Abstract
A Pb(II)-DNAzyme is used in an amperometric method for the determination of Pb(II). The method is based on two feedback processes. In the first, the Pb(II)-DNAzyme initiates a reaction in presence of Pb(II) in a micro-tube to release a linear DNA (S1). In the second, the S1 triggers the recycling amplification between two types of hairpin-shaped DNA templates (H1 and H2) which consist of a primer sequence and a Pb(II)-DNAzyme substrate sequence. The Pb(II)-DNAzyme has excellent cleavage specificity toward the substrate sequence in S1 that combined firstly with H1 and then is linked to H2. This process will connect H1 and H2. After hybridization with H1 and H2 to form two DNA complexes, S1 is released and initiates the next recycling process. This results in efficient amplification. A glassy carbon electrode (GCE) was immersed into solution of HAuCl4 to electrodeposit a layer of gold nanoparticles. This is followed by the assembly of the hairpin probe H1 on the GCE. In addition, a nanohybrid consisting of 3, 4, 9, 10-Perylenetetracarboxylic acid (PTCA) and nitrogen-doped graphene (NG) was loaded with electroactive thionine (Thi) and gold to form nanoparticles of type NG-PTCA-Thi-Au. This is responsible for generating the amperometric signal (best measured at around −0.30 V vs. SCE) and also acts as the reducing agent for synthesizing the NG-PTCA-Thi-Au nanohybrid. H2 is immobilized on NG-PTCA-Thi-Au to form a new tracer label. The concentration of Pb(II) in a solution can be quantified by determination of the amount of cleaved S1. The method has high sensitivity and selectivity for Pb(II). The detection limit is 0.42 pM (S/N = 3), and the detection range extends from 1 pM to 1000 nM.

Schematic representation of electrodes for the determination of the lead ions (Pb2+). The sensor is using Pb2+-DNAzyme assisted recycling amplification based on hairpin assembly on a composite prepared from nitrogen doped graphene, perylenetetracarboxylic anhydride, thionine and gold nanoparticles (NG-PTCA-Thi-Au). This versatile platform expands studies on the detection of heavy metal ions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lead ion (Pb2+) can accumulate in the body to cause various sorts of disorders [1, 2]. Even low-dose of Pb2+ has strong toxicity to affect the infants’ and children’s growth, mental development [3]. It is of great significance to monitor the contamination levels of Pb2+, especially the trace Pb2+ in the environment. Conventional methods for detecting Pb2+ include atomic absorption spectroscopy (AAS), atomic fluorescence spectrometry (AFS) and inductively coupled plasma mass spectrometry (ICP-MS) [4]. These methods have been used in accurate detection of Pb2+, but they need expensive equipment or complicated sample preparations [5]. Thus, it is necessary to develop an approach based on fewer samples, more convenient operations and higher sensitivity for Pb2+ detection and analysis.
Electrochemical method has received particular attention because of its sensitivity, selectivity, low cost, easy operation, short response time and minimal sample preparation [6]. Generally, enzyme-assisted amplification technologies such as polymerase chain reaction (PCR), rolling circle amplification (RCA) and loop-mediated isothermal amplification (LAMP) have been widely used in the construction methods for detecting many target analyses [7]. However, multiple assay steps and exogenous reagents obviously increase the time, cost and complexity in studies [8]. In addition, protein enzyme required in these methods with inherent instability can cause these methods susceptibility to environments, which limits their applications [9]. To overcome these shortcomings, researchers have struggled for developing many enzyme-free amplification approaches, such as entropy-driven catalysis, hybridization chain reaction (HCR) and catalytic hairpin assembly (CHA) [10]. CHA has arrested considerable attention owing to its high sensitivity and simplicity. In the CHA procedure, the single-stranded DNA (ssDNA) with a complementary sequence of hairpin-shaped capture probe spontaneously triggers this process by opening and hybridizing two hairpin-shaped probes, accompanied by the release of ssDNA for initiating the next cycle [11]. In this case, CHA will go through several rounds of ssDNA recycling, which means a significant detection signal can be generated with only a super-low dose of target and also efficiently increase the sensitivity of the method. The cleaved substrate fragment (S1) of 8–17 DNAzyme with the existence of Pb2+ can act as a ssDNA for triggering CHA process. Therefore, it is promising for construction of a method for Pb2+ detection.
To further improve the sensitivity of the method, various nanomaterials with excellent properties are extensively utilized to amplify the signals [12]. Nitrogen-doped graphene (NG), nitrogen doping in graphene, is considered as a desirable material with good electrical conductivity and high specific surface area, making it a good platform for nanomaterial loading [13, 14]. However, NG exhibits poor dispersion and easy aggregation, which restricts its further application in methods. The reported literature to get high dispersion of NG was to adjust the pH of the NG solution to alkaline by the addition of NaOH, which may damage the structure and decrease the electronic conductivity of NG [15, 16]. To prevent this problem, 3, 4, 9, 10-Perylenetetracarboxylic acid (PTCA), a perylene derivative that contains abundant polycarboxylic sites as a water-soluble molecule, was used to improve the dispersion of NG [17]. Specially, PTCA possesses large specific surface area, excellent stability and desirable electrical conductivity to functionalize NG via π stacking and hydrophobic interactions without further destroying the conjugated π-system of NG [18, 19]. In addition, the active sites and effective areas of NG-PTCA have been increased, which can sufficiently load electroactive Thionine (Thi) through π stacking and benefit the growth of Au nanoparticles (AuNPs) [20]. Specifically, Thi plays an electroactive material for the following electrochemical signal generation and acts as a reducing agent for the reduction of HAuCl4 [21]. Significantly, the NG-PTCA-Thi-Au nanocomposite is successfully designed and used for assembling hairpin-shaped signal probes (H2) to obtain a new NG-PTCA-Thi-Au-H2 nanocomposite (tracer label) through strong Au-S bonds [22]. In this regard, the new tracer label is ideally applied to the further signal amplification and sensitivity enhancement of the method.
We designed a dual signal amplification strategy for detecting Pb2+ by using NG-PTCA-Thi-Au as a signal tag and CHA as a signal amplifier. The cleavage-induced capability of 8–17 DNAzyme with the existence of Pb2+ was utilized for ensuring the specificity of method [23]. It is consisting of a substrate chain (S) and a catalytic chain (C) [24]. In the presence of Pb2+, the S was cleaved into two fragments at the cleavage site [25]. The cleaved S1 can trigger the CHA process. It can open the hairpin capture probes (H1) by hybridization reaction, exposing the concealed sequence of H1, which sequentially opened H2 to form H1-S1-H2 hybrid complex. Then, the S1 was released to initiate the next cycle, leading to accumulation of tracer label. As a result, the signal increased by using the dual signal amplification strategies of S1 recycle and the plentifully captured tracer label. This method displayed excellent sensitivity, specificity and good consistent with the conventional ICP-MS method when used for quantitative detection of Pb2+ in environment water samples.
Experiment
Materials and reagents
Nitrogen-doped graphene (NG) was purchased from Nanjing Xianfeng Nano (Nanjing, China, https://www.xfnano.com/). Thionine acetate (Thi), 3,4,9,10-Perylenetetracarboxylic dianhydride (C24H8O6, PTCDA) were acquired from Alfa Aesar (China, https://www.alfa.com/) Chemicals Co., Ltd. Tetrachloroaurate (III) acid (HAuCl4·4H2O), 6-mercapto-1-hexanol (MCH), potassium ferrocyanide [K4Fe(CN)6], potassium ferricyanide [K3Fe(CN)6] were supplied by Sigma-Aldrich (St. Louis, USA, http://www.sigmaaldrich.com). HPLC-purified oligonucleotides were purchased from Sangon Biotech Co., Ltd. (Shanghai, China, http://www.sangon.com/). The DNA sequences are listed in Table S1.
Buffers involved in this study were as follows: diluted solutions of substrate strand and catalytic strand prepared using 1 × TAE buffer (40 mM Tris-acetate and 1 mM EDTA, pH 8.0) containing 20 mM MgCl2. Hybridization buffer containing 20 mM Tris-HCl (pH 7.4), 10 mM TCEP, 10 mM NaCl and 1 mM MgCl2 was used to dilute hairpin-shaped captured probes, whereas the hybridization buffer without TCEP was used to dilute hairpin-shaped signal probe. 0.1 M phosphate buffered saline (PBS, pH 7.4) for square wave voltammetry (SWV) determination was prepared by mixing 10 mM Na2HPO4, 10 mM KH2PO4, and 2 mM KCl. All reagents were of analytical grade and used without further purification. Ultrapure water (>18.2 MΩ·cm, USA, http://www.emdmillipore.com) obtained from a Mill-Q purification system was used throughout the experiment.
Apparatus and characterization
All electrochemical measurements were performed on a CHI 660D electrochemical workstation (Shanghai Chenhua instrument, China, http://www.chinstruments.com), including cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS) and SWV. The electrochemical detection was carried out in a three-electrode system which used modified glassy carbon electrode (GCE, Φ = 4 mm) as the working electrode, saturated calomel electrode (SCE) as the reference electrode and platinum wire as auxiliary electrode. The morphological structure of materials were shown using transmission electron microscopy (TEM, Hitachi Limited, Japan, http://www.hitachi.com) and Field emission scanning electron microscopy (FE-SEM, Oxford x-max50 microscope, Japan, http://www.hitachi.com). Elemental composition of the nanomaterials was determined using X-ray photoelectron spectroscopy (XPS, VG Scientic ESCALAB 250 spectrometer, USA, http://www.thermofisher.com) coupled with Energy dispersive X-ray spectroscopy (EDS, JEOLJSM-6700F, Japan, http://jeol.bioon.com.cn). Chemical construction was analyzed by Ultraviolet visible absorption spectra (UV-vis, UV-2450 spectrophotometer, Shimazu, Japan, https://www.shimadzu.com.cn/).
Preparation of perylenetetracarboxylic acid (PTCA)
PTCA was prepared from PTCDA using modified reported method [26]. Typically, 100 mg PTCDA was initially dissolved in 10 mL 0.1 M NaOH at 80 °C for stirring 1 h until completely hydrolyzed. Then, 0.1 M HCl was dropwise added in the mixture and maintained at weak acidic pH. Next, the red deposits were centrifuged and washed with ultrapure water. Finally, the resultant PTCA was dried under vacuum to get the powder.
Preparation of NG-PTCA
The NG-PTCA was synthesized by a π stacking approach. NG was dispersed in ultrapure water to ultrasonic treatment in an ice bath for 90 min in order to exfoliate with impurities. Afterwards, 1 mg PTCA was added into the 1 mg· mL−1 of NG aqueous dispersion with the assistance of ultrasonication for 5 h. Then, the resulting mixture was stirred continuously for 12 h at 40 °C. The product was subsequently collected by centrifugation and purified by repeated washing using ultrapure water. By the way, we obtained NG-PTCA, which was then dried under vacuum at 60 °C and stored for further use.
Preparation of NG-PTCA-Thi-Au
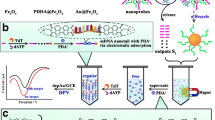
The preparation process for the synthesis of NG-PTCA-Thi-Au is shown in Scheme 1a. Firstly, the dispersion of NG-PTCA was treated with ultrasound for 30 min. Then 0.5 mM Thi was added to 1 mL NG-PTCA (1 mg·mL−1) to get the NG-PTCA-Thi mixture. After reacting for a while, 25 μL 1% HAuCl4·4H2O was added by dropwise into the NG-PTCA-Thi dispersion and stirred vigorously overnight at room temperature (RT) in the dark, making HAuCl4 completely react with NG-PTCA-Thi mixture to grow Au nanoparticles (AuNPs). Finally, the products were collected by centrifugation at 9000 rpm for 15 min and washed with ultrapure water for at least two times. Then the resultant NG-PTCA-Thi-Au was dispersed in 1 mL ultrapure water for further use.
Preparation of the tracer label (or tracer label 1)
The preparation process for tracer label is shown in Scheme 1a. H1, H2 and H3 were heated at 95 °C for 5 min and gradually cooled down to form the stem-loop structures. 1 mL NG-PTCA-Thi-Au dispersion and 200 μL H2 (or H3) labeled at the 5’-SH were mixed together with gently stirring for 12 h to sufficiently immobilize H2 (or H3) onto the surface of the nanocomposites through strong Au-S bonds. Then the tracer label (or tracer label 1) was acquired using centrifugation and washing to remove unbound H2 (or H3). Ultimately, the tracer labels were dispersed in 1 mL hybridization buffer without TCEP and kept at 4 °C until use.
Preparation of 8–17 DNAzyme
2 μM substrate strand of 8–17 DNAzyme was heated in a water bath (95 °C) for 5 min and then cooled down to room temperature at least 2 h. Next, the same volume of catalytic strand was added into the substrate strand and incubated in a water bath (65 °C) for 10 min. After the hybridization reaction, the DNA mixture was gradually cooled down to room temperature to form the 8–17 DNAzymes. For catalytic cleavage reaction, various concentrations of Pb2+ were added into 10 μL DNAzymes and reacted at 37 °C in the incubator for 40 min (the optimized incubation time). This reaction can produce three fragments. The cleaved fragment (S1) was complementary of partial fragment of H1.
Catalytic hairpin assembly (CHA) agarose gel electrophoresis assays
To verify the feasibility of CHA, the 10% native polyacrylamide gel electrophoresis (PAGE) was used. In the gel electrophoresis assay, the loading sample was mixed with 1 μL DNA sample and 1 μL 36% glycerol. The gel electrophoresis was run at 90 V for 40 min in 1 × Tris-borate-EDTA (TBE) buffer. After running, the gel was placed in a container with a sufficient amount of the 3 × SolarGelRed dye and agitated at room temperature for 30 min. The dyed gel was illuminated with UV light and photographed with ChampChemi.
Detection of Pb2+ in real water samples
Real water samples from waste water (pond water, river water and tap water) were used to evaluate the practical performance of the system. We have taken 50 ml of the waste water samples in each sampling point. Water samples were centrifuged for 10 min at 12,000 rpm and filtered to remove impurities and large particles.
Fabrication of electrochemical method
As is shown in Scheme 1b, the method was fabricated for Pb2+ detection. Prior to fabrication, the GCE was polished with 0.3 and 0.05 μm alumina slurries and following sonication in ethanol and ultrapure water for 3 min until it showed mirrorlike surface. The 8–17 DNAzyme was cleaved in a micro-tube, generating a new sequence S1 for subsequent use. To fabricate the method, AuNPs were first deposited on the cleaned GCE via in-situ electrochemical deposition with a constant potential at −0.2 V for 30 s in 1% HAuCl4 solution (Note: the deposition time of AuNPs was optimized). Then, the modified electrodes were blocked with 10 μL 0.5 mM MCH for 30 min at RT to eliminate nonspecific binding sites. After rinsing, 10 μL 2 μM H1 was self-assembled on the surface of AuNPs via the Au-N bond for 2 h at 37 °C. The electrodes were incubated with different concentrations of S1 for 2 h at 37 °C. Then the 10 μL tracer labels of NG-PTCA-Thi-Au-H2 bioconjugates were cast onto the rinsed electrodes and incubated for another 2 h at 37 °C. Afterwards, the electrodes were rinsed with ultrapure water and for electrochemical detection.
Electrochemical measurement
The SWV signal was originated from Thi on tracer label and recorded in the potential range from - 0.55 to - 0.15 V in 5 mL of PBS (pH 7.4) at room temperature, which was used for quantitative detection of Pb2+.
Results and discussion
Characterization of different nanomaterials
To verify the successful synthesis of NG-PTCA-Thi-Au with good morphology, we used FE-SEM and TEM to investigate the nanostructures. As is shown in Fig. 1a and Fig. S1A, the NG exhibits a typical crumpled and wrinkled structure, consisting of some lamellate and tulle-like parts, indicating that the NG is well maintained with two-dimensional structure and high surface/volume ratio. In Fig. 1b and Fig. S1B, PTCA shows a rod-like structure with length of up to about 1 μm, distributing relatively homogeneously, which is in agreement with the reported literature [27]. Following the growth of PTCA on the NG, the nanocomposites reacted with Thi and HAuCl4. The products revealed that well-dispersed AuNPs were formed on the surface of NG-PTCA, suggesting the successful synthesis of NG-PTCA-Thi-Au (Fig. 1c and Fig. S1C). In addition, we recorded the reaction process using UV-Vis spectra and the results are shown in Fig. 1d. For NG, no absorption peaks are observed (curve a). After PTCA being adsorbed on the surface of NG, the characteristic peaks of PTCA (curve b) are all observed in the spectrum of the NG-PTCA composites and shifted to much lower wavenumbers (curve c). The two UV–vis absorption peaks at around 440 and 467 nm are red-shifted, which may be attributed to the π stacking and hydrophobic forces between PTCA and NG. After modification with Thi and AuNPs, the characteristic peaks of NG-PTCA-Thi-Au ranged from 400 to 550 nm and from 560 to 650 nm are observed in the spectra. It is corresponded to the peaks of NG-PTCA, Thi and AuNPs [22, 28]. PTCA in an orange-red color dispersed well in water (Fig. 1d, inset b). After PTCA adsorbed on the NG, the aggregated black NG sheets changed to the uniform dark brown suspension, which was stable for a few weeks at room temperature (Fig. 1d, inset a and c).
To confirm the formation of NG-PTCA-Thi-Au, EDS and XPS were used to investigate elemental compositions. As is shown in Fig. 1e, signature C, N, O, S and Au peaks were obtained in the EDS image, confirming that the C, N, O, S and Au elements were existed in the nanocomposite. The Au4f, C1s, N1 s, O1s and S2p peaks appeared in the XPS survey spectrum (Fig. 1f), the inset in Fig. 1F represents the XPS signature of the different elements of NG-PTCA-Thi-Au, further proving the existence of Thi and AuNPs on the surface of NG-PTCA. Favorably, the above results proved that NG-PTCA-Thi-Au nanocomposites were successfully prepared.
Feasibility of the catalytic hairpin assembly (CHA) strategy
In order to confirm the feasibility of CHA, agarose gel electrophoresis analysis experiments were performed. As is shown in Fig. S3, lane a is 20 bp marker and lanes b and c represent H1 and H2 respectively. Although H1 and H2 contain complementary sequences, their mixture does not show the change of band positions after incubation for 2 h (lane d), implying no hybridization between the two hairpin probes. When S1 was added into the mixture of H1 and H2 (lane e), a new band with relative less migration than that of H1–H2 complexes appeared. This may be because more bases than H1 and H2 are produced after the reaction of three DNAs, thus suggesting that S1 can trigger the CHA to form a H1–H2 complex. Also, the bands corresponding to H1 and H2 are probably the incomplete reaction of them. The number of bases of S1 is less combined with less dye, which makes it impossible to be observed after image formation. As expected, these results reflected the successful dynamic assembly catalyzed by S1. In addition, the respective data and Figure of “amplified effect of CHA” are described in Electronic Supporting Material (Fig. S2).
Characterization of stepwise fabrication of the method
CV and EIS are effective ways to probe the interface properties of surface-modified electrodes. The results are shown in Fig. 2. In Fig. 2a, after the deposition of AuNPs on the electrode surface (curve b), a sharply increasing current was observed compared with the bare electrode (curve a), attributing to the fact that AuNPs with high electrical conductivity and large surface area can efficiently accelerate the electron transfer. When the MCH (curve c) was incubated, the peak current response decreased due to the blocking effect of the MCH. Additionally, the peak current was further decreased after immobilization with H1 (curve d), ascribing to the electrostatic repulsion between the negative charge of the phosphate skeleton of DNA probe and the [Fe(CN)6]3−/4- ions. After that, while S1 (curve e) and tracer labels (curve f) were captured on the electrode, respectively, the current response decreased further for the same reason.
CV results were further demonstrated by EIS characterization. EIS was used to monitor the fabrication processes and probe the interfacial properties of electrodes in the presence of [Fe(CN)6]3−/4 [29]. As is shown in Fig. 2b, after the electro-deposition of AuNPs (curve b), it exhibited extremely low resistance due to enhanced electron transfer ability. After blocked by MCH (curve c), the electrical resistance increased due to its electronic hindrance. Then, when H1 was immobilized on the electrode surface (curve d), the electrostatic interaction due to the negative charge carried by its phosphate backbone led to a further increase in electrical resistance. Subsequently, a gradual increase in electric resistance was observed when S1 (curve e) and tracer labels (curve f) were immobilized respectively. Both of them were caused by an increase in the amount of the phosphoric acid skeleton and then the electrostatic repulsion was further increased. These results were consistent with the results of the CV, indicating that the fabrication of the method was successful.
Optimization of the experimental conditions
The following parameters were optimized: (a) MCH immobilization time; (b) assembly time and concentration for H1; (c) 8–17 DNAzyme cleavage time; (d) AuNPs deposition time. Respective data and Figures are given in the Electronic Supporting Material (Fig. S4). The following experimental conditions were found to give best results: (a) optimal MCH immobilization time: 30 min; (b) the optimized assembly time and concentration for H1: 60 min and 2.0 μM; (c) optimum cleavage time: 40 min; (d) optimum AuNPs deposition time: 30 s.
Analytical performance
To investigate the electrochemical performance of the method, it was employed to detect various Pb2+ concentrations at the optimal conditions. As is shown in Fig. 3a, the SWV responses increased as the Pb2+ concentrations rose. The calibration plot is illustrated in Fig. 3b, for quantitative determination of Pb2+, showing an increase in current response directly proportional to the logarithm of Pb2+ concentrations within the range of 1 pM to 1000 nM. The linear regression equation is Y = 0.48586*log CPb2++1.145, with a linear correlation coefficient of 0.99901. The limit of detection (LOD) was calculated as 0.42 pM at a signal-to-noise ratio of 3. Comparing the analytical performance with previous reported method to detect Pb2+ listed in Table S2, the method with a wider linear range and a lower LOD is superior because it exhibits higher sensitivity.
a SWV curves of the method incubated with different concentration of Pb2+ (from a to i): 0 pM, 1 pM, 10 pM, 100 pM, 1 nM, 10 nM, 100 nM, 500 nM and 1000 nM at an operating potential of −0.3 V (vs. SCE). b Calibration plot of the method for different concentrations of Pb2+ detection (n = 3). c Selectivity of the method toward Pb2+ by comparing it to the interference metal ions (Cu2+, Mg2+, Cd2+, Sn2+, Co2+, Tl3+, Bi3+, Ca2+, Hg2+, Zn2+ and Fe3+) (1 μM), a mixture (Mix) of these interfering metal ions (1.0 μM) and Pb2+ (100 pM) and zero analyte (Blank). (D) Reproducibility of five different electrodes modified with 100 pM of Pb2+
Selectivity, stability and reproducibility of the method
The selectivity of the method was evaluated by testing different interfering ions at 1 μM such as Cu2+, Mg2+, Cd2+, Sn2+, Co2+, Tl3+, Bi3+, Ca2+, Hg2+, Zn2+ and Fe3+. The mixture of these ions with 100 pM Pb2+ had been tested under the same conditions. As is shown in Fig. 3c, in contrast to the blank test, the SWV peak current of the relative metal ions was close to the blank group and far from the peak current of Pb2+ and mixture. The results indicate that the method is highly selective for Pb2+ detection and can’t recognize other ions.
In Fig. 3d, the reproducibility of the method was investigated using five independent electrodes under the same conditions to analyze the same concentration of Pb2+ (100 pM). All electrodes presented similar electrochemical responses and showed the relative standard deviation (RSD) of 2.8% suggesting that the method has appreciable reproducibility.
The stability of the method was measured by recording the current signals of the electrodes kept at 4 °C every 7 days. Fig. S5 revealed that, in the presence of Pb2+, the current signal retained about 88.4% of the initial value after 28 days. This result indicates that the method has good stability.
Application for the analysis of real samples
Detection performance in water samples
To analyze the applicability of the method towards Pb2+, the standard addition method was applied. Samples were spiked with 10 pM, 1 nM, and 100 nM Pb2+ in water samples. The concentration of Pb2+ in the test samples was quantitatively determined via the calibration plot by SWV and listed in Table 1. The data in Table 1 presents that the recovery is from 99.00% to 106.90% and the RSD is from 0.84% to 1.08%, indicating that the method has a prospect for application in real samples.
Detection of Pb2+ in real samples
Further to investigate the feasibility of the method, real samples from waste water (pond water, river water and tap water) were measured by both the method and ICP-MS method without further pretreatment. The experimental data is shown in Table 2 and analyzed using Statistical Product and Service Solutions version 20 (SPSS 20.0). A paired t-test was conducted to compare the method with ICP-MS and no statistically significant differences was found (P = 0.624 > 0.05, P < 0.05 was considered statistically significant). The above results clearly demonstrate that the method is capable of detecting Pb2+ in the environment with excellent detection performances.
Conclusion
The contribution of this study is to introduce a dual signal amplification strategy based on CHA combining with NG-PTCA-Thi-Au for the sensitive detection of Pb2+. 8–17 DNAzyme is regarded as the recognition element of Pb2+ to guarantee the specificity of the method. Additionally, it has been successfully applied in real samples. Featuring a high sensitivity, excellent specificity, good stability and acceptable reproducibility, the method has the potential to precisely detect Pb2+ in complex environmental water samples. We believe that this method can extend a versatile platform to analyze other metal ions by changing the corresponding dependent DNAzyme. However, the small sample size of this study argues for more samples to demonstrate the feasibility of the method for detecting Pb2+. Therefore, our future research will focus on collecting enough samples to verify the feasibility of this method considering practical application.
References
Roushani M, Baghelani YM, Mavaei M, Abbasi S, Mohammadi SZ (2018) Preparation of modified magnetic cobalt nanoparticles as a new magnetic sorbent for the Preconcentration and determination of trace amounts of Lead ions in environmental water and soil (air-dust) samples. Commun Soil Sci Plant Anal 49:645–657. https://doi.org/10.1080/00103624.2017.1417419
Singh P, Mittal LS, Kumar K, Sharma P, Bhargava G, Kumar S (2018) Multifunctional metallo-supramolecular interlocked hexagonal microstructures for the detection of lead and thiols in water. Chem Commun (Camb) 54:9482–9485. https://doi.org/10.1039/c8cc05814h
Needleman H (2004) Lead poisoning. Annu Rev Med 55:209–222. https://doi.org/10.1146/annurev.med.55.091902.103653
Li J, Lu Y (2000) A highly sensitive and selective catalytic DNA biosensor for Lead ions. J Am Chem Soc 122:10466–10467. https://doi.org/10.1021/ja0021316
Lu Y, Li X, Wang G, Tang W (2013) A highly sensitive and selective optical sensor for Pb2+ by using conjugated polymers and label-free oligonucleotides. Biosens Bioelectron 39:231–235. https://doi.org/10.1016/j.bios.2012.07.045
Zhu D, Liu W, Zhao D, Hao Q, Li J, Huang J, Shi J, Chao J, Su S, Wang L (2017) Label-free electrochemical sensing platform for MicroRNA-21 detection using Thionine and gold nanoparticles co-functionalized MoS2 Nanosheet. ACS Appl Mater Interfaces 9:35597–35603. https://doi.org/10.1021/acsami.7b11385
Li Y, Yu C, Yang B, Liu Z, Xia P, Wang Q (2018) Target-catalyzed hairpin assembly and metal-organic frameworks mediated nonenzymatic co-reaction for multiple signal amplification detection of miR-122 in human serum. Biosens Bioelectron 102:307–315. https://doi.org/10.1016/j.bios.2017.11.047
Xie H, Chai Y, Yuan Y, Yuan R (2017) Highly effective molecule converting strategy based on enzyme-free dual recycling amplification for ultrasensitive electrochemical detection of ATP. Chem Commun (Camb) 53:8368–8371. https://doi.org/10.1039/c7cc03497k
Liu S, Wang Y, Ming J, Lin Y, Cheng C, Li F (2013) Enzyme-free and ultrasensitive electrochemical detection of nucleic acids by target catalyzed hairpin assembly followed with hybridization chain reaction. Biosens Bioelectron 49:472–477. https://doi.org/10.1016/j.bios.2013.05.037
Huang J, Wu J, Li Z (2016) Molecular beacon-based enzyme-free strategy for amplified DNA detection. Biosens Bioelectron 79:758–762. https://doi.org/10.1016/j.bios.2016.01.014
Zang Y, Lei J, Ling P, Ju H (2015) Catalytic hairpin assembly-programmed porphyrin-DNA complex as Photoelectrochemical initiator for DNA biosensing. Anal Chem 87:5430–5436. https://doi.org/10.1021/acs.analchem.5b00888
Zhou Q, Lin Y, Lin Y, Wei Q, Chen G, Tang D (2016) Highly sensitive electrochemical sensing platform for lead ion based on synergetic catalysis of DNAzyme and au-Pd porous bimetallic nanostructures. Biosens Bioelectron 78:236–243. https://doi.org/10.1016/j.bios.2015.11.055
Roushani M, Shahdost-Fard F (2019) Applicability of AuNPs@N-GQDs nanocomposite in the modeling of the amplified electrochemical ibuprofen aptasensing assay by monitoring of riboflavin. Bioelectrochemistry 126:38–47. https://doi.org/10.1016/j.bioelechem.2018.11.005
Chen M, Hou C, Huo D, Bao J, Fa H, Shen C (2016) An electrochemical DNA biosensor based on nitrogen-doped graphene/au nanoparticles for human multidrug resistance gene detection. Biosens Bioelectron 85:684–691. https://doi.org/10.1016/j.bios.2016.05.051
Wu J, He J, Zhang C, Chen J, Niu Y, Yuan Q, Yu C (2018) PdPt nanoparticles anchored on the N-G with the integration of PANI nanohybrids as novel redox probe and catalyst for the detection of rs1801177. Biosens Bioelectron 102:403–410. https://doi.org/10.1016/j.bios.2017.11.054
Rezaei B, Jamei HR, Ensafi AA (2018) An ultrasensitive and selective electrochemical aptasensor based on rGO-MWCNTs/chitosan/carbon quantum dot for the detection of lysozyme. Biosens Bioelectron 115:37–44. https://doi.org/10.1016/j.bios.2018.05.012
Gan S, Zhong L, Engelbrekt C, Zhang J, Han D, Ulstrup J, Chi Q, Niu L (2014) Graphene controlled H- and J-stacking of perylene dyes into highly stable supramolecular nanostructures for enhanced photocurrent generation. Nanoscale 6:10516–10523. https://doi.org/10.1039/c4nr02308k
Feng L, Chen Y, Ren J, Qu X (2011) A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials 32:2930–2937. https://doi.org/10.1016/j.biomaterials.2011.01.002
Chang Y, Xie S, Chai Y, Yuan Y, Yuan R (2015) 3,4,9,10-Perylenetetracarboxylic acid/o-phenylenediamine nanomaterials as novel redox probes for electrochemical aptasensor systems based on an Fe3O4 magnetic bead as a nonenzymatic catalyst. Chem Commun (Camb) 51:7657–7660. https://doi.org/10.1039/c5cc00684h
Li F, Yang H, Shan C, Zhang Q, Han D, Ivaska A, Niu L (2009) The synthesis of perylene-coated graphene sheets decorated with au nanoparticles and its electrocatalysis toward oxygen reduction. J Mater Chem 19:4022. https://doi.org/10.1039/b902791b
Han J, Ma J, Ma Z (2013) One-step synthesis of graphene oxide-thionine-au nanocomposites and its application for electrochemical immunosensing. Biosens Bioelectron 47:243–247. https://doi.org/10.1016/j.bios.2013.03.032
Su S, Zou M, Zhao H, Yuan C, Xu Y, Zhang C, Wang L, Fan C, Wang L (2015) Shape-controlled gold nanoparticles supported on MoS(2) nanosheets: synergistic effect of thionine and MoS(2) and their application for electrochemical label-free immunosensing. Nanoscale 7:19129–19135. https://doi.org/10.1039/c5nr05614d
Yun W, Cai D, Jiang J, Zhao P, Huang Y, Sang G (2016) Enzyme-free and label-free ultra-sensitive colorimetric detection of Pb(2+) using molecular beacon and DNAzyme based amplification strategy. Biosens Bioelectron 80:187–193. https://doi.org/10.1016/j.bios.2016.01.053
Zhang J, Tang Y, Teng L, Lu M, Tang D (2015) Low-cost and highly efficient DNA biosensor for heavy metal ion using specific DNAzyme-modified microplate and portable glucometer-based detection mode. Biosens Bioelectron 68:232–238. https://doi.org/10.1016/j.bios.2015.01.001
Xue S, Jing P, Xu W (2016) Hemin on graphene nanosheets functionalized with flower-like MnO2 and hollow AuPd for the electrochemical sensing lead ion based on the specific DNAzyme. Biosens Bioelectron 86:958–965. https://doi.org/10.1016/j.bios.2016.07.111
Yang Z, Jiang W, Liu F, Zhou Y, Yin H, Ai S (2015) A novel electrochemical immunosensor for the quantitative detection of 5-hydroxymethylcytosine in genomic DNA of breast cancer tissue. Chem Commun (Camb) 51:14671–14673. https://doi.org/10.1039/c5cc05921f
Lei YM, Wen RX, Zhou J, Chai YQ, Yuan R, Zhuo Y (2018) Silver ions as novel Coreaction accelerator for remarkably enhanced Electrochemiluminescence in a PTCA-S2O8(2-) system and its application in an ultrasensitive assay for mercury ions. Anal Chem 90:6851–6858. https://doi.org/10.1021/acs.analchem.8b01018
Xu LL, Zhang W, Shang L, Ma RN, Jia LP, Jia WL, Wang HS, Niu L (2018) Perylenetetracarboxylic acid and carbon quantum dots assembled synergistic electrochemiluminescence nanomaterial for ultra-sensitive carcinoembryonic antigen detection. Biosens Bioelectron 103:6–11. https://doi.org/10.1016/j.bios.2017.10.035
Hu C, Yang DP, Xu K, Cao H, Wu B, Cui D, Jia N (2012) Ag@BSA core/shell microspheres as an electrochemical interface for sensitive detection of urinary retinal-binding protein. Anal Chem 84:10324–10331. https://doi.org/10.1021/ac3023795
Acknowledgements
We are grateful for the financial support from the National Natural Science Foundation of China (No. 31571554), and the Outstanding Graduate Student Cultivation Program of Chongqing Medical University (No. BJRC201915).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human or animal subjects.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 7163 kb)
Rights and permissions
About this article
Cite this article
Ma, Y., Yu, C., Yu, Y. et al. DNAzyme assisted recycling amplification method for ultrasensitive amperometric determination of lead(II) based on the use of a hairpin assembly on a composite prepared from nitrogen doped graphene, perylenetetracarboxylic anhydride, thionine and gold nanoparticles. Microchim Acta 186, 677 (2019). https://doi.org/10.1007/s00604-019-3790-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3790-1