hybridization efficiency. From the data obtained it can be easily concluded that compared with other previously reported methods which have their inherent limitations as discussed in preceding section 1 and other works reported based on hybridization this genosensor shows a wider detection range, better sensitivity and low detection limit (cf. Table S3 and S4).
Selectivity study
The pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode was found to be selective towards different target DNA sequences such as non-complementary, one base mismatch and complementary sequences. The selectivity was studied using EIS technique (Fig. S9). There was only slight increase in the Rct value after incubating the bioelectrode with non-complementary DNA sequence (844 Ω) (Fig. S9 curve (ii)) as compared to pDNA (curve (i)). This is very obvious since the non-complementary DNA base sequences do not match with the pDNA base sequence and no hybridization would lead to a negligible change in the charge distribution of electrode surface and therefore very less change in Rct value. When the pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode was incubated with one-base mismatch DNA sequence (curve iii), there was a slight increase in the Rct value in comparison to pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode which can be clearly seen by the semicircular diameter of the curve (i) and curve (iii) of Fig. S9. This might be due to partial hybridization of pDNA with bases of one-base mismatch DNA as a result of which there is increase in negative charge at the electrode surface. The negatively charged electrode surface now applied a blocking effect i.e. repulsion towards the redox ions. While on exposure of pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode with cDNA there is complete hybridization of bases takes place leading to increase in negative charge and higher blocking effect towards the redox ions (curve iv) as can be ascertained from high Rct value.
Specificity study
The specificity of pDNA/APTMS-ZnO/c-GNF/ITO bioelectrode was checked towards the isolated DNA from the panel strains of E. coli and various other water-borne pathogens such as Salmonella Typhimurium, Neisseria meningitides and Klebsiella pneumonia using EIS study (Fig. S10). There was a marked increase in Rct value when the bioelectrode was incubated with clinical E. coli strains indicating the complete hybridization. Whereas, negligible change was recorded when the electrodewas incubated with other pathogenic stains S. Typhimurium, N. meningitides and K. pneumonia. While the strains of other bacteria such as S. Typhimurium do not affect the hybridization of the bioelectrode with the strains of E. coli revealing the specificity of the fabricated biosensor.
The reusability of the fabricated electrode was evaluated by immersing the electrode in a buffer solution (pH 8.0) containing Tris buffer (10 mM) and EDTA (1 mM) at 100 °C for 5 min, followed by cooling in an ice bath for about 25 min. This process completely removes the cDNA via thermal denaturation. The Rct value of the nucleic acid sensor was found to decrease after each regeneration process (5 cycles) with a total loss of 10.42% of the original Rct value. This reduction in signal may be due to surface fouling during the regeneration process indicating that the biosensor can be used for atleast 5 times (data not shown).
Conclusion
In summary, the work emphasizes the synthesis of APTMS functionalized ZnO-carboxylated graphene nanoflakes composite using wet chemical route and its electrophoretic deposition on ITO supported electrode at 120 V. This fabricated platform was immobilized with probe DNA sequences for the selective determination of E. coli O157:H7 DNA using impedemetric technique. The proposed E. coli O157:H7 DNA sensor retained its activity for 5 repetitive uses with a loss of approximately 10% activity. The lowest concentration of E. coli O157:H7 DNA detection using the fabricated biosensor was found to be 0.1 fM. The sensing platform can be interestingly used for the detection of other water borne pathogenic DNA sequences. Besides all this, efforts are being made to develop affinity based biosensors (using antibody–antigen interactions) to allow the detection of E. coli O157:H7 cells directly from water samples.
References
Pandey CM, Dewan S, Chawla S, Yadav BK, Sumana G, Malhotra BD (2016) Controlled deposition of functionalized silica-coated zinc oxide nano-assemblies at the air/water interface for blood cancer detection. Anal Chim Acta 937:29–38
Zhang P, Wang S (2014) Designing fractal nanostructured biointerfaces for biomedical applications. Chem phys chem 15:1550–1561
Xu Q, Wang L, Lei J, Deng S, Ju H (2013) Platinum nanodendrite functionalized graphene nanosheets as a non-enzymatic label for electrochemical immunosensing. J Mater Chem B 1:5347–5352
Wang J (2005) Nanomaterial-based electrochemical biosensors. Analyst 130:421–426
Pan C, Zhu J (2009) The syntheses, properties and applications of Si, ZnO, metal, and heterojunction nanowires. J Mater Chem 19:869–884
Rahman MM, Ahammad AJS, Jin JH, Ahn SJ, Lee JJ (2010) A comprehensive review of glucose biosensors based on nanostructured metal-oxides. Sensors 10:4855–4886
Solanki PR, Kaushik A, Agrawal VV, Malhotra BD (2011) nanostructured metal oxide-based biosensors. NPG Asia Mater 3:17–24 (2011)
Chai G, Lupan O, Chow L, Heinrich H (2009) Crossed zinc oxide nanorods for ultraviolet radiation detection. Sens Actuators A: Physical 150:184–187
Mehrabian M, Azimirad R, Mirabbaszadeh K, Afarideh H, Davoudian M (2011) UV detecting properties of hydrothermally synthesized ZnO nanorods. Physica E Low dimens Syst Nanostruct 43:1141–1145
Ali SMU, Nur O, Willander M, Danielsson B (2010) A fast and sensitive potentiometric glucose microsensor based on glucose oxidase coated ZnO nanowires grown on a thin silver wire. Sensors Actuators B Chem 145:869–874
Fulati A, Ali SMU, Asif MH, Alvi NH, Willander M, Brännmark C, Stralfors P, Börjesson SI, Elinder F, Danielssond B (2010) An intracellular glucose biosensor based on nanoflake ZnO. Sensors Actuators B Chem 150:673–680
Mohammed AM, Ibraheem IJ, Obaid AS, Bououdina M (2017) Nanostructured ZnO-based biosensor: DNA immobilization and hybridization. Sens Biosensing Res 15:46–52
Ahmad R, Tripathy N, Jang NK, Khang G, Hahn YB (2015) Fabrication of highly sensitive uric acid biosensor based on directly grown ZnO nanosheets on electrode surface. Sens. Actuators B Chem 206:146–151
Tak M, Gupta V, Tomar M (2014) Flower-like ZnO nanostructure based electrochemical DNA biosensor for bacterial meningitis detection. Biosens Bioelectron 59:200–207
Chen X, Yan H, Shi Z, Feng Y, Li J, Lin Q, Wang X, Sun W (2017) A novel biosensor based on electro-co-deposition of sodium alginate-Fe3O4-graphene composite on the carbon ionic liquid electrode for the direct electrochemistry and electrocatalysis of myoglobin. Polym Bull 74:75–90
Salih E, Mekawy M, Hassan RYA, El-Sherbiny IM (2016) Synthesis, characterization and electrochemical-sensor applications of zinc oxide/graphene oxide nanocomposite. J Nanostructure Chem 6:137–144
Tiwari JN, Tiwari RN, Kim KS (2012) Zero-dimensional, one-dimensional, two-dimensional and three-dimensional nanostructured materials for advanced electrochemical energy devices. Prog Mater Sci 57:724–803
Pokropivny VV, Skorokhod VV (2007) Classification of nanostructures by dimensionality and concept of surface forms engineering in nanomaterial science. Mater Sci Eng C 27:990–993
Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M (2009) Understanding biophysicochemical interactions at the nano–bio interface. Nat Mater 8:543–557
Loo AH, Bonanni A, Pumera M (2013) Thrombin aptasensing with inherently electroactive graphene oxide nanoplatelets as labels. Nanoscale 5:4758–4762
Chang H, Wang Y, Li J (2011) Electrochemical DNA sensors: from nanoconstruction to biosensing. Curr Org Chem 15:506–517
Chen A, Chatterjee S (2013) Nanomaterials based electrochemical sensors for biomedical applications. Chem Soc Rev 42:5425–5438
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM (2011) Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis 17:7–15
Tiwari I, Singh M, Pandey CM, Sumana G (2015) Electrochemical detection of a pathogenic Escherichia coli specific DNA sequence based on a graphene oxide–chitosan composite decorated with nickel ferrite nanoparticles. RSC Adv 5:67115–67124
Jaiswal N, Pandey CM, Soni A, Tiwari I, Rosillo-Lopez M, Salzmann CG, Malhotra BD, Sumana G (2018) Electrochemical genosensor based on carboxylated graphene for detection of water-borne pathogen. Sensors Actuators B Chem 275:312–321
Xu M, Wang R, Li Y (2017) Electrochemical biosensors for rapid detection of Escherichia coli O157:H7. Talanta 162:511–522
Wang J, Li S, Zhang Y (2010) A sensitive DNA biosensor fabricated from gold nanoparticles, carbon nanotubes, and zinc oxide nanowires on a glassy carbon electrode. Electrochim Acta 55:4436–4440
Solanki PR, Kaushik A, Chavhan PM, Maheshwari SN, Malhotra BD (2009) Nanostructured zirconium oxide based genosensor for Escherichia coli detection. Electrochem Commun 11:2272–2277
Yang Y, Wang Z, Yang M, Li J, Zheng F, Shen G, Yu R (2007) Electrical detection of deoxyribonucleic acid hybridization based on carbon-nanotubes/nano zirconium dioxide/chitosan-modified electrodes. Anal Chim Acta 584:268–274
Rosillo-Lopez M, Lee TJ, Bella M, Harta M, Salzmann CG (2015) Formation and chemistry of carboxylic anhydrides at the graphene edge. RSC Adv 5:104198–104202
Wu C, Qiao X, Chen J, Wang H, Tan F, Li S (2008) A novel chemical route to prepare ZnO nanoparticles. Mater Lett 60:1828–1832
Zhu J, He J (2012) Facile synthesis of Graphene-wrapped honeycomb MnO2 Nanospheres and their application in Supercapacitors. Appl Mater Interfaces 4:1770–1776
Ivers-Tiffée E, Weber A, Schichlein H (2010) Electrochemical impedance spectroscopy. Handbook of Fuel Cells, John Wiley Sons Ltd Chichester
Khan SB, Faisal M, Rahman MM, Jamal A (2011) Low-temperature growth of ZnO nanoparticles: Photocatalyst and acetone sensor. Talanta 85:943–949
Acknowledgements
N.J. and S.S. are thankful to CSIR, India for the award of SRF and RA, respectively. C.M.P. acknowledges the Department of Science and Technology, New Delhi, India for DST-INSPIRE Faculty Award (DST/INSPIRE/04/2015/000932). We thank Dr. Gajjala Sumana (CSIR-National Physical Laboratory) for interesting discussions. N.J. and I.T. are sincerely thankful to Prof. O.N Srivastava (Dept. of Physics, I.Sc. B.H.U) and Prof. Rajiv Prakash (IIT B.H.U) for providing TEM and SEM facilities respectively. B.D.M. thanks Science & Engineering Research Board (Govt. of India) for the award of a Distinguished Fellowship (SB/S9/YSCP/SERB-DF/2018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1346 kb)
Rights and permissions
About this article
Cite this article
Jaiswal, N., Pandey, C.M., Solanki, S. et al. An impedimetric biosensor based on electrophoretically assembled ZnO nanorods and carboxylated graphene nanoflakes on an indium tin oxide electrode for detection of the DNA of Escherichia coli O157:H7. Microchim Acta 187, 1 (2020). https://doi.org/10.1007/s00604-019-3921-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3921-8