Abstract
The authors have developed a competitive immunoassay for the model pesticide triazophos (TRIAZ). The method is based on the use of only one monoclonal antibody immobilized on multi-labeled gold nanoparticle (AuNP) probes. The immunoassay uses two sets of probe. The first is a multi-labeled AuNP probe that carries monoclonal antibody (mcAb), single-stranded DNA (ssDNA) and horse radish peroxidase (HRP). The second is a magnetic microparticle (MMP) probe that is obtained by coating MMPs with ovalbumin coupled to TRIAZ. Free TRIAZ and MMP-immobilized TRIAZ compete for binding to the mcAb on the surface of the AuNPs. Because TRIAZ is a rather small molecule, it cannot be bound by two antibodies. The competitive immunoassay overcomes the limitations of small molecule detection using one kind of mcAb only. Sensitive transduction of the immunoreaction is accomplished by enzymatically catalyzed amplified. TRIAZ was quantified by a conventional ELISA and by the immunoassay presented here. Both method are highly sensitive and well reproducible. The new assay has a linear response in the 15 ng L−1 to 40 μg L−1 TRIAZ concentration range, and a 14 ng L−1 limit of detection which make it more sensitive than the ELISA. The recovery rate in case of spiked samples ranges from 78.4 to 105%, and the RSD is <20%. A good correlation was further established between the results of the immunoassay and those of GC-MS analysis.

Schematic of a competitive colorimetric triazophos immunoassay employing magnetic microspheres (black color) and multi-labeled gold nanoparticles (red color). The assay overcomes the obstacles in pesticide detection and shows higher sensitivity than the conventional ELISA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Modern agriculture has benefited from the use of agrochemicals, including herbicides, fungicides and insecticides. The productive capacity of agriculture may decrease by approximately 10–15% if these compounds are not used [1]. However, these agrochemicals can also create environmental concerns due to the formation of toxic metabolites and chemical persistence. Pesticides have been detected in surface, ground water and soil. And they can contaminate agricultural products and drinking water. Triazophos [diethyl o-(1-phenyl-1 h-1,2,4-triazol-3-yl) phosphorothioate] (TRIAZ) is a broad-spectrum organ phosphorus insecticide pest control. The stability and slow degradation of TRIAZ result in a health risk to non-target species and humans. The residue of TRIAZ has raised concern in food and the environment safety [2, 3]. Various detection techniques and analytical methods have been developed to detect pesticide residue [4], such as gas chromatography detection (GC), gas chromatography-mass spectrophotometry detection (GC-MS), thin-layer chromatography detection (TLC) and liquid chromatography-mass spectrometry detection (LC-MS). However, these methods require expensive instruments, technical expertise, consumption of time and large sample volume, complicated sample preparation, thus limiting their application, especially in developing countries [5].
Immunoassays are one of the most important methods for the determination of pollutants [6,7,8,9]. Among the numerous kinds of immunoassays, the enzyme-linked immunosorbent assays (ELISA) show reproducible results, accurate quantity and high selectivity for the detection of pesticides on the microwell plate based platform (Scheme 1a). Now, the maximum residue levels (MRLs) as defined in many countries for pesticides residue becomes lower and lower, which are less than their respective lowest detection limits (LOD) in various samples. New methods have been developed by using analytical techniques such as chemoluminescence, electrochemistry, fluorescence and optical spectroscopy to achieve highly sensitive and selective detection [10,11,12]. In this context, we have reported a rapid immunoassay based on immunomagnetic-bead (IMB-ELISA) for TRIAZ analysis in our previous work (Scheme 1b) [3]. The IMB-ELISA was designed based on immune magnetic-bead detection tethered with mcAb. IMB-ELISA takes advantage of immune magnetic-bead with mcAb for generation of detection. The detection limit for IMB-ELISA was 0.10 ng mL−1 and 0.22 ng mL−1 for ELISA. The sensitivity of the IMB-ELISA was not significantly improved compared with the ELISA.
a Schematic illustration of the conventional ELISA for detection triazophos. b Schematic illustration of the IMB-ELISA for detection triazophos. c Schematic illustration of the competitive colorimetric immunoassay employing magnetic microspheres and multi-labeled gold nanoparticles for detection triazophos
Nanotechnology as the rapidly emerging research field is more and more attractive for researchers in development of new analytical methods [13,14,15]. One major property of the nanomaterials is that it can be bonded with many chemical groups. So the nanomaterials can be labeled by many biomolecule including antibody, enzyme, oligonucleotides, fluorescence, etc. [16,17,18,19,20,21]. The conjugated compounds have wide pH stability, high ionic strength and ionic stability, which provides a very good basis for immunoassay. In particular, gold nanoparticles are widely used in various research fields. Tang et al. [22,23,24,25,26,27] did a comprehensive study of nano-material’s chemical and physical properties including self-assembly, Au nano-material and DNA assemblies, preparation, optical activity, and chiral properties, which can be applied in the fields of chemical sensing, optical devices, and chiral nanocatalysis. These methods have great significance in practical applications and fundamental research.
The Bio-bar-code immunoassay based on nanotechnology has attracted substantial research interest in the analytical fields. The barcode immunoassay is a sensitive strategy that takes advantage of short oligonucleotides as surrogate targets in biological detection [16]. Mirkin et al. [28] established a bio-barcode assay to quantify prostate-specific antigen (PSA) based on nanoparticles and the sensitivity of this method was higher than the conventional assays. Zhang et al. [29] developed a sensitive label-free bio-barcode assay for quantitative detection of two nucleic acid targets (HTLV-I and HTLV-II) simultaneously. Liu et al. [30] introduced a highly sensitive method for protein detection based on a novel enzyme-labeled gold nanoparticle (AuNP) probe. Unfortunately, most of the present bio-bar-code reaction systems were applied to detect nucleic acids, protein and few reports were focused on the pesticide residue detection. One major reason is ascribed to the structure of pesticide molecule, as small molecule are unable to be combined with two antibodies due to steric hindrance. In order to solve this problem, the competition mode replacing double antibodies sandwich mode of Enzyme-linked immunosorbent assays (ELISA) can be used for reference. In 2015, our group developed a bio-barcode amplification immunoassay using Real-time quantitative PCR for the detection of triazophos [31]. We find although the bio-barcode immunoassay using Real-time quantitative PCR has high sensitivity, it needed more operation steps, cost higher, and has a higher demand to experiment operator. So it is hard to popularize.
In the end, combining these useful properties of bio-bar-code immunoassay with the ability of the IMB-ELISA system, we here report a highly sensitive immunoassay based on multi-labeled gold nanoparticle probes using only one monoclonal antibody for detection pesticides residue (Scheme 1c). Monoclonal antibody (mcAb) was assembled primarily onto the surface of gold nanoparticles by electrostatic interaction. Single-stranded DNA can be bonded to the gold nanoparticles based on Au-S covalent bond formation. The single-stranded DNA was labeled with biotin. The HRP molecule can be bound to the surface of the gold nanoparticle by the single-stranded DNA and streptavidin-biotin. The magnetic microparticle (MMP) probe was functionalized with ovalbumin coupled with the pesticide hapten. When the target pesticide was present in the test solution, target pesticide and pesticide hapten labeled on the surface of MMPs competed with mcAb labeled on the surface of AuNPs. The complex can be easily separated from a solution using a magnet. Then the substrate was added for enzyme-catalyzed color reaction, the absorbance was measured. The absorbance value can indirectly reflect the target pesticide. We achieved greater amplified sensitivity using the multi-labeled AuNPs probes. The detection limit of this method for TRIAZ was 14.5 ng L−1.
Materials and methods
Reagents and chemicals
Sodium Citrate, Chloroauric acid (HAuCl4·4H2O), 1-Ethyl-3-[3-(dimethylamino) propyl] carbodiimide (EDC), 2-(N-morpholino) ethanesulfonic acid (MES), N-hydroxysuccinimide (NHS), polyethylene glycol 20,000 (PEG20000), ovalbumin (OVA), streptavidin-horseradish peroxidase and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO, USA) (www.sigmaaldrich.com). Triazophos (TRIAZ) standard was obtained from Dr. Ehrenstorfer (Augsburg, Germany) (www. ehrenstorfergmbh.com). H2SO4, Octadecyl (C18) and primary secondary amine (PSA) were purchased from Bonna-Agela Technologies (Tianjin, China). 3,3′,5,5′-Tetramethylbenzidine (TMB) was purchased from TransGen biotech(Beijing, China)(www.transgen.com.).Carboxyl-functionalized MMPs were from Invitrogen (Grand Island, NY) (www.thermofisher.com), HPLC-grade acetonitrile and methanol were purchased from Thermo Fisher Scientific (MA, USA) (www.thermofisher.com). 96-Well polystyrene microtiter plates were obtained from Costar Inc. (Cambridge, MA) (www.costar.com). Ultrapure water was obtained by a Milli-Q RC apparatus. All oligonucleotides used in this work were synthetized by Sangon Co., Ltd. (Shanghai, China) (www.sangon.com). The base sequence was 5′-SH-(A10)-CGCAGTTAACACGAGGCAATCATAC-(A10)-biotin-3′. The hapten of TRIAZ (THBu) and monoclonal antibodies (mcAb) were obtained from Zhejiang University (http://www.zju.edu.cn).
Buffers and solutions
The following buffers were prepared in our research work: (1) A phosphate-buffered saline (PBS; 0.01 mol L−1 phosphate buffer +0.15 mol L−1 NaCl, pH 7.4) was used for the dilution of antibodies and in the preparation of standard solutions reproduce; (2) A Carbonate buffer (CBS; 0.1 mol L−1 carbonate buffer, pH 9,6)was used as the coating buffer; (3) 2% BSA (m/v) in PBS was utilized as the blocking buffer; (4) a PBS buffer containing 0.05% Tween-20 was used for washing buffer; (5) a PBS buffer containing 0.01% Tween-20, and 0.5% PEG 20000 was used for storage buffer.
Multi-labeled AuNPs probe preparation
The synthetic method of citrate-stabilized AuNPs is listed in the supporting information. We followed a previously reported procedure to prepare the multi-labeled AuNP probes, with a few modifications [5, 30]. In brief, 1 mL of AuNPs was adjusted to pH 8.5 and incubated with 11.0 μg of mcAb of TRIAZ for 30 min under gentle shaking. The adjustment PBS buffer was added to obtain a final concentration of 0.01 mol L−1. Subsequently, ssDNA (final concentration, 1.5 μmol L−1) was added and incubated for 16 h at 4 °C. Then, 2 mol L−1 NaCl was mixed in five steps and a final concentration of 0.15 mol L−1 NaCl was obtained in 40 h. Next, the mixture of 5% PEG 20000 and 10% BSA were added to obtain a final concentration (BSA of 0.5% and PEG 20000 of 0.5%). After a 1 h incubation, the solution was repeatedly centrifuged to remove the excesses mcAb and ssDNA. The compound were resuspended with 1 mL of storage buffer. Then, we added 5 μL of streptavidin-HRP (90 μmol L−1) to mixture solutions containing AuNP, the ssDNA molecule and mcAb for 2 h incubation at room temperature. The solution was repeatedly centrifuged to remove the unbound streptavidin-HRP. The final solution was resuspended in 1 mL storage buffer and stored at 4 °C.
Preparation of MMP probes
The MMP probes were prepared according to the manufacture’s protocol with slight modification. In brief, carboxyl-functionalized MMPs was activated by EDC and NHS solutions. Then the supernatant in the tube was removed in a magnet for 2 min. Then 800 μg of the THBu-OVA conjugates (The preparation was listed in supporting information) was added and diluted in MES buffer pH 6.0 to a total volume 500 μL. After incubation over night at room temperature, the tube was on a magnet for 2 min to remove the supernatant. Finally the immune magnetic beads was resuspended in PBS with 0.1% Tween-20 and 0.1% BSA to give the wanted concentration.
Sample preparation and pretreatment method
Pesticide free samples (oranges, apples, cabbage and rice) purchased from local markets in Beijing were spiked following the principle described by our group [3].10 g of pesticide free homogeneous samples (apple, cabbage, orange and rice) were respectively weighed into 50 mL plastic centrifuge tube. An appropriate volume of TRIAZ standard solutions in methanol were added to the samples for the standard curve and recovery studies. The fortification level of recovery assay was 10, 50, 100 μg.kg−1 according to the MRLs’ performance and the CAC (Codex Alimentarius Commission). The spiked samples were left alone for 24 h. The sample pretreatment method was carried out according to QuEChERS (quick, easy, cheap, effective, rugged, and safe) method. In brief, 10 mL of acetonitrile was added to the plastic centrifuge tube and the mixture was vigorous stirring for 30s. Then we added 1 g of sodium chloride and 4 g of anhydrous magnesium sulfate with vigorously shaking, tubes were centrifuged for 5 min at 5000 rpm min−1. 2 mL of supernatant was transferred to 5 mL centrifuge tube containing 50 mg of PSA and 50 mg of C18. Then, the mixture was centrifuged at 10,000 rpm min−1 for 5 min and1 mL of the supernatant was transferred to a microcentrifuge tube. After evaporation using nitrogen-blow, the residue was re-dissolved in PBS containing 5% methanol and quantitated by the competitive colorimetric immunoassay and conventional ELISA.
The competitive colorimetric immunoassay detection
In a typical assay, 50 μL of multi-labeled AuNP probes diluted in probe dilution buffer were transfered to the Eppendorf tubes (EP tubes). TRIAZ standards solutions in 5% (v/v) methanol-PBS (20 μL) at known concentrations ranging from 10.0 ng L−1 to 40 μg L−1 or samples (20 μL) and MMP probe solutions (20 μL) diluted in probe dilution buffer were added to the EP tubes, respectively. The mixtures were incubated at 37 °C for 30 min. TRIAZ hapten labeled on the surface of MMPs were able to compete with TRIAZ for the binding sites on the surface of multi-labeled AuNPs probe in the process of immunoreaction. Then the complexes were separated on a magnet and washed four times with 100 μL of PBS solution. Finally, 100 μL of substrate solution (TMB-H2O2) was added to each well with gentle shaking. After reaction for 15 min at 37 °C, 100 μL of terminal liquid (0.5 mol L−1 H2SO4) was added. Every well’s absorbance was measured in wave length of 450 nm.
Conventional ELISA detection
We performed a conventional ELISA according to our previous work [2, 3]. Briefly, 96-well microplates were coated with 100 μL mcAb of TRIAZ (4.53 μg mL−1) at 37 °C for 2 h. The unoccupied binding sites were blocked with 2% BSA in PBS (300 μL/well). After incubation for 30 min, the microplates were washed again as described previously. A total of 50 μL/well of TRIAZ samples or standards and 50 μL/well of THBu-HRP (0.4 μg mL−1) were added to the wells. The plates were washed after 1 h incubation at 37 °C. Finally, 100 μL of substrate solution (TMB-H2O2) was added to each well with gentle shaking. After reaction for 15 min at 37 °C, 100 μL of terminal liquid (0.5 mol L−1 H2SO4) was added. Every well’s absorbance was measured in wave length of 450 nm.
Results and discussion
Choice of materials
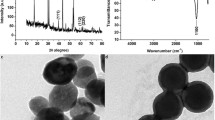
Gold particles are widely used because of their simple preparation methods, good stability, a large surface-to-volume ratio, high surface reaction activity, strong adsorption ability and favorable biocompatibility to biomolecule. Therefore, many laboratories use AuNPs to improve the detection sensitivity. The monoclonal antibody was immobilized on to the surface of AuNPs by electrostatic interaction. The AuNPs coupled with ssDNA was based on Au-S covalent bond formation. The process was simple. The size of AuNPs in the paper was 13 nm. In order to simplify the separation, we immobilized the OVA-hapten on the surface of carboxyl functionalized magnetic microparticles. The reaction complex can be easily separated from the non-magnetic component, driven by the applied magnetic field force, and the operation is efficient.
Optimization of method
The following parameters were characterized: (a) conjugation of probes to AuNPs by UV-Vis spectroscopy and Transmission Electron Microscope (TEM), optimized: (a) Concentration of Streptavidin-HRP; (b) antibody concentration; (c) hapten concentration. Respective data and Figures are given in the Electronic Supporting Material (Figs. S1, S2, Table S1). We found the following experimental conditions to give best results: (a) 90 μmol•L−1 streptavidin-HRP concentration (b) 1:100 MMPS dilution; (c) 1:20 AuNPs dilution;
Analytical performance of the competitive colorimetric immunoassay
On the basis of the competitive immunoassay with multi-labled AuNP probe and MMP probe, the competitive colorimetric immunoassay using only one monoclonal antibody was developed for sensitive detection of the target pesticide under optimal conditions. In addition to the competitive colorimetric immunoassay, the target pesticide was also simultaneously detected by conventional ELISA. Figure 1 shows the absorbance at 450 nm for the detection of TRIAZ. The absorbance at 450 nm depends on the concentration of TRIAZ and decreases proportionally with TRIAZ concentrations. The OD value of the competitive colorimetric immunoassay was 2–5 folds higher than the conventional ELISA. Figure 2 shows the standard curve between inhibition rate and logarithmic TRIAZ concentration. The competitive colorimetric immunoassay and conventional ELISA had a dynamic linear range of 15.0 ng L−1 to 40.0 μg L−1 and 140.0 ng L−1 to 40.0 μg L−1, respectively. The regression equation of competitive colorimetric immunoassay for the quantification of TRIAZ was y = 0.2419 x − 0.1814 (R2 = 0.98). The detection limit IC10 (10% inhibition in the maximal OD value) and IC50 (50% inhibition of the maximum OD value) were 14.5 ng L−1 and 604.3 ng L−1, respectively. The regression equation of the conventional ELISA was y = 0.3326 x − 0.6094 (R2 = 0.99). The detection limit and IC50 were 135.8 ng L−1 and 2.2 μg L−1, respectively. Each concentration was parallel measured for 5 times. Both two methods have a good linear response. And the competitive colorimetric immunoassay is much lower than the conventional ELISA and previously reported assays (Table 1). A large number of ssDNA linking with horseradish peroxidase by the biotin-strepavidin system were decorated on the AuNPs surface leading to successful signal enhancement. In addition, nanoparticles were shown to enhance the activity of the enzymes, which also further improved the high sensitivity of the immunoassay based on multi-labeled AuNP probe [30, 32, 33].
a Standard curve of the competitive colorimetric immunoassay in the TRIAZ concentration range from 15.0 ng L−1 to 40.0 μg L−1. The error bars were based on five duplicate experiments. b Standard curve of the conventional ELISA in the TRIAZ concentration range from 140.0 ng L−1 to 40.0 μg L−1. The error bars were based on five duplicate experiments
Note: “I%” represents the inhibition rate, “ODx” is the signal related with the concentration of TRIAZ standard, “ODmax” represents the maximum signal (the concentration of TRIAZ was zero), and “ODmin” represents the signal of blank wells (no antibody).
Immunoassay validation
To validate the feasibility of the competitive colorimetric immunoassay in practical analysis, recovery assay were conducted. Four kinds of blank samples (orange, cabbage, apple and rice) were spiked with the TRIAZ standard solutions. The spiking level were 10 μg kg−1, 50 μg kg−1, and 100 μg kg−1. All the samples were simultaneously determined by the competitive colorimetric immunoassay and the conventional ELISA. As shown in Table S2, the mean recovery rates with the competitive colorimetric immunoassay were determined to range from 78.4% to 105.1% for TRIAZ, and the RSD (n = 5) ranged from 8.09% to 19.1%. The mean recovery rates with the conventional ELISA were determined to range from 80.4% to 96.0% for TRIAZ, and the RSD (n = 5) ranged from 6.9% to 16.8%. The results indicate that this method meets the requirements for pesticide residue analysis and there was no significant difference between the two methods in the quantitative determination of TRIAZ. The results also demonstrate that the developed immunoassay presents higher sensitivity and good reproducibility in the analysis spiked samples.
Confirmatory test by GC-MS method analysis
The confirmatory test was investigated with GC-MS method analysis. Cabbage and rice samples spiked with three level (10 μg kg−1, 50 μg kg−1, and 100 μg kg−1) for five replicate measurements were used to evaluate the correlation between the competitive colorimetric immunoassay and GC-MS for determination TRIAZ. The GC-MS method and the competitive colorimetric immunoassay were simultaneously used to measure the concentrations of TRIAZ. The correlation regression equation of cabbage was y = 1.0574 x + 1.225 (R2 = 0.9642), the correlation regression equation of rice was y = 0.9946 x + 1.2949 (R2 = 0.9903). The results of TRIAZ detection in rice and apple samples using the competitive colorimetric immunoassay and GC-MS, which present a very good correlation with linear regression analysis. Therefore, the competitive colorimetric immunoassay is a credible immunoassay for detection of TRIAZ.
Conclusions
In this study, the TRIAZ was quantified with higher sensitivity and good reproducibility by the competitive colorimetric immunoassay. The detection limit and IC50 were 14.5 ng L−1 and 604.3 ng L−1, respectively, with a working range of 15.0 ng L−1 to 40.0 μg L−1. By comparing with the conventional ELISA, the competitive colorimetric immunoassay was more sensitive. The competitive colorimetric immunoassay in the manuscript has provided a model for the detection of small molecules. As a new strategy for the detection of small molecular analyte in samples, the competitive colorimetric immunoassay is accurate and sensitive, which is promising for the rapid detection and screening of pesticide residue in environment, agricultural products and food safety.
References
Shim WB, Yang Z, Kim J, Choi JG, Je JH, Kang SJ, Kolosova AY, Eremin SA, Chung DH (2006) Immunochromatography using colloidal gold−antibody probe for the detection of atrazine in water samples. J Agric Food Chem 54:9728–9734. doi:10.1021/jf0620057
Liang CZ, Jin RY, Gui WJ, Zhu GN (2007) Enzyme-linked immunosorbent assay based on a monoclonal antibody for the detection of the insecticide Triazophos: assay optimization and application to environmental samples. Environ Sci Technol 41:6783–6788. doi:10.1021/es070828m
Du P, Jin M, Yang L, Du X, Chen G, Zhang C, Jin F, Shao H, She Y, Wang S, Zheng L, Wang J (2015) A rapid immunomagnetic-bead-based immunoassay for triazophos analysis. RSC Adv 5(99):81046–81051. doi:10.1039/c5ra15106f
Grimalt S, Dehouck P (2016) Review of analytical methods for the determination of pesticide residues in grapes. J Chromatogr A 1433:1–23. doi:10.1016/j.chroma.2015.12.076
Yang G, Zhuang H, Chen H, Ping X, Bu D (2015) A gold nanoparticle based immunosorbent bio-barcode assay combined with real-time immuno-PCR for the detection of polychlorinated biphenyls. Sensors Actuators B Chem 214:152–158. doi:10.1016/j.snb.2015.02.128
Duan H, Chen X, Xu W, Fu J, Xiong Y, Wang A (2015) Quantum-dot submicrobead-based immunochromatographic assay for quantitative and sensitive detection of zearalenone. Talanta 132:126–131. doi:10.1016/j.talanta.2014.08.076
Huang X, Chen R, Xu H, Lai W, Xiong Y (2016) Nanospherical brush as catalase container for enhancing the detection sensitivity of competitive Plasmonic ELISA. Anal Chem 88(3):1951–1958. doi:10.1021/acs.analchem.5b04518
Mariana MS, Mayorga Martinez CC, Watanabe T, Ivandini TA, Honda Y, Pino F, Nakata K, Fujishima A, Einaga Y, Merkoçi A (2016) Microfluidic platform for environmental contaminants sensing and degradation based on boron-doped diamond electrodes. Biosens Bioelectron 75:365–374. doi:10.1016/j.bios.2015.08.058
Wang X, Mu Z, Shangguan F, Liu R, Pu Y, Yin L (2014) Rapid and sensitive suspension array for multiplex detection of organophosphorus pesticides and carbamate pesticides based on silica-hydrogel hybrid microbeads. J Hazard Mater 273:287–292. doi:10.1016/j.jhazmat.2014.03.006
Li J, Song S, Liu X, Wang L, Pan D, Huang Q, Zhao Y, Fan C (2008) Enzyme-based multi-component optical Nanoprobes for sequence- specific detection of DNA hybridization. Adv Mater 20(3):497–500. doi:10.1002/adma.200701918
Zhou W, Gao X, Liu D, Chen X (2015) Gold nanoparticles for in vitro diagnostics. Chem Rev 115(19):10575–10636. doi:10.1021/acs.chemrev.5b00100
Han KC, Yang EG, Ahn DR (2012) A highly sensitive, multiplex immunoassay using gold nanoparticle-enhanced signal amplification. Chem Commun 48(47):5895–5897. doi:10.1039/c2cc31659e
Lan M, Guo Y, Zhao Y, Liu Y, Gui W, Zhu G (2016) Multi-residue detection of pesticides using a sensitive immunochip assay based on nanogold enhancement. Anal Chim Acta 938:146–155. doi:10.1016/j.aca.2016.07.044
Zhang W, Asiri AM, Liu D, Du D, Lin Y (2014) Nanomaterial-based biosensors for environmental and biological monitoring of organophosphorus pesticides and nerve agents. TrAC Trends Anal Chem 54:1–10. doi:10.1016/j.trac.2013.10.007
Arduini F, Cinti S, Scognamiglio V, Moscone D (2016) Nanomaterials in electrochemical biosensors for pesticide detection: advances and challenges in food analysis. Microchim Acta 183(7):2063–2083. doi:10.1007/s00604-016-1858-8
Demers LM, Mirkin CA, Mucic RC, Reynolds RA, Letsinger RL, Elghanian R, Viswanadham G (2000) A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal Chem 72:5535–5541. doi:10.1021/ac0006627
Oh BK, Nam JM, Lee SW, Mirkin CA (2006) A fluorophore-based bio-barcode amplification assay for proteins. Small 2(1):103–108. doi:10.1002/smll.200500260
Zhou Y, Tian XL, Li YS, Pan FG, Zhang YY, Zhang JH, Yang L, Wang XR, Ren HL, Lu SY, Li ZH, Chen QJ, Liu ZS, Liu JQ (2011) An enhanced ELISA based on modified colloidal gold nanoparticles for the detection of Pb(II). Biosens Bioelectron 26(8):3700–3704. doi:10.1016/j.bios.2011.02.008
Zhou Y, Tian XL, Li YS, Zhang YY, Yang L, Zhang JH, Wang XR, Lu SY, Ren HL, Liu ZS (2011) A versatile and highly sensitive probe for hg(II), Pb(II) and cd(II) detection individually and totally in water samples. Biosens Bioelectron 30(1):310–314. doi:10.1016/j.bios.2011.08.034
Li YS, Meng XY, Zhou Y, Zhang YY, Meng XM, Yang L, Hu P, Lu SY, Ren HL, Liu ZS, Wang XR (2015) Magnetic bead and gold nanoparticle probes based immunoassay for beta-casein detection in bovine milk samples. Biosens Bioelectron 66:559–564. doi:10.1016/j.bios.2014.12.025
Du L, Ji W, Zhang Y, Zhang C, Liu G, Wang S (2015) An ultrasensitive detection of 17beta-estradiol using a gold nanoparticle-based fluorescence immunoassay. Analyst 140(6):2001–2007. doi:10.1039/c4an01952k
Han B, Zhu Z, Li Z, Zhang W, Tang Z (2014) Conformation modulated optical activity enhancement in chiral cysteine and au nanorod assemblies. J Am Chem Soc 136(46):16104–16107. doi:10.1021/ja506790w
Li Z, Zhu Z, Liu W, Zhou Y, Han B, Gao Y, Tang Z (2012) Reversible plasmonic circular dichroism of au nanorod and DNA assemblies. J Am Chem Soc 134(7):3322–3325. doi:10.1021/ja209981n
Liu W, Zhu Z, Deng K, Li Z, Zhou Y, Qiu H, Gao Y, Che S, Tang Z (2013) Gold nanorod@chiral mesoporous silica core-shell nanoparticles with unique optical properties. J Am Chem Soc 135(26):9659–9664. doi:10.1021/ja312327m
Zhou Y, Yang M, Sun K, Tang Z, Kotov N (2010) Similar topological origin of chiral centers in organic and nanoscale inorganic structures: effect of stabilizer chirality on optical isomerism and growth of CdTe nanocrystals. J Am Chem Soc 132:6006–6013. doi:10.1021/ja906894r
Zhu Z, Liu W, Li Z, Han B, Zhou Y, Gao Y, Tang Z (2012) Manipulation of collective optical activity in one-dimensional Plasmonic assembly. ACS Nano 6(3):2326–2332. doi:10.1021/nn2044802
Zhou Y, Zhu Z, Huang W, Liu W, Wu S, Liu X, Gao Y, Zhang W, Tang Z (2011) Optical coupling between chiral biomolecules and semiconductor nanoparticles: size-dependent circular dichroism absorption. Angew Chem 50(48):11456–11459. doi:10.1002/anie.201103762
Nam JM, Thaxton CS, Mirkin CA (2003) Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 301(5641):1884–1886. doi:10.1126/science.1088755
Zhang X, Su H, Bi S, Li S, Zhang S (2009) DNA-based amplified electrical bio-barcode assay for one-pot detection of two target DNAs. Biosens Bioelectron 24(8):2730–2734. doi:10.1016/j.bios.2008.12.032
Liu M, Jia C, Huang Y, Lou X, Yao S, Jin Q, Zhao J, Xiang J (2010) Highly sensitive protein detection using enzyme-labeled gold nanoparticle probes. Analyst 135(2):327–331. doi:10.1039/b916629g
Du P, Jin M, Chen G, Zhang C, Jiang Z, Zhang Y, Zou P, She Y, Jin F, Shao H, Wang S, Zheng L, Wang J (2016) A competitive bio-barcode amplification immunoassay for small molecules based on nanoparticles. Sci Rep 6:38114. doi:10.1038/srep38114
Pratibha P, Surinder PS, Sunil KA, Vinay G, Monika D, Sukhvir S, Bansi DM (2007) Application of Thiolated gold nanoparticles for the enhancement of glucose oxidase activity. Langmuir 23(6):3333–3337. doi:10.1021/la062901c
Chen ZJ, Ou XM, Tang FQ, Jiang L (1996) Effect of nanometer particles on the adsorbability and enzymatic activity of glucose oxidase. Colloids Surf B 7:173–179. doi:10.1016/0927-7765(96)01291-X
Li H, Xie T, Ye L, Wang Y, Xie C (2017) Core-shell magnetic molecularly imprinted polymer nanoparticles for the extraction of triazophos residues from vegetables. Microchim Acta 184(4):1011–1019. doi:10.1007/s00604-017-2096-4
Ju KJ, Feng JX, Feng JJ, Zhang QL, Xu TQ, Wei J, Wang A-J (2015) Biosensor for pesticide triazophos based on its inhibition of acetylcholinesterase and using a glassy carbon electrode modified with coral-like gold nanostructures supported on reduced graphene oxide. Microchim Acta 182(15–16):2427–2434. doi:10.1007/s00604-015-1584-7
Li H, Xie C, Li S, Xu K (2012) Electropolymerized molecular imprinting on gold nanoparticle-carbon nanotube modified electrode for electrochemical detection of triazophos. Colloids Surf B 89:175–181. doi:10.1016/j.colsurfb.2011.09.010
Bhamore JR, Ganguly P, Kailasa SK (2016) Molecular assembly of 3-mercaptopropinonic acid and guanidine acetic acid on silver nanoparticles for selective colorimetric detection of triazophos in water and food samples. Sensors Actuators B Chem 233:486–495. doi:10.1016/j.snb.2016.04.111
Acknowledgments
This study was supported by the National Natural Science Foundation (31671938), Chinese Public Interest Industrial Science & Technology Project (201203094), and National Key Foundation for Exploring Scientific Instrument (2013YQ140371).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOC 569 kb)
Rights and permissions
About this article
Cite this article
Du, P., Jin, M., Chen, G. et al. Competitive colorimetric triazophos immunoassay employing magnetic microspheres and multi-labeled gold nanoparticles along with enzymatic signal enhancement. Microchim Acta 184, 3705–3712 (2017). https://doi.org/10.1007/s00604-017-2365-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2365-2