Abstract
The authors describe an integrated biosensor for amperometric DNA detection of Dengue virus in real time. Cu2CdSnS4 (CCTS) quaternary alloy nanostructures were successfully synthesized, deposited on an oxygen-etched silicon substrate (O2/Si) via spin coating, and annealed at 400 °C. The nanostructures were investigated by using UV-vis spectroscopy, X-ray diffraction, atomic force microscopy and scanning electron microscopy. Interdigitated electrodes were fabricated using silver as a metal contact deposited on the CCTS/O2/Si substrate using a thermal evaporator vacuum coater and a hard mask. The quaternary alloy acts as a support for immobilization of a Dengue-specific DNA probe that is employed as the recognition element. Single-stranded DNA in concentrations from 100 f. to 10 nM were successfully recognized via amperometry, typically at a working voltage of 1.5 V. The lower detection limit is 17 nM. Sensitivity is found to be 24.2 μA nM−1 cm−2. The biosensor is inexpensive, fast, highly sensitive, and has low power consumption.

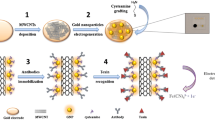
Schematic illustration of IDEs fabrication steps. (a) Si substrate (b) O2/Si, (c) CCTS/O2/Si, (d) Ag/CCTS/O2/Si, (e) cross section view of the sample.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dengue virus belongs to the Flaviviridae family comprising 5 serotypes (DENV-1 through DENV-5). The disease is transmitted by Aedes aegypti mosquito which feeds on human blood. Dengue virus has a spherical nucleocapsid with approximate size of 50–60 nm. The virus contains a single-stranded RNA of 11,000 nucleotides in length [1].
Due to the limited sensitivity of existing methods, there is a need to detect the disease in early stages; one way of accomplishing that is to use biosensors. Advances in nanotechnology have motivated researchers to develop biosensors in different areas including medicine and biomolecular detection of DNA [2], bacteria [3], RNA [4], protein [5], enzyme [6], glucose [7] and pH sensing [8]. DNA biosensors usually rely on the immobilization of a single-stranded DNA (ssDNA) probe onto a surface, which permits it to bind its complementary DNA target sequence by hybridization [9]. The method of transduction depends on the type of physicochemical change resulting from the sensing event whereas transduction of hybridization can be measured optically [10], electrochemically [11], or mass-sensitive [12]. DNA can be immobilized on surfaces using different methods; physical adsorption, covalent immobilization and streptavidin-biotin. 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) converts the carboxyl group into O-acylisourea intermediate that reacts with the amine group resulting in a covalent bonding between biomolecules and the solid surface. Moreover, aminated-DNA probes can be immobilized on the carboxylate-terminated 4-aminobenzoic acid using EDC and N-hydroxysulfosuccinimide (NHS) [13]. Many biosensors require attaching a label to the target for easy detection; these labels can be magnetic beads, fluorophores or active enzymes [14].
Researchers are actively pursuing new methods to design and fabricate biosensors based on interdigitated electrodes (IDEs), which are the most common sensing capacitive structure since they are more favored and highly effective in acquiring effective dielectric properties [15]. IDEs are usually considered as the major part for sensing operation, where electrical signals generated by the sensing material have to be detected by IDEs [16]. The fabrication process of IDEs by photolithography was dominant in the near past and consists of different steps including photoresist coating, exposure and etching. This process requires the use of special equipment [17] thus, alternative approaches were attempted utilizing techniques such as screen printing [18], inkjet-printing [19], microfabrication [20] and stamp method [21]. These techniques are widely used in many sensing applications in bacteria [22], gas [23], pressure [24] and humidity [25].
At the DNA level, there is a need for nanostructures that can bind DNA probe strands and act as a recognition element in the sensing device. The I2-II-IV-VI4 (I = Cu,Ag; II = Zn,Cd; I = Si,Ge,Sn; VI = S,Se) series of quaternary alloys have attracted significant interest due to their appropriate energy band gaps (Eg); 1.1–1.5 eV, high optical absorption coefficient; 104-105 cm−1 and high abundant elements. Cu2CdSnS4 (CCTS) alloy nanostructures have been synthesized using solvothermal method [26], pulsed laser deposition [27], microwave irradiation method [28], spray pyrolysis deposition [29], co-sputtering deposition [30] and facile synthesis [31]. Using ultrasonic cleaning in synthesizing CCTS quaternary alloy nanostructures, via spin coating technique, introduces more peaks, where the crystallite size increases under high annealing temperature [32].
Herein, CCTS alloy nanostructures were synthesized and then deposited, using spin coating, on O2 /Si at annealing temperature; 400 °C. The optical, structural, morphological and topographical properties of the alloy were investigated thoroughly. The design and fabrication of Ag interdigitated electrodes is described in details, and its morphological properties were characterized. The electrical properties of the DNA probe and different concentrations of Dengue DNA serotype-2 were identified.
Experimental
Synthesis of CCTS solution
All chemicals and solvents were purchased from Sigma-Aldrich (USA, http://www.sigmaaldrich.com); the precursor solutions of CCTS alloy nanostructures were prepared from copper (II) chloride dihydrate (0.6 M), cadmium (II) chloride dihydrate (0.8 M), tin (II) chloride dihydrate (0.8 M), thiourea (0.8 M), 2-methoxyethanol and monoethanolamine. The solvent and stabilizer were 2-methoxyethanol and monoethanolamine, respectively. The molar ratios of Cu, Cd, Sn and S in the solution were 2:1:1:4.
The precursors were mixed using a magnetic stirrer for 3 h (1500 RPM) at 50 °C to dissolve the metals completely, which forms a yellow and transparent solution. Moreover, the solution has been covered with parafilm and placed inside ultrasonic cleaner partially filled with deionized water at 50 °C, and left for 30 min to enhance the structural properties of the solution. It is worth mentioning here, that the molarity of copper (0.6 M) was chosen based on a previous study [33].
Solution deposition using spin-coating technique
Si n-type substrate with an orientation of <100> was cleaned by immersing it in standard cleaning solutions (RCA 1 and RCA 2) successively to remove residual organic, ionic and metallic contamination as shown in Fig. S1a. Next, a layer of O2 was etched on top of the Si substrate by exposing it to a temperature of 1000 °C for 1 h under a flow of O2 gas using diffusion furnace (MODU-LAB, USA, http://www.modu-lab.com) as depicted in Fig. S1b. A few drops of the resulting solution were deposited, using plastic dropper, on the O2 /Si substrate. A spin coater (Laurell WS-400B, USA, http://www.laurell.com) was used at a rotation speed of 2500 RPM for 30 s to deposit the solution on the substrate, followed by drying the substrate at 80 °C for 1 min. The spin coating and drying processes were repeated 15 times to obtain a uniform nanostructure layer with a proper thickness at the top of O2 /Si substrate as shown in Fig. S1c. Subsequently, the substrate was annealed at 400 °C for 1 h under N2 gas flow to keep the deposited nanostructure coherent, and then it was cooled to room temperature. This deposition process yielded a CCTS/O2 /Si sample. The annealing temperature of 400 °C was chosen based on a previous study [32].
Fabrication of ag interdigitated electrodes
For the deposition of effective interdigitated electrodes, a hard mask made from chrome was used since it is more favored than conventional lithography method due to its simplicity and the ease of application. Silver (Ag) was used for the deposition of interdigitated electrodes with a thickness of 200 nm on CCTS/O2/Si using a thermal evaporator (PVD) vacuum coater which lead to the formation of a substrate with 4 layers; Ag/CCTS/O2/Si as depicted in Fig. S1d. Moreover, Fig. S1e clearly shows the cross section view of the final sample. According to the specifications of the PVD machine, the thickness was proportional to the weight of raw silver used in the process hence, there is a direct correlation between weight and thickness. The thickness of IDE’s was measured and displayed on the PVD indicator.
Immobilization and hybridization of dengue DNA
All DNA oligonucleotides of Dengue serotype-2 were purchased from Sigma-Aldrich. Totally, 7 different oligonucleotides were prepared, which included the capture DNA probe and 6 targets with different concentrations. The sequence of the capture DNA probe, modified with carboxylic group at 5’end, was 5′-COOH-ATGAAGCTGTAGTCTCACTGGAAGG-3′; the target sequence was 5′-CCTTCCAGTGAGACTACAGCTTCAT-3′. In this assay we used 4 Ag/CCTS/O2/Si substrates. Firstly, 6 different concentrations were prepared from the specific target DNA sequence; 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM. The medium used to bind DNA molecules to the surface of the deposited CCTS nanostructures was (3-Aminopropyl)triethoxysilane (APTES), which was in very small quantities to covalently bond them. Secondly, a 100 μL of 49% deionized water, 49% Ethanol and 2% APTES was prepared and used as a primary solution followed by adding few drops from this solution onto the fingers of the electrodes. The sample was left covered and incubated for 1 h, then excess solution was sucked by pipetting followed by one wash with water. Finally, a manual dryer was used to dry the sample properly. Thirdly, 50% of EDC and 50% of NHS (100 μL total volume) were mixed together and then pipetted on the fingers of the electrodes. The sample was left intact for 10 min followed by sucking excess fluid, followed a quick wash with few drops of water and then drying. Fourthly, a few drops of the DNA probe were dropped on the fingers of the electrodes and the sample was left intact to incubate for 1 h followed by sucking excess DNA probe, washing with water, and then drying. Electrical properties of the DNA probe were identified after that. Dengue DNA concentrations similar to those above in step four (from 100 f. to 10 nM) were used but with an incubation period of 10 min. Electrical properties were evaluated for every concentration using Keithley 6487 and Alpha-A. It is worth mentioning, the resulted data have been generated in two forms; text and image files. The used voltage in the assay was from −3 to 3 V whereas the used frequency was from 1 MHz to 1 Hz.
The optical properties of CCTS have been tested using UV-vis spectrometer (Perkin Elmer Lambda 35, USA, http://www.perkinelmer.com). The reflection spectra were recorded over the wavelength range 200–1000 nm and the energy band gap has been identified. Also, the structural properties have been investigated via X-ray diffractometer (Bruker D2 PHASER, Germany, https://www.bruker.com), which record the intensity as a function of Bragg’s angle in 2θ range from 10o- 60° using Cu kα(λ = 1.5406 Å). The major diffraction peaks have been recorded and attributed to the corresponding planes. The topography was characterized by Atomic Force Microscopy (AFM) (SPA 400, Seiko Instruments Inc., Japan, http://www.sii.co.jp/en) with a scanning area of 10 μm × 10 μm and a scanning rate of 1 Hz, whereas, the surface morphology was investigated using Scanning Electron Microscopy (SEM) (JSM-6010LV, USA, http://www.jeolusa.com) with a magnification factor of ×2000. Electrical properties of DNA probe and the different concentrations of the Dengue DNA serptype-2 are characterized by using (Keithley 6487, Tektronix, USA, http://www.tek.com) and (Alpha-A High-performance Modular Measurement System, novocontrol, Germany, http://www.novocontrol.de).
Results and discussion
Choice of materials
The selection of the proper elements for making a quaternary alloy should be carefully considered. The high cost of indium and gallium obstructs further development of CuIn1-xGaxSe2 (CIGS). Also, Cu and Zn have similar ionic radii and this causes antisite defects in Cu2ZnSnS4 (CZTS). Not to mention the use of copper because of its high conductivity of electric charges, the expensive and scarce indium element was replaced with cadmium; gallium was replaced with Sn, and Se with the abundant and cheap S.
CCTS analysis and characterization
Optical properties
Reflectometry was performed at room temperature using UV-vis spectroscopy at the range of 200–1000 nm to investigate the reflection spectra. According to Tauc formula for direct band gap semiconductors, the energy band gap of CCTS alloy nanostructures can be estimated using the following:
where α is the absorption coefficient, A is a constant, E g is the energy band gap, v is incident photon frequency and h is the Planck’s constant. The energy band gap is estimated from absorption spectra (αhv)2 versus hv by extrapolating the straight line portion of the curve to zero absorption coefficient as shown in Fig. S2. The energy band gap was found to be 1.35 eV, which was in good agreement with previous reports [26–33].
Structural properties
The CCTS alloy nanostructures were investigated by XRD as shown in Fig. S3. XRD pattern provided information about the crystalline structure of the nanoparticles as well as the crystallite size. After studying the patterns, four major diffraction peaks appeared at 2θ = 28.7°, 29.57°, 47.77° and 55.95, which were attributed to 200, 112, 204 and 116 planes, respectively. All resulting peaks were matched with the tetragonal stannite structure of CCTS corresponding to the standard (ICDD PDF2008, 00–029-0537). It was noticed that the main peak was at 112 plane. The lattice constants a and c were calculated from XRD pattern of the 112 plane, using the eqs. (S1 to S7) listed in the electronic supplementary material according to the tetragonal crystal system. Also, Table S1 displays the values of a and c along with other structural parameters.
Topographical and morphological studies
The surface topography of CCTS was characterized. Fig. S4 shows 2-D and 3-D images with an area of 10 μm × 10 μm at 1 Hz scan rate. The measured roughness was 15.7 nm. The SEM image shown in Fig. S5 depicted the surface morphology of CCTS. A closer look at the image shows that the sample has a needle’s shape.
Interdigitated electrodes characterization
Silver (Ag) fabricated IDEs were deposited on CCTS/O2/Si substrate using thermal evaporator that formed a pair of interdigitated electrodes with 4 fingers each. Ag was selected for fabricating electrodes because it has a very high electrical conductivity compared to other elements. The IDEs comprised 8 fingers which were 3.75 mm long (L), 0.25 mm wide (W) and 0.25 mm gap (G) between two adjacent interdigitated electrodes as shown in Fig. S6a. Moreover, the thickness of the fingers was 200 nm. Furthermore, the microscopic image captured by the polarizing microscope (Olympus BX51P, USA, http://www.olympus-ims.com) displayed part of IDEs and the scattered black spots confirmed the existence of CCTS alloy nanostructures as depicted in Fig. S6b. Lastly, an image of the whole sample with Ag IDEs is shown in Fig. S6c.
Electrical properties of the biosensor
Electrical properties at different Dengue DNA serotype-2 concentrations were investigated first using Keithley 6487 with two-point probes. When applying voltage ranging from −3 to 3 V, the results indicated that the highest current was 4 μA at DNA probe followed by 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM concentrations. The current was lowest (1 μA) at 10 nM. As shown in Fig. 1a, a semi-linear relationship was identified as Schottky barrier formed at a metal–semiconductor junction. IDEs have metallic electrodes on Si substrate, with a thin insulator of O2, therefore, the I–V curves showed finite currents at zero voltage. This was caused by the charging effect on the electrical double layer capacitance and the existence of the quaternary alloy between the O2 layer and IDEs. It worth mentioning here that an inverse relationship exists between concentration and current. Ibraheam et al. [34] have reported Dengue DNA type 2 biosensor based Cu2Zn1−xCdxSnS4 quinternary alloy nanostructures. Dielectric measurements of current to voltage (I–V) were taken with recorded readings from −6 V to 6 V, the results indicated that the highest was at DNA probe followed by 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM which is in agreement with this research. As depicted in Fig. 1b, the highest resistance was 5 MΩ at 10 nM followed by 1 nM, 100 pM, 10 pM, 1 pM, 100 f. and DNA probe. The resistance was 1 MΩ at DNA probe. Of these figures, decreasing the concentration will lead to decreasing the resistance and vise-versa. These results were consistent with the results of the I-V test. It was noticed that applying zero voltage will force the resistance to drop to zero for all concentrations. Interestingly, when the gap between two adjacent interdigitated electrodes increased, the resistance also increased. This was due to charge carriers having to travel a larger distance. As illustrated in Fig. 1c, the highest conductance was 1.2 μS at DNA probe followed by 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM, whereas 10 nM being the lowest (0.4 μS). Collectively, our results corroborated the R-V test and the inverse relationship between the resistance and conductance. Electrical power is the rate at which electrical energy is transferred by an electric circuit and it is measured in watt (W), one joule per second, and it results from multiplying voltage (V) by current (I). As shown in Fig. 1d, the highest power of 11 μW was obtained at the DNA probe followed by 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM- power was lowest (3 μW) at 10 nM. These results are consistent with the results of the I-V test in that the current has a proportional relationship with the power.
The second set of measurements were implemented by Alpha-A to investigate a different group of electrical parameters as a function of frequency. The capacitance increases with decreasing the frequency in an inverse relationship, as shown in Fig. 2a, the capacitance for most concentrations started from 0.0009 μF at 1 MHz and continued to increase as frequency decreased until it reached 1 Hz. As shown in the internal inset of Fig. 2a, the highest capacitance was 0.04 μF at DNA probe followed by 100 fM, 1 pM, 100 pM, 10 pM, 1 nM and 10 nM. The capacitance was 0.0075 μF at 10 nM. Despite the current findings, capacitance can be increased by increasing the number of fingers as well as increasing the thickness of the electrodes. On the other hand, electrical impedance is a measure of the opposition by a circuit to a current when the voltage is applied. Impedance possesses both magnitude and phase, unlike resistance, which has only magnitude. Fig. 2b shows the impedance at different Dengue DNA serotype-2 concentrations as a function of frequency. The impedance for most concentrations started from 0.17 KΩ at 1 MHz and continued to increase as frequency decreased until it reached 1 Hz. As shown in the internal inset of Fig. 2b, the highest impedance was 5.25 MΩ at 10 nM followed by 1 nM, 10 pM, 100 pM, 1 pM, 100 f. and DNA probe. The impedance was 1 MΩ in DNA probe. Moreover, impedance increases as frequency decreases, and our results corroborate this; whereas 10 nM DNA concentration was the highest in the impedance test it yielded the lowest outcome in the capacitance test. The conductivity for most concentrations started from 2.5 μS/m at 1 MHz and continued to decrease as frequency decreased until it reached 1 Hz except for the DNA probe, which started from 0.55 μS/m. As shown in Fig. 2c, there is a significant overlap between the different concentrations, and most of them have almost the same amount of conductivity except DNA probe which has a lower conductivity than the others.
Fig. S7a displays the permittivity of the different concentrations of Dengue DNA serotype-2 as a function of frequency. The permittivity for most of the concentrations started from 8 F/m at 1 MHz and continued to increase with decreasing frequency until reaching 1 Hz. As shown in the internal inset of Fig. S7a, the highest permittivity was 1400 F/m at DNA probe followed by 100 fM, 1 pM, 10 pM, 100 pM, 1 nM and 10 nM. The permittivity was 270 F/m at 10 nM. On the other hand, Fig. S7b shows the modulus for different concentrations of Dengue DNA serotype-2 as a function of frequency. The modulus for most of the concentrations started from 0.12 m/F at 1 MHz and continued to increase with the decrease of the frequency up to 1 Hz. There was a great amount of overlapping between the different concentrations albeit the same amount of modulus.
The third set of measurements were recorded using Alpha-A High to find capacitance and impedance as a function of voltage. Fig. 3a shows the capacitance at different concentrations of Dengue DNA serotype-2 as a function of voltage; most of the concentrations fluctuated between 0.001 μF and 0.0025 μF. It is worth mentioning, there is a slight increase in capacitance as voltage increases. Moreover, the highest capacitance was obtained at DNA probe followed by 1 pM, 100 fm, 10 pM, 100 pM, 1 nM and 10 nM. Finally, Fig. 3b shows the impedance at different concentrations of Dengue DNA serotype-2 as a function of voltage; most of the concentrations fluctuated between 0.4 MΩ and 1.4 MΩ. There was a slight decrease in impedance as voltage increased. Moreover, the highest impedance was at 10 nM followed by 1 nM, 10 pM, 100 pM, 100 fM, 1 pM and DNA probe.
Performance of the biosensor
The development of analytical method is always evaluated in terms of performance and effectiveness. In this study, sensitivity, limit of detection (LOD) and limit of quantification (LOQ) are important factors in method validation. The sensitivity can be evaluated by measuring the slope of the resulting current versus DNA concentration. Sensitivity of the biosensor is calculated using the following equation;
There are 3 common methods for the estimation of detection and quantitation limit; Visual definition, calculation from the signal-to-noise ratio and calculation from the standard deviation of the concentrations, which was used in the current study. The calibration equation was established as a function of DNA concentration; y = −0.0604× + 1.0167 where y is the transduced current and x is DNA concentration. As shown in Fig. 4, the linear regression, R2 = 0.5059 and the slope m = 0.0604 μA nM−1 were also calculated. The calculated sensitivity of the biosensor was 24.2 μA nM−1 cm−2 which is considered as a high sensitivity. The standard deviation of the current was 0.34 μA and the calculated LOD of the biosensor was 16.9 nM, and accordingly, the calculated LOQ of the biosensor was 56.3 nM. To compare with the present biosensor, various types of DNA biosensors have been reported. Nuzaihan et al. [35] have achieved Dengue virus DNA detection using silicon nanowire biosensor by molecular gate control which demonstrated the device sensitivity of 45 μA nM−1 cm−2.
Conclusions
CCTS alloy nanostructures were successfully synthesized and deposited on O2 /Si substrate via spin coating technique at an annealing temperature of 400 °C. UV-vis measurement demonstrated that energy band gap was 1.35 eV. XRD patterns revealed the presence of four peaks; the highest corresponded to (112) plane whereas, crystallite size was 20.4 nm. The morphological and topographical studies indicated the presence of a needle’s shape, whereas the measured roughness was 15.7 nm. Ag interdigitated electrodes were fabricated on CCTS/O2/Si using a hard mask and thermal evaporator. There was a clear difference between the different concentrations of DNA used in all tests. The current study revealed a proportional relationship with the voltage which is explained as Schottky barrier hence. On the other hand, capacitance and impedance displayed an inverse relationship, whereas, they are affected slightly by changing the voltage. Moreover, Conductivity increased with increasing frequency. It is worth mentioning here, longitudinal design parameters, such as the spacing between fingers, fingers width and fingers thickness are very important factors when designing biosensors. The calculated sensitivity of the biosensor was 24.2 μA nM−1 cm−2, whereas the calculated LOD and LOQ were 16.9 and 56.3 nM, respectively. Overall, we have designed, fabricated and characterized a small and low-cost sensor that has a high sensitivity as well as an adequate LOD and LOQ. The results showed that the fabricated biosensor is promising for detecting the Dengue DNA serotype-2.
References
Du Y, Guo S, Dong S, Wang E (2011) An integrated sensing system for detection of DNA using new parallel-motif DNA triplex system and graphene-mesoporous silica-gold nanoparticle hybrids. Biomaterials 32:8584–8592
Chao J, Zhu D, Zhang Y et al (2016) DNA nanotechnology-enabled biosensors. Biosens Bioelectron 76:68–79
Mallén-Alberdi M, Vigués N, Mas J et al (2016) Impedance spectral fingerprint of E. coli cells on interdigitated electrodes: a new approach for label free and selective detection. Sens Bio-Sensing Res 1:100–106
Cheen OC, Gopinath SCB, Perumal V et al (2017) Aptamer-based Impedimetric determination of the human blood clotting factor IX in serum using an interdigitated electrode modified with a ZnO Nanolayer. Microchim Acta 184:117–125
Gopinath SCB, Perumal V, Kumaresan R et al (2016) Nanogapped dielectric impedance surface for sensitive detection of mycobacterium tuberculosis 16 kDa antigen. Microchim Acta 183:2697–2703
Amine A, Arduini F, Moscone D, Palleschi G (2016) Recent advances in biosensors based on enzyme inhibition. Biosens Bioelectron 76:180–194
Gopalan AI, Lee KP, Ragupathy D et al (2009) An electrochemical glucose biosensor exploiting a polyaniline grafted multiwalled carbon nanotube/perfluorosulfonate ionomer-silica nanocomposite. Biomaterials 30:5999–6005
Deng ZJ, Morton SW, Bonner DK et al (2015) A plug-and-play ratiometric pH-sensing nanoprobe for high-throughput investigation of endosomal escape. Biomaterials 51:250–256
Sassolas A, Leca-bouvier BD, Blum LJ (2008) DNA biosensors and microarrays. Chem Rev 108:109–139
Tereshchenko A, Bechelany M, Viter R et al (2016) Optical biosensors based on ZnO nanostructures: advantages and perspectives. A review. Sensors Actuators B Chem 229:664–677
Song Y, Luo Y, Zhu C et al (2016) Recent advances in electrochemical biosensors based on graphene two-dimensional nanomaterials. Biosens Bioelectron 76:195–212
Guo Y, Wang Y, Yang G et al (2016) MicroRNA-mediated signal amplification coupled with GNP/dendrimers on a mass-sensitive biosensor and its applications in intracellular microRNA quantification. Biosens Bioelectron 85:897–902
Nimse SB, Song K, Sonawane MD et al (2014) Immobilization techniques for microarray: challenges and applications. Sensors (Switzerland) 14:22208–22229
Daniels JS, Pourmand N (2007) Label-free impedance biosensors: opportunities and challenges. Electroanalysis 19:1239–1257
Rivadeneyra A, Fernández-Salmerón J, Agudo-Acemel M et al (2016) Printed electrodes structures as capacitive humidity sensors: a comparison. Sensors Actuators A Phys 244:56–65
Igreja R, Dias CJ (2004) Analytical evaluation of the interdigital electrodes capacitance for a multi-layered structure. Sensors Actuators A Phys 172:392–399
Kitsara M, Goustouridis D, Chatzandroulis S et al (2007) Single chip interdigitated electrode capacitive chemical sensor arrays. Sensors Actuators B Chem 127:186–192
Manjakkal L, Cvejin K, Kulawik J et al (2014) Fabrication of thick film sensitive RuO2-TiO2 and ag/AgCl/KCl reference electrodes and their application for pH measurements. Sensors Actuators B Chem 204:57–67
Pavinatto FJ, Paschoal CWA, Arias AC (2015) Printed and flexible biosensor for antioxidants using interdigitated ink-jetted electrodes and gravure-deposited active layer. Biosens Bioelectron 67:553–559
Zeng Z, Long X, Zhou H et al (2015) On-chip interdigitated supercapacitor based on nano-porous gold/manganese oxide nanowires hybrid electrode. Electrochim Acta 163:107–115
Chou K, Lee C (2014) Fabrication of silver interdigitated electrode by a stamp method. Adv Mater Sci Eng 2014:1–5
Yu J, Liu Z, Liu Q et al (2009) A polyethylene glycol (PEG) microfluidic chip with nanostructures for bacteria rapid patterning and detection. Sensors Actuators A 154:288–294
Brandenburg A, Kita J, Groß A, Moos R (2013) Novel tube-type LTCC transducers with buried heaters and inner interdigitated electrodes as a platform for gas sensing at various high temperatures. Sensors Actuators B Chem 189:80–88
Korostynska O, Arshak A, Arshak K, Morris D (2011) Investigation into real-time pressure sensing properties of SnO 2, TiO2, and TiO2/ZnO thick films with interdigitated electrodes. Mater Sci Eng B Solid-State Mater Adv Technol 176:1297–1300
Lei S, Deng C, Chen Y, Li Y (2011) A novel serial high frequency surface acoustic wave humidity sensor. Sensors Actuators A Phys 167:231–236
Cui Y, Wang G, Pan D (2012) Synthesis and photoresponse of novel Cu2CdSnS4 semiconductor nanorods. J Mater Chem 22:12471–12473
Vanalakar SA, Agawane GL, Shin SW et al (2015) A review on pulsed laser deposited CZTS thin films for solar cell applications. J Alloys Compd 619:109–121
Guan H, Shi Y, Hou H et al (2014) Quaternary Cu2CdSnS4 nanoparticles synthesised by microwave irradiation method. Micro Nano Lett 9:251–252
Nie L, Liu S, Chai Y, Yuan R (2015) Spray pyrolysis deposition and photoresponse of Cu2CdSnS4 thin films. J Anal Appl Pyrolysis 112:363–368
Guo H, Li Y, Fang X et al (2016) Co-sputtering deposition and optical-electrical characteristic of Cu2CdSnS4 thin films for use in solar cells. Mater Lett 162:97–100
Li C, Cao M, Huang J et al (2015) Facile synthesis of ultra-long Cu2CdSnS4 nanowires with wurtzite-derived structure. Mater Lett 140:170–173
Abu Odeh A, Al-Douri Y, Ayub RM, Ibraheam AS (2016) Ultrasonic effect on optical, structural, topographical and morphological studies of Cu2CdSnS4 quaternary alloy nanostructures. J Alloys Compd 686:883–895
Odeh AA, Al-Douri Y, Ayub RM et al (2016) Optical analysis of lens-like Cu2CdSnS4 quaternary alloy nanostructures. Appl Phys A Mater Sci Process 122:888–902
Ibraheam AS, Al-Douri Y, Gopinath SCB, Hashim U (2016) A novel quinternary alloy (Cu2Zn1−xCdxSnS4) nanostructured sensor for biomedical diagnosis. Mater Res Express 3:85022
Nuzaihan MMN, Hashim U, Md Arshad MK et al (2016) Electrical detection of dengue virus (DENV) DNA oligomer using silicon nanowire biosensor with novel molecular gate control. Biosens Bioelectron 83:106–114
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOCX 708 kb)
Rights and permissions
About this article
Cite this article
Odeh, A.A., Al-Douri, Y., Voon, C.H. et al. A needle-like Cu2CdSnS4 alloy nanostructure-based integrated electrochemical biosensor for detecting the DNA of Dengue serotype 2. Microchim Acta 184, 2211–2218 (2017). https://doi.org/10.1007/s00604-017-2249-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2249-5