Abstract
The authors are presenting a rapid method for the determination of theophylline using unique non-crosslinking gold nanoparticle (AuNP) aggregation. An RNA aptamer against theophylline is firstly split into two RNA fragments which then interact with bare AuNPs. The two RNA probes cause an enhancement of the salt tolerance of AuNPs. However, in the presence of theophylline, the RNA probes form a complex with theophylline so that less RNA probes are available to protect the AuNPs from salt-induced aggregation. Theophylline induced aggregation of AuNPs is accompanied by a color change from red to blue. The color change can be detected visually and via UV-vis absorptiometry by ratioing the absorbances at 650 and 520 nm. The ratio increases linearly in the 0.1 to 20 μM theophylline concentration range, with a 67 nM limit of detection. The method is highly sensitive and selective.

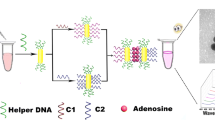
Single-stranded split RNA aptamers (R1 and R2) protect gold nanoparticles (AuNPs) from salt-induced non-crosslinking aggregation. After recognition of theophylline by the RNA probe, the unprotected AuNPs aggregate and undergo a color change from red to blue, and this is used to quantify the theophylline concentration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gold nanoparticles (AuNPs) have attracted great attention and show many distinct physical and chemical properties, such as localized surface plasmon resonance (LSPR) effect [1], photothermal effect [2], good electrical conductivity [3], huge surface-to-volume ratio [4], ease of biofunctionalization [5, 6] and so on. Taking advantage of LSPR effect of AuNPs, a number of label-free colorimetric biosensors have been developed [7,8,9,10]. AuNPs based colorimetric assays are becoming conventional systems for bioanalysis [11,12,13,14,15,16]. Nevertheless, there is still much room for improvement and innovation by further combining other nanomaterials or developing novel biomolecule binding events on the nanostructured metallic surface.
Theophylline belongs to methylxanthine family, which is considered as a crucial molecule for physiological processes in human metabolism [17]. It has been commonly used in the clinic for the treatment of asthma, chronic obstructive pulmonary disease (COPD) and neonatal apnea [18]. However, the serum therapeutic range is quite narrow. High concentrations of theophylline may cause lethal or neurological damages [19]. Therefore, there is an urgent need to develop convenient and fast detection methods to monitor theophylline level. So far, great efforts have been made [20,21,22,23]. For example, Stone’s group immobilized two DNA probes on the surface of AuNPs, which was linked by the aptamer against theophylline. The AuNPs aggregates were disassociated when exposed to theophylline. By monitoring the peak and intensity of UV-vis absorption spectrum, the concentration of theophylline was determined [24]. Zhang and coworkers combined the fluorescence quenching ability of AuNPs and RNA aptamer based theophylline recognition. The labelled fluorophore on the RNA was localized on the surface of AuNPs after the formation of RNA aptamer-theophylline complex. The decreased fluorescence was used to represent initial theophylline concentration [25]. Although current methods may meet the requirement of high sensitivity, the detection procedures of most methods are always complicated. In addition, expensive materials (e.g., modified nucleic acids) and instruments (e.g., fluorescence spectrophotometer) may be required.
In this contribution, we have succeeded the development of a novel method for rapid detection of theophylline. The employed recognition element of split RNA aptamers are unmodified, which makes the detection method cost-effective and convenient. On the other hand, non-crosslinking AuNPs aggregation is involved. Compared with commonly used crosslinking AuNPs aggregation, the reaction is much succinct and faster. In addition, the method is sensitive and selective. The presence of theophylline also can be determined with bare eyes. Therefore, it meets the requirements of point-of-care testing (POCT). Furthermore, the colorimetric strategy is believed to be applicable to other aptamer-based detection methods by altering the nucleic acid sequences on the AuNPs.
Experimental
Materials and instruments
Gold (III) chloride trihydrate (HAuCl4•3H2O), trisodium citrate, diethypyrocarbonate (DEPC), ethylenediaminetetraacetic acid (EDTA) were purchased from Sigma (http://www.sigmaaldrich.com/). Theophylline, caffeine, theobromine, guanine and adenine were obtained from Solarbio Science &Technology Co., Ltd. (http://solarbio.bioon.com.cn/). Human serum samples were collected from local hospitals with the approval by the Medical Ethics Committee (Suzhou, China). All other chemicals were of analytical grade and used as received. Double-distilled water was used to prepare all solutions. The resistance was 18 MΩ cm and the water was also treated with DEPC. RNA probes were synthesized and purified by Takara Biotechnology Co., Ltd. (http://www.takarabiomed.com.cn/). The sequences were listed as follows:
-
RNA probe 1: 5’-AAAGGCGAUACCAGCCGAAA-3’
-
RNA probe 2: 5’-GGCCCUUGGCAGCGUCAAAA-3’
UV-vis absorption spectra were measured by a Synergy HT multifunction microplate reader (BioTek Instruments, Inc., USA). Transmission electron micrographs were recorded by a FEI Tecnai G20 transmission electron microscope (TEM) (FEI Company, USA). Dynamic light scattering (DLS) characterization of AuNPs were performed using a Zetasizer Nano ZS90 (Malvern Instruments, UK). Photographs were taken with a mobile phone (ZTE Blade A2 Plus).
Optimization of NaCl concentration
Freshly prepared AuNPs were kept stable for at least 3 months. However, after the introduction of certain amount of salt like NaCl, non-crosslinking aggregation of AuNPs occurred immediately. The color and UV-vis absorption spectrum were changed accordingly. The concentration of NaCl was a critical parameter, thus it was optimized. First, standard solutions of NaCl with a series of concentrations were prepared. Next, 0.5 mL of concentrated AuNPs were mixed with 0.5 mL of NaCl with various concentrations. UV-vis absorption spectra of the mixtures were then measured from 350 to 700 nm. The absorbance values at 520 nm and 650 nm were recorded (A 520 and A 650). The ratios of R 650/520 (A 650/A 520) were calculated, which were related to the NaCl concentration.
Colorimetric analysis of theophylline
AuNPs were mixed with RNA probe 1 (5 μM) and RNA probe 2 (5 μM), respectively. The two solutions (AuNPs-R1 and AuNPs-R2) were then purified by centrifuging at 10, 000 rpm for 20 min to remove excess RNA, respectively. Next, 245 μL of AuNPs-R1 and 245 μL of AuNPs-R2 were blended. The mixture was then treated with 10 μL of theophylline with various concentrations. Subsequently, UV-vis absorption spectra were measured to check the salt-tolerances of the solutions in the presence of NaCl (70 mM). Specifically, the absorbance values at 520 nm and 650 nm were obtained for the calculation of R 650/520. The relationship between this parameter and the concentration of initial theophylline was then studied.
Interference investigation and serum sample analysis
To verify the selectivity of this colorimetric method, interfering molecules such as caffeine, theobromine, guanine and adenine with similar chemical structures were employed. Some common biomolecules like bovine serum albumin (BSA) and duplex DNA were also checked. These molecules interacted with AuNPs-R1 and AuNPs-R2 instead of target theophylline. Briefly, 490 μL of the mixture of AuNPs-R1 and AuNPs-R2 was blended with 10 μL of the interferents with the final concentration of 100 μM. After treated with 70 mL NaCl, corresponding UV-vis absorption spectra were recorded. R 650/520 values were calculated, which were then compared with that of theophylline (10 μM).
To examine the utility of this colorimetric method in complex biological fluids, human serum samples were challenged for theophylline analysis. Two independent serum samples were collected and diluted for 10 times. After that, 5, 10, 15 μM theophylline were spiked, which were then detected in the colloid system in the presence of 70 mM NaCl. The concentrations of theophylline were obtained and recoveries and relative standard deviations (RSDs) were calculated.
Results and discussion
Working principle
Schematic illustration of the working mechanism for the detection of theophylline is shown in Scheme 1. A 33-mer sequence of the theophylline aptamer is splitted into two shorter fragments, which are made up to 20-mer (RNA probe 1 and RNA probe 2) [26]. The binding property is not affected. The two split RNA aptamers are able to remain stable during the experimental process with shorter RNA fragments. Single-stranded RNA probes interact with AuNPs via their exposed nitrogen-containing bases, which can protect AuNPs from salt-induced non-crosslinking aggregation [27]. Therefore, the mixture of AuNPs-R1 and AuNPs-R2 shows high salt-tolerance and the UV-vis absorption spectra barely change after treated with NaCl of relatively high concentration. However, in the presence of target theophylline, RNA probe 1, RNA probe 2 and theophylline self-assemble into an intact RNA aptamer-theophylline complex through a substantial change in the secondary structure. As a result, without the protection of RNA probes, AuNPs cannot resist the salt-induced aggregation any more, which is reflected by the color changes and UV-vis absorption spectra. A colorimetric strategy for the detection of theophylline is thus established.
Salt-induced non-crosslinking AuNPs aggregation
TEM images are taken to characterize the size and distribution state of AuNPs. Bare AuNPs disperse well in water with the diameter around 13 nm (Fig. S1a). The negatively surface charge of bare AuNPs is due to the capped trisodium citrate (Fig. S2). However, if salt is added to the colloid system, the outer shell of the electrical double layer of AuNPs is compressed. The electrostatic repulsions among AuNPs are thus decreased, which leads to the non-crosslinking aggregation (Fig. S1b) [28]. The sizes of dispersed and aggregated AuNPs are well distinguished by DLS characterizations (Fig. S3). Moreover, since LSPR of AuNPs is strongly correlated with their distribution state, UV-vis absorption spectrum may also be used to indicate the changes. As shown in Fig. 1, bare AuNPs have an obvious absorbance peak at 520 nm (curve a). If the nanoparticles aggregate after adding salts, the peak at 520 nm decreases sharply and the absorbance at 650 nm increases significantly (curve b). The phenomenon is in good accordance with previous reports [29]. Nevertheless, with the existence of RNA molecules like RNA probe 1 on the surface of AuNPs, salt-induced aggregation can be effectively inhibited even with high levels of salt. The UV-vis absorption spectrum is restored to that of AuNPs without salt (curve c). The results indicate that by monitoring the changes of UV-vis absorption spectra, the salt-tolerance of AuNPs adjusted by RNA on their surface can be well determined.
Preliminary experiments
Since both of the absorbance values at 520 nm and 650 nm change, we have used the value at 650 nm to that at 520 nm (R 650/520) to reveal the distribution state of AuNPs [30]. We have obtained the R 650/520 values of bare AuNPs in the presence of NaCl with a series of concentrations. With the increase of NaCl, the AuNPs aggregate more drastically. As a result, the larger R 650/520 value will be (Fig. 2). The relationship between NaCl concentration and R 650/520 is fitted with a Boltzmann sigmoid equation as follows:
in which y is R 650/520, x is the NaCl concentration (mM). When the AuNPs were treated with NaCl of the concentration of x 0 (51.7018 mM), the slope of the calibration curve is the largest. In another word, R 650/520 is most sensitive at this point to the changes of NaCl concentration. In addition, when NaCl concentration is increased to 70 mM, the R 650/520 value reaches a plateau, which is used as the optimized NaCl concentration in the following experiments. The concentration of RNA probes used to protect AuNPs from salt-induced aggregation is also investigated. The optimized concentration is 5 μM after comparing the values of R 650/520 (Fig. S4).
Color changes
We have observed the color changes of AuNPs after different treatments. AuNPs before and after mixing with RNA probes or further theophylline exhibit wine-red color, indicating that AuNPs disperse well in water (Fig. 3a–e). After further incubating with NaCl, bare AuNPs aggregate and the color turns to blue (Fig. 3f). In the presence of RNA probe 1, the AuNPs color does not change, demonstrating the excellent salt-tolerance of RNA protected AuNPs (Fig. 3g). However, in the colloid system of AuNPs-R1 and AuNPs-R2, theophylline is able to remove the RNA probes on the surface of AuNPs, forming RNA aptamer-theophylline complex. The remained AuNPs cannot resist salt-induced aggregation any more, thus the color turns to blue (Fig. 3h). Since the aptamer is splitted into two fragments, none of the RNA probe can interact with theophylline alone. AuNPs-R1 or AuNPs-R2 colloid system does not respond to theophylline, and the solution colors remain wine-red (Fig. 3i–j). These color changes demonstrate that the strategy works well.
Solution colors of a AuNPs, b AuNPs-R1, c mixture of AuNPs-R1, AuNPs-R2 and theophylline, d AuNPs-R1 and theophylline, e AuNPs-R2 and theophylline. f–j are the corresponding top cases after adding NaCl. The concentration of AuNPs is 5 nM; the concentration of NaCl is 70 mM; the concentration of theophylline is 50 μM
Sensitive detection of theophylline
Standard theophylline solutions with a series of concentrations are prepared which are added to the mixture of AuNPs-R1 and AuNPs-R2. Then, the solutions are treated with NaCl with the concentration of 70 mM. UV-vis absorption spectra are recorded which are shown in Fig. 4a. With the increase of theophylline level, more RNA probes are stripped, and the salt-tolerance of AuNPs decreases, which can be reflected by the decrease of A 520 and the increase of A 650. The calibration curve representing the relationship between R 650/520 and theophylline is depicted in Fig. 4b and the linear relationship is established between 0.1 and 20 μM. The linear fitting equate is as follows:
in which, y stands for R 650/520, x is the concentration of theophylline (μM). Previous report has indicated that the safe theophylline concentration in the plasma of medicinal theophylline users is no larger than 111 μM [31]. The range of this method is quite acceptable for theophylline assay in real samples. Moreover, a quite low limit of detection (LOD) is calculated to be 0.067 μM. The analytical performances are excellent compared with most previously reported methods (Table S1).
a UV-vis absorption spectra of the mixture of AuNPs-R1 and AuNPs-R2 after treated with different amount of theophylline in the presence of NaCl. b Calibration curve of theophylline detection with a series of concentrations. Inset shows the linear relationship between R 650/520 and the concentration of theophylline. Error bars represent standard deviations of three measurements
Selectivity investigation
To investigate the selectivity of the theophylline detection method, we have employed several molecules with similar chemical structures as interferents including caffeine, theobromine, guanine, and adenine. We also compared the response of this method to common biomolecules like BSA and duplex DNA. The R 650/520 values in the cases of 10 μM theophylline and 100 μM interferents are compared in Fig. 5. Only in the presence of target theophylline, significant increase R 650/520 value is observed. Caffeine, theobromine, guanine, adenine, BSA and DNA with 10 times higher concentrations only contribute limited R 650/520 values. The results demonstrate the high selectivity of the method.
Determination of theophylline in human serum samples
To demonstrate the practical application of the present colorimetric method, the concentrations of theophylline are determined in human serum samples. The samples are firstly diluted for 10 times and different amount of theophylline are added to the solutions. Then, the samples are measured by the method. By comparing with the standard curve achieved in Fig. 4, the concentrations of added theophylline are calculated. As listed in Table 1, the values are quite consistent. In addition, the recoveries are among 99 and 107%, and all RSDs are less than 5%, verifying the method may have good potential utility in clinical screening applications. One can also distinguish the existence of theophylline in serum samples with bare eyes (Fig. S5).
Conclusions
In summary, we have successfully fabricated a novel detection method for theophylline analysis with high sensitivity. Non-crosslinking AuNPs aggregation phenomenon is involved in the strategy and the presence of target theophylline can be distinguished even visually. The satisfactory linear concentration range makes the method suitable for therapeutically relevant theophylline levels application. Although the introduction of RNA molecules may demand the use of RNase-free water and pipette tips, the split RNA aptamers promise the high selectivity. The method does not respond to interfering molecules like caffeine even with quite high concentrations. Moreover, the operation is quite convenient. The detection procedure only involves mixing the split aptamers, AuNPs, samples and NaCl solution. Thus, a convenient protocol is presented for fast and cost-effective detection of theophylline in serum samples. In addition, the strategy can also be expanded to other analytical applications of RNA aptamers and AuNPs.
References
Ghosh SK, Pal T (2007) Interparticle coupling effect on the surface plasmon resonance of gold nanoparticles: from theory to applications. Chem Rev 107:4797–4862
Abadeer NS, Murphy CJ (2016) Recent progress in cancer thermal therapy using gold nanoparticles. J Phys Chem C 120:4691–4716
Miao P, Wang BD, Han K, Tang YG (2014) Electrochemical impedance spectroscopy study of proteolysis using unmodified gold nanoparticles. Electrochem Commun 47:21–24
Miao P, Ning LM, Li XX (2013) Gold nanoparticles and cleavage-based dual signal amplification for ultrasensitive detection of silver ions. Anal Chem 85:7966–7970
Ghosh P, Han G, De M, Kim CK, Rotello VM (2008) Gold nanoparticles in delivery applications. Adv Drug Deliv Rev 60:1307–1315
Ross MB, JC K, Vaccarezza VM, Schatz GC, Mirkin CA (2015) Nanoscale form dictates mesoscale function in plasmonic DNA-nanoparticle superlattices. Nat Nanotechnol 10:453–458
Haghnazari N, Alizadeh A, Karami C, Hamidi Z (2013) Simple optical determination of silver ion in aqueous solutions using benzo crown-ether modified gold nanoparticles. Microchim Acta 180:287–294
Lin CY, CJ Y, Lin YH, Tseng WL (2010) Colorimetric sensing of silver(I) and mercury(II) ions based on an assembly of tween 20-stabilized gold nanoparticles. Anal Chem 82:6830–6837
Liu P, Yang XH, Sun S, Wang Q, Wang KM, Huang J, Liu JB, He LL (2013) Enzyme-free colorimetric detection of DNA by using gold nanoparticles and hybridization chain reaction amplification. Anal Chem 85:7689–7695
Miao P, Tang YG, Mao ZQ, Liu YZ (2017) Adamantane derivatives functionalized gold nanoparticles for colorimetric detection of miRNA. Part Part Syst Charact 34:1600405
Cui L, Li YY, Lu MF, Tang B, Zhang CY (2018) An ultrasensitive electrochemical biosensor for polynucleotide kinase assay based on gold nanoparticle-mediated lambda exonuclease cleavage-induced signal amplification. Biosens Bioelectron 99:1–7
Liu Q, Na WD, Wang L, Su XG (2017) Gold nanocluster-based fluorescent assay for label-free detection of protein kinase and its inhibitors. Microchim Acta 184:3381–3387
Su S, Cao WF, Liu W, Lu ZW, Zhu D, Chao J, Weng LX, Wang LH, Fan CH, Wang LH (2017) Dual-mode electrochemical analysis of microRNA-21 using gold nanoparticle-decorated MoS2 nanosheet. Biosens Bioelectron 94:552–559
Zhang J, Liu Y, Lv J, Cao Y, Li GX (2015) Dipeptidyl peptidase-IV activity assay and inhibitor screening using a gold nanoparticle-modified gold electrode with an immobilized enzyme substrate. Microchim Acta 182:281–288
Yin YM, Cao Y, Xu YY, Li GX (2010) Colorimetric immunoassay for detection of tumor markers. Int J Mol Sci 11:5078–5095
Miao P, Yang DW, Chen XF, Guo ZZ, Tang YG (2017) Voltammetric determination of tumor necrosis factor-alpha based on the use of an aptamer and magnetic nanoparticles loaded with gold nanoparticles. Microchim Acta 184:3901–3907
Ito K, Lim S, Caramori G, Cosio B, Chung KF, Adcock IM, Barnes PJ (2002) A molecular mechanism of action of theophylline: induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci U S A 99:8921–8926
Mastiholimath VS, Dandagi PM, Jain SS, Gadad AP, Kulkarni AR (2007) Time and pH dependent colon specific, pulsatile delivery of theophylline for nocturnal asthma. Int J Pharm 328:49–56
Ferapontova EE, Gothelf KV (2009) Optimization of the electrochemical RNA-aptamer based biosensor for theophylline by using a methylene blue redox label. Electroanalysis 21:1261–1266
Wang Y, Wu T, Bi CY (2016) Simultaneous determination of acetaminophen, theophylline and caffeine using a glassy carbon disk electrode modified with a composite consisting of poly(alizarin violet 3B), multiwalled carbon nanotubes and graphene. Microchim Acta 183:731–739
Yang YJ, Guo LL, Zhang WQ (2016) The electropolymerization of CTAB on glassy carbon electrode for simultaneous determination of dopamine, uric acid, tryptophan and theophylline. J Electroanal Chem 768:102–109
Sato Y, Zhang YS, Nishizawa S, Seino T, Nakamura K, Li MJ, Teramae N (2012) Competitive assay for theophylline based on an abasic site-containing DNA duplex aptamer and a fluorescent ligand. Chem Eur J 18:12719–12724
Wang J, Cheng WB, Meng FY, Yang M, Pan Y, Miao P (2018) Hand-in-hand RNA nanowire-based aptasensor for the detection of theophylline. Biosens Bioelectron 101:153–158
Chavez JL, Lyon W, Kelley-Loughnane N, Stone MO (2010) Theophylline detection using an aptamer and DNA-gold nanoparticle conjugates. Biosens Bioelectron 26:23–28
Jiang HY, Ling K, Tao XJ, Zhang QQ (2015) Theophylline detection in serum using a self-assembling RNA aptamer-based gold nanoparticle sensor. Biosens Bioelectron 70:299–303
Ling K, Jiang HY, Li Y, Tao XJ, Qiu C, Li FR (2016) A self-assembling RNA aptamer-based graphene oxide sensor for the turn-on detection of theophylline in serum. Biosens Bioelectron 86:8–13
Wang LH, Liu XF, XF H, Song SP, Fan CH (2006) Unmodified gold nanoparticles as a colorimetric probe for potassium DNA aptamers. Chem Commun: 3780–3782. https://doi.org/10.1039/b607448k
Cumberland SL, Strouse GF (2002) Analysis of the nature of oxyanion adsorption on gold nanomaterial surfaces. Langmuir 18:269–276
Li RD, Yin BC, Ye BC (2016) Ultrasensitive, colorimetric detection of microRNAs based on isothermal exponential amplification reaction-assisted gold nanoparticle amplification. Biosens Bioelectron 86:1011–1016
Zhu XL, Li YX, Yang JH, Liang ZQ, Li GX (2010) Gold nanoparticle-based colorimetric assay of single-nucleotide polymorphism of triplex DNA. Biosens Bioelectron 25:2135–2139
Mitenko PA, Ogilvie RI (1973) Rational intravenous doses of theophylline. N Engl J Med 289:600–603
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant no. 81771929 and 31400847), China Postdoctoral Science Foundation (Grant no. 2017 M611911) and the Science and Technology Program of Suzhou (Grant no. SYG201605).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(PDF 663 kb)
Rights and permissions
About this article
Cite this article
Ma, X., Guo, Z., Mao, Z. et al. Colorimetric theophylline aggregation assay using an RNA aptamer and non-crosslinking gold nanoparticles. Microchim Acta 185, 33 (2018). https://doi.org/10.1007/s00604-017-2606-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-017-2606-4